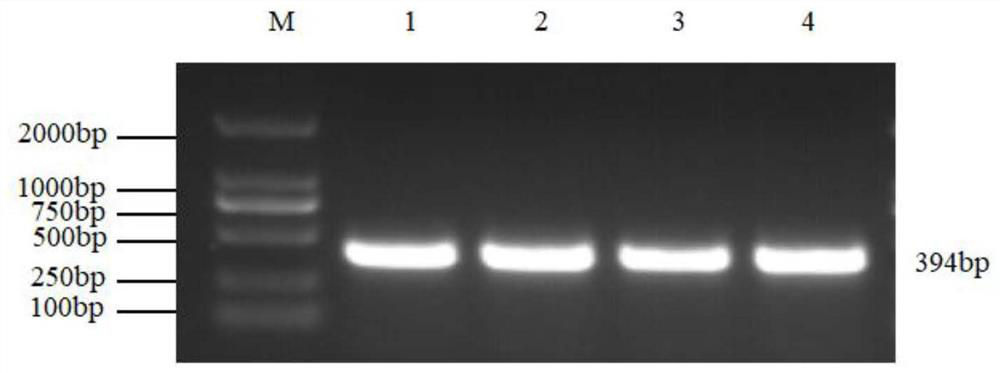
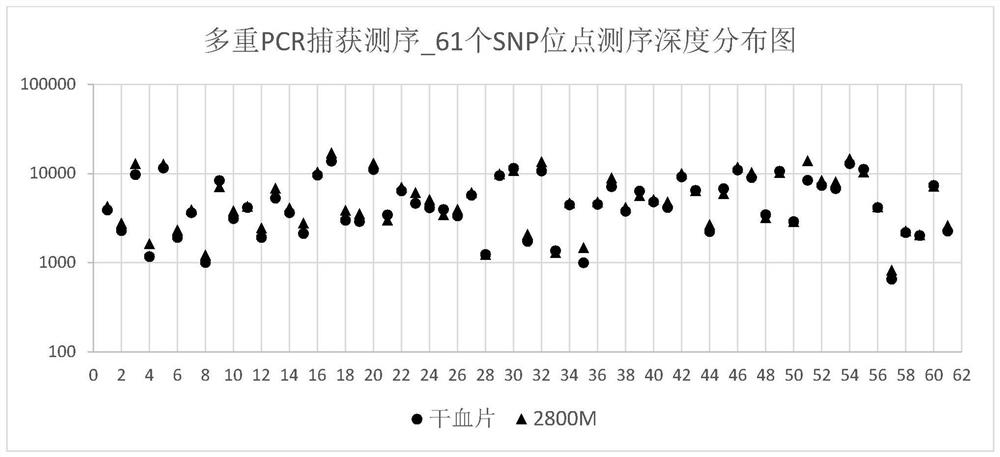
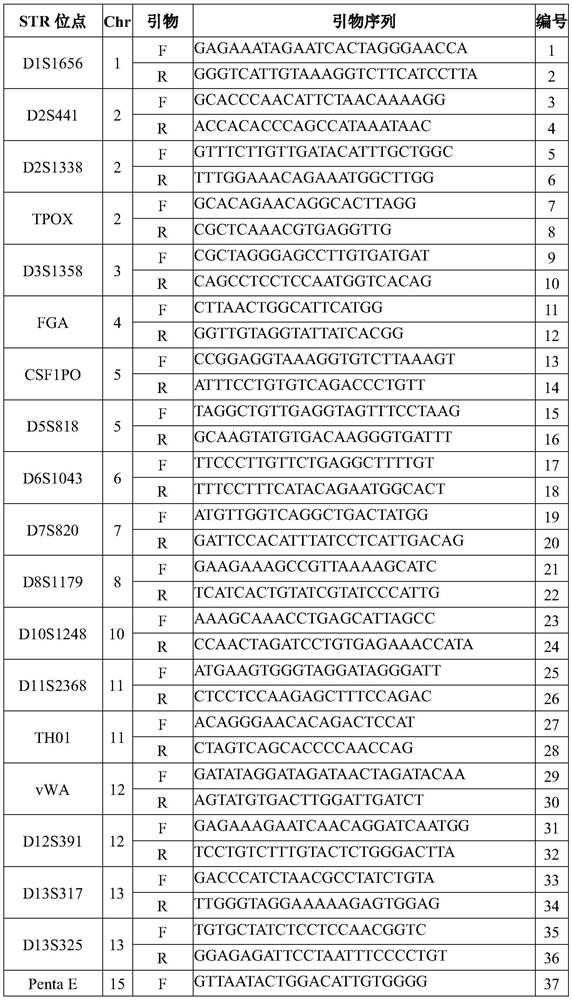
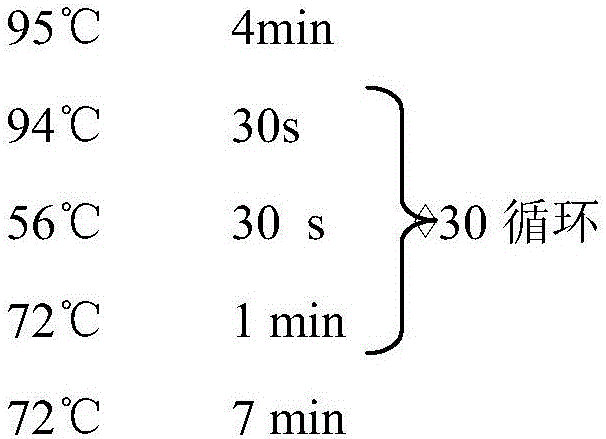Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
45 results about "Locus (genetics)" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000.
SNP (single nucleotide polymorphism) marker for evaluating growth performance of ctenopharyngodonidella, primer and evaluation method
ActiveCN106755527AEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationGenetic exchangeIntein
The invention provides an SNP (single nucleotide polymorphism) marker (comprising SNP1 and SNP4) for evaluating growth performance of ctenopharyngodonidella and a primer pair for amplifying a gene segment where an SNP locus is positioned by virtue of methods of molecular genetics and molecular biology. SNP1 is positioned at the 940th locus in a promoter of a complete sequence of a MyoD (myogenic determining factor) gene of the ctenopharyngodonidella, and a base T is inserted or deficient at the position; SNP4 is positioned in a 107th locus in a first intron of the complete sequence of the MyoD gene of the ctenopharyngodonidella, and a base at the position is A or T. The growth performance of the ctenopharyngodonidella is determined by detecting haplotypes of the two SNP loci. The adopted haplotypes are mutated according to bases generated in the MyoD gene, so that genetic exchange and further phenotype verification are avoided. By virtue of the haplotypes of the SNP marker, rapidly growing ctenopharyngodonidella can be simply and rapidly identified, and rapidly growing ctenopharyngodonidella of a new line can also be guided to be bred.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Rapid identification of powdery mildew resistance Lotus corniculatus genes utilizing comparative genomics
InactiveCN102703463AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyGenetics genomics
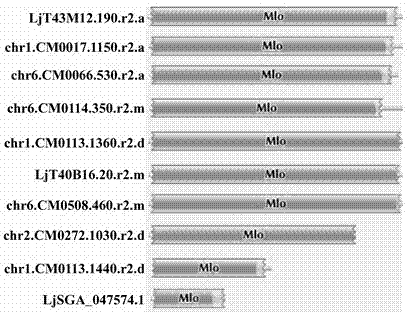
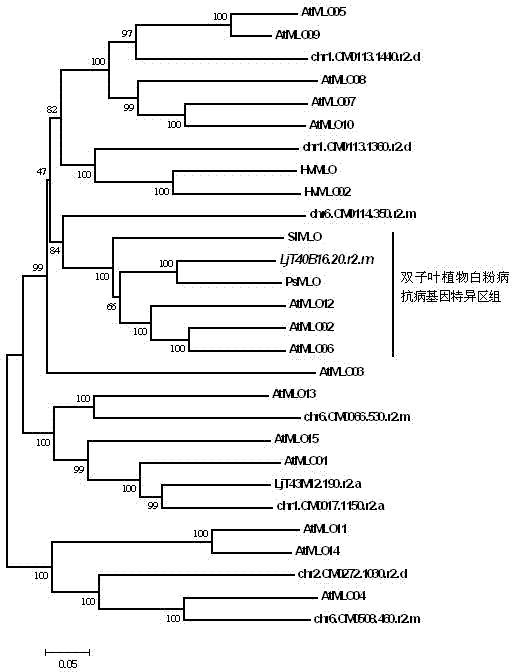
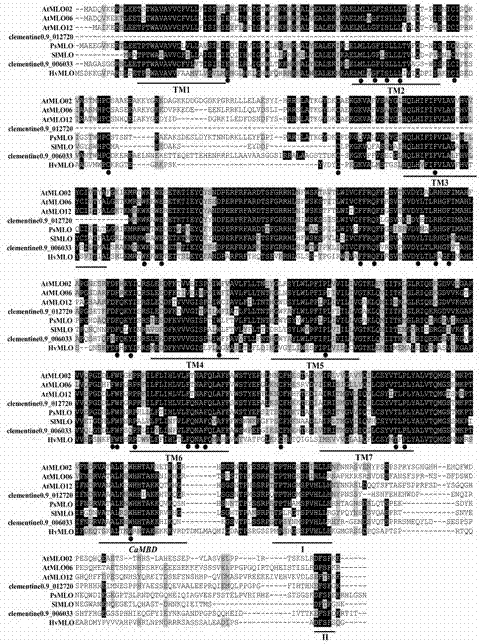
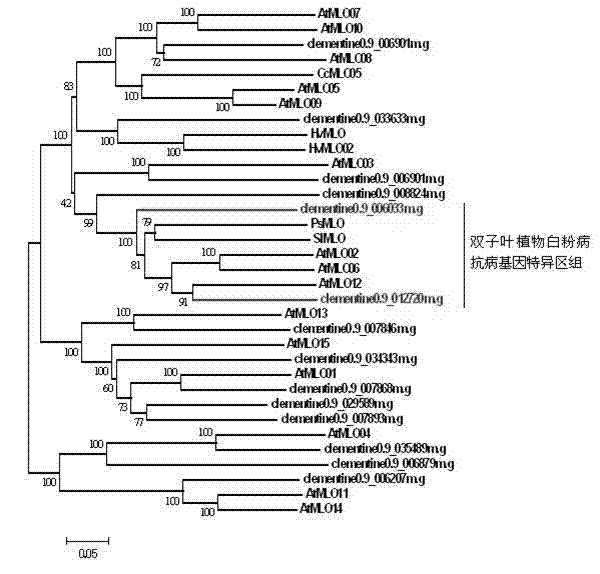
The invention relates to rapid identification of powdery mildew resistance Lotus corniculatus genes utilizing comparative genomics, relates to knowledge of subjects such as plant comparative genomics, genetics and bioinformatics, and belongs to the technical field of plant biotechnology. The rapid identification of powdery mildew resistance Lotus corniculatus genes utilizing the comparative genomics mainly includes the steps of firstly, downloading full genomic sequences of Lotus corniculatus and collecting MLO (mildew resistance locus o) genes; secondly, identifying the MLO genes; thirdly, identifying phylogenetic relationship of the MLO genes; and fourthly comparing the MLO powdery mildew resistance genes. By the rapid identification, mining cycle of the powdery mildew resistance genes is shortened, and the powdery mildew resistance genes can be identified quickly. Corresponding co-separation functional marks (SNP (single nucleotide polymorphism), Scar and the like) can be developed through candidate powdery mildew resistance genes identified, the rapid identification is also available for molecular marker-assisted selection of the powdery mildew resistance genes, and accuracy is high. The rapid identification can also be used with other molecular markers for resistance genes to create multiresistance breeding materials, and accordingly breeding period is shortened, breeding efficiency is improved, and basis for elaborating powdery mildew resistance Lotus corniculatus molecular mechanisms is laid.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Rapid identification of MLO (mildew resistance locus o) powdery mildew resistance Poncirus trifoliata genes
InactiveCN102703464AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyRapid identification
The invention relates to rapid identification of powdery mildew resistance Poncirus trifoliata genes, relates to knowledge of subjects such as plant comparative genomics, genetics and bioinformatics, and belongs to the technical field of plant biotechnology. The rapid identification mainly includes the steps of firstly, downloading full genomic sequences of Poncirus trifoliate and collecting MLO genes; secondly, identifying the MLO genes; thirdly, identifying phylogenetic relationship of the MLO genes; and fourthly, comparing the MLO powdery mildew resistance genes. By the rapid identification, mining cycle of the powdery mildew resistance Poncirus trifoliata genes is shortened effectively, and the powdery mildew resistance genes can be identified quickly. Corresponding co-separation functional marks (SNP (single nucleotide polymorphism), Scar and the like) can be developed through candidate powdery mildew resistance genes identified, the rapid identification is also available for molecular marker-assisted selection of the powdery mildew resistance genes, and accuracy is high. The rapid identification can also be used with other resistance gene molecular markers to create multiresistance breeding materials, breeding period is shortened, breeding efficiency is improved, and basis for elaborating powdery mildew resistance Poncirus trifoliata molecular mechanisms is laid.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Rapid identification of anti-powdery-mildew gene of Brachypodium distachyon
InactiveCN102703462AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyBrachypodium sylvaticum
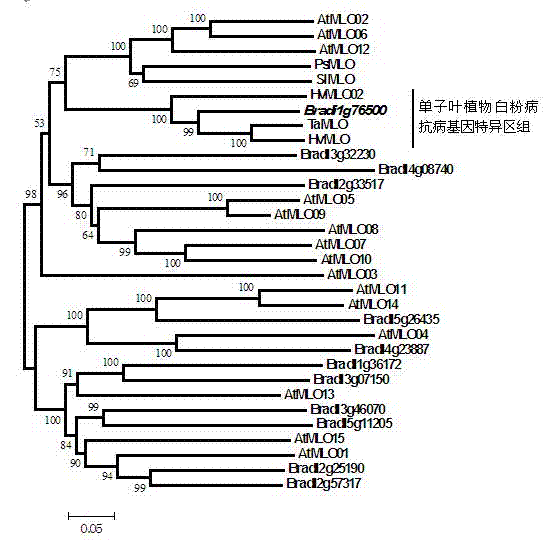
The invention relates to rapid identification of anti-powdery-mildew gene of Brachypodium distachyon, relates to the knowledge of plant comparative genomics, genetics, biological information and other subjects, and belongs to the scientific field of plant biotechnology. The rapid identification of anti-powdery-mildew gene of Brachypodium distachyon comprises the main steps of: (1) downloading full genome sequences of Brachypodium distachyon and collecting MLO (mildew resistance locus o) type gene; (2) identifying the MLO type gene; (3) analyzing the MLO type gene historical development relation; and (4) comparing the MLO type powdery mildew gene. According to the rapid identification of anti-powdery-mildew gene of Brachypodium distachyon provided by the invention, the excavation period of the anti-powdery-mildew gene of Brachypodium distachyon is effectively shortened, and the rapid identification of the powdery mildew gene is facilitated; the identified powdery mildew gene can be used for developing corresponding coseparation functional markers (SNP (single nucleotide polymorphism), SCAR (sequence-characterized amplified region), and the like), and can also be rapidly used for assisted selection of molecule markers of anti-powdery-mildew gene, and the accuracy is high; development of multi-resistance breeding materials can be carried out by combining with other anti-disease gene molecule markers, the breeding time is shortened and the breeding efficiency is improved; and foundation is established for expounding the molecular mechanism of the anti-powdery-mildew gene of Brachypodium distachyon.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
SNP (Single Nucleotide Polymorphism) molecular marker capable of influencing wool shearing amount of alpine merino and application of SNP molecular marker
ActiveCN114214428AImproving Breeding AccuracyImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideMolecular genetics
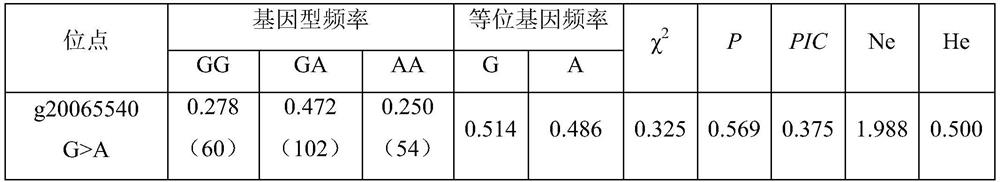
The invention belongs to the technical field of molecular genetics, and particularly relates to an SNP (Single Nucleotide Polymorphism) molecular marker influencing the wool shearing amount of alpine merino and application. The SNP molecular marker is located at the 20065540th nucleotide site G / A mutation on the fifth chromosome of the international sheep reference genome Oarv4.0 version. The invention further relates to a specific primer pair for detecting the SNP molecular marker by using a PCR technology, a kit containing the primer pair and a nucleotide polymorphism detection method, and compared with a traditional detection method, the technology has the advantages of high accuracy, high detection speed, low cost, easiness in result judgment and the like. SNP locus detection is used for carrying out early selection of the shearing amount of the high-mountain merino sheep, shortening the breeding period and accelerating the breeding process, a high-mountain merino sheep shearing amount early selection technology is established, the breeding time of the excellent character of the shearing amount of the high-mountain merino sheep is shortened, the breeding cost is reduced, and the application value is very high.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
SNP (Single Nucleotide Polymorphism) molecular marker for influencing wool length character of alpine merino and application of SNP molecular marker
ActiveCN114214426AImproving Breeding AccuracyThe detection process is fastMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
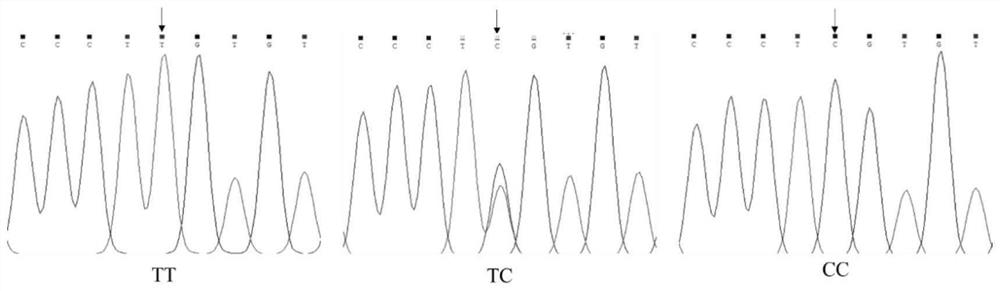
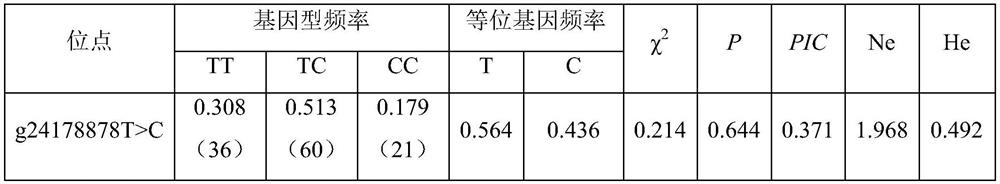
The invention belongs to the technical field of molecular genetics, and particularly relates to an SNP molecular marker influencing the wool length character of alpine merino and application. The SNP molecular marker is located at the 24178878th nucleotide site T / C mutation on the 22nd chromosome of the international sheep reference genome Oarv4.0 version. The invention further relates to a specific primer pair for detecting the SNP molecular marker by using a PCR technology, a kit containing the primer pair and a nucleotide polymorphism detection method, and compared with a traditional detection method, the technology has the advantages of high accuracy, high detection speed, low cost, easiness in result judgment and the like. SNP locus detection is used for carrying out early selection of the high-mountain merino wool length character, shortening the cultivation period and accelerating the cultivation process, an early selection technology of the high-mountain merino wool length character is established, the breeding time of the high-mountain merino wool length character is shortened, the breeding cost is reduced, and the application value is very high.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
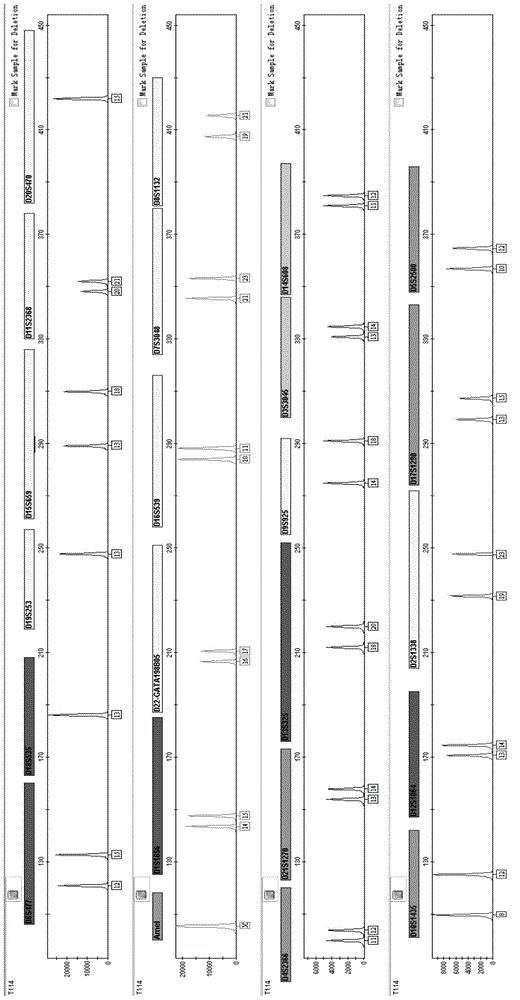
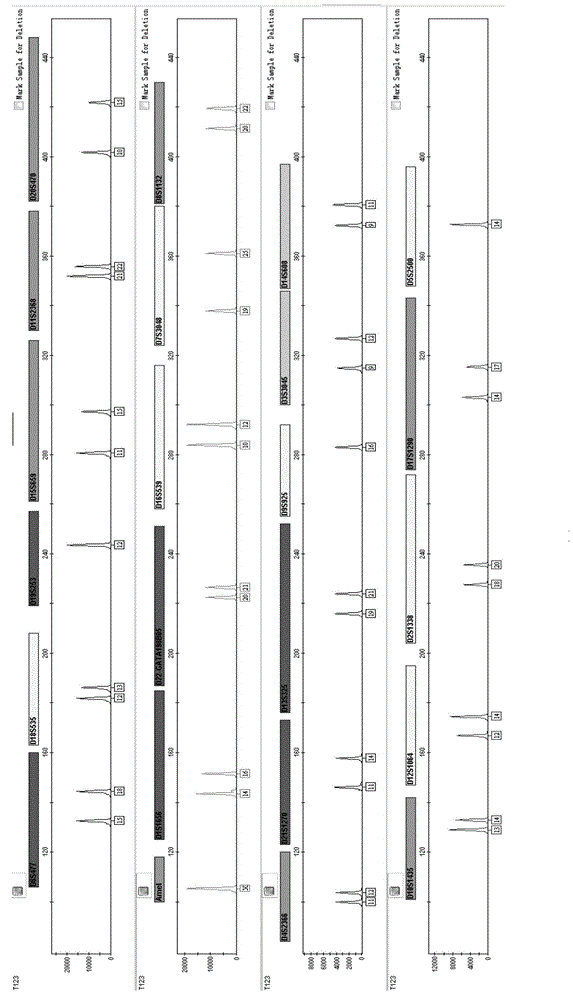
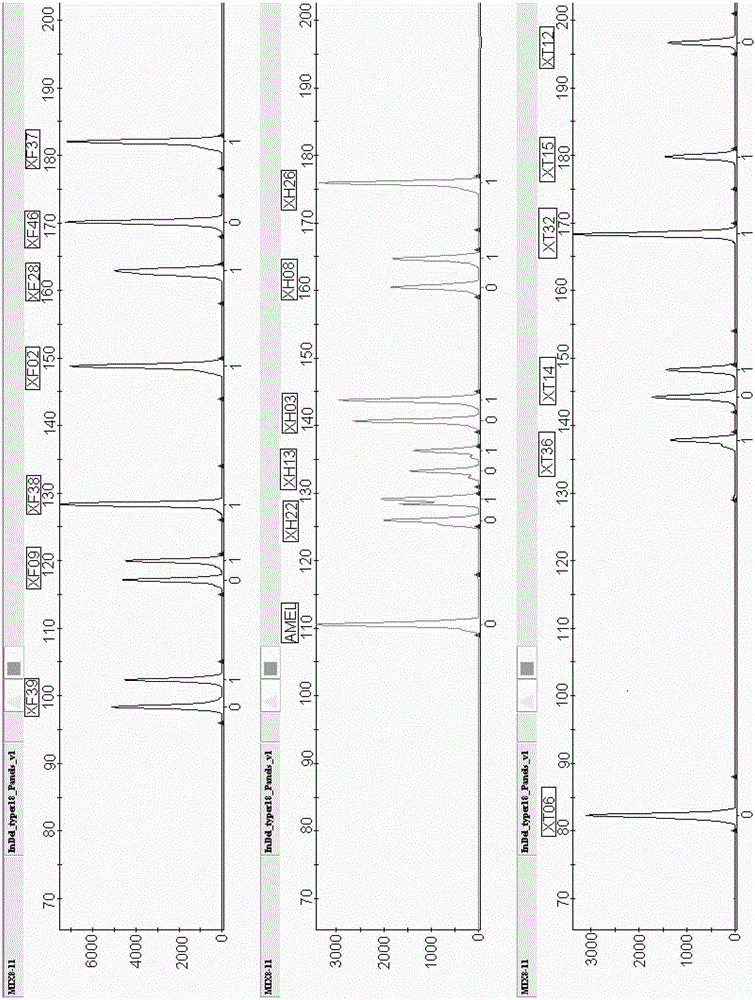
Fluorescently-labeled X-InDel locus composite amplification system and application thereof
ActiveCN104131067AHigh sensitivityHigh individual recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceInsertion deletionDNA paternity testing
The invention provides a fluorescently-labeled X-InDel locus composite amplification system. By means of the system, eighteen insertion-deletion genetic polymorphism loci on an X chromosome can be compositely amplified, wherein the eighteen loci are respectively labeled with three fluoresceins FAM, HEX and TAMRA. The composite amplification system is high in sensitivity and individual identification rate, is high in polymorphism, is good in stability and repeatability, is accurate in typing results and can satisfy a practical requirement. A kit can be manufactured on the basis of the composite amplification system, can be used for paternity test, dyad paternity test, grandparent and grandchild test, sibling test and individual identification. New technologies can be provided for the fields such as anthropology, medical genetics and the like.
Owner:ACADEMY OF FORENSIC SCIENCE
SNP molecular marker influencing weaning weight character of merino and application of SNP molecular marker
ActiveCN113584183AImproving Breeding AccuracyImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceNucleotide
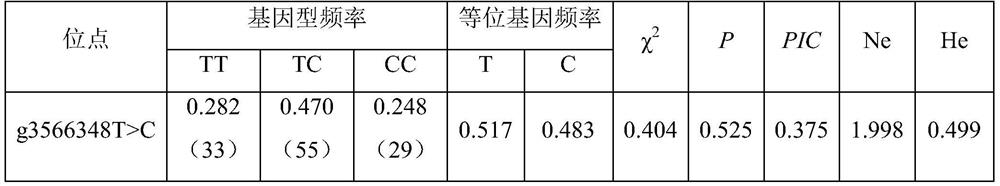
The invention belongs to the technical field of molecular genetics, and particularly relates to an SNP molecular marker influencing weaning weight character of merinos and application. The SNP molecular marker is located at the 3566348 nucleotide site T / C mutation on the 23<rd> chromosome of the international sheep reference genome Oar_v4.0 version. The invention further relates to a specific primer pair for detecting the SNP molecular marker by using a PCR technology, a kit containing the primer pair, and a nucleotide polymorphism detection method. Compared with a traditional detection method, the technology has the advantages of high accuracy, high detection speed, low cost, easiness in result judgment and the like. SNP locus detection is used for carrying out early selection of the weaning weight characters of high-mountain merinos, shortening the breeding period and accelerating the breeding process, an early selection technology of the weaning weight characters of the high-mountain merinos is established, the breeding time of the excellent weaning weight characters of the high-mountain merinos is shortened, the breeding cost is reduced, and the application value is very high.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
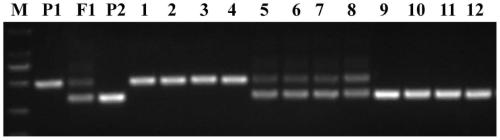
Primer group, kit and method for DNA file library building
ActiveCN112592981AMeet the application of personal identificationPrevent extractionMicrobiological testing/measurementLibrary creationMultiplexY-Chromosome Genes
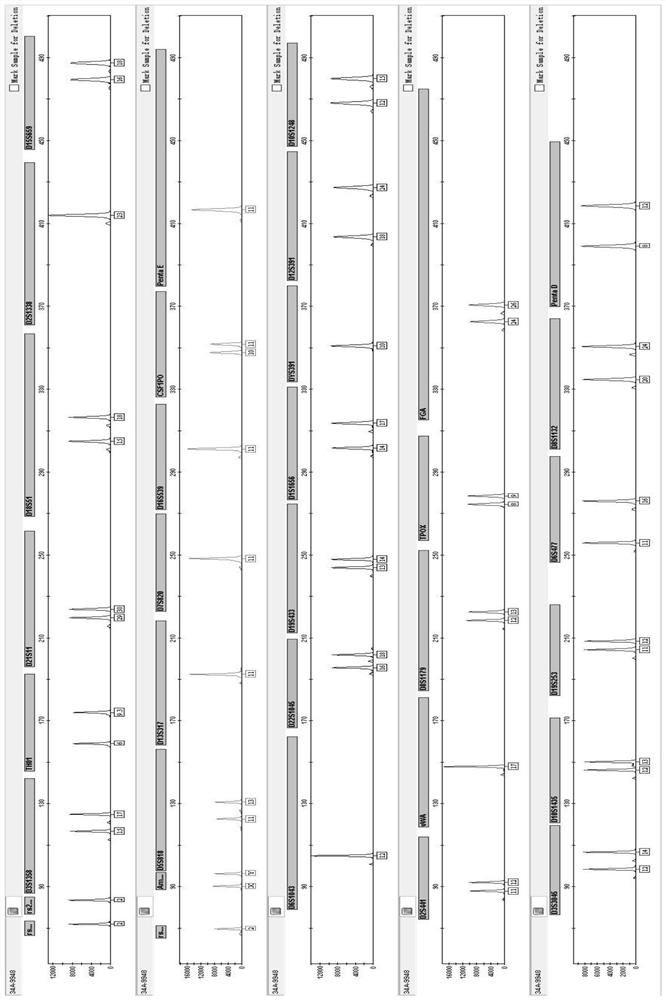
The invention discloses a primer group, a kit and a method for DNA file library building. The primer group aims at 25 autosomal STR gene loci, 1 X and Y chromosome homologous gene locus, AMEL, 27 Y-STR gene loci, 25 X-STR gene loci, 33 global population common autosomal SNP loci, 28 East Asia population common Y chromosome SNP loci and 3 mitochondrial hypervariable regions. Through advanced screening, the loci can meet various judicial or forensic identification purposes, the cumulative individual recognition rate reaches 0.99999999999999, and the individual recognition application of ChineseHan nationality and most people in the world can be met. Primers are as shown in SEQ ID NO.:1-284. The primer group can be directly used for multiplex PCR, and the genetic marker information of a detected material can be obtained to the maximum extent only by one round of experiment in combination of the multiplex PCR and next-generation sequencing technology. The primer group is suitable for application scenes of newborn birth medicine proving DNA file library building, paternity test, DNA comparison, population genetics research and the like, is also suitable for severely degraded and decayed sample detection, and has important value.
Owner:GUANGZHOU JINGKE DX CO LTD +1
Reagent and kit for detecting 13 CODIS STR gene loci of trace mixed human source DNA sample and application thereof
InactiveCN107557481AReach suitabilityNo knock-on effectMicrobiological testing/measurementCommercial kitAmelogenin
The invention discloses a reagent and a kit for detecting 13 CODIS STR gene loci of a trace mixed human source DNA sample and an application thereof. The reagent comprises amplification primers for 13CODIS STR gene loci and a sex gene locus, wherein the 13 CODIS STR gene loci are separately TPOX, DS31358, FGA, D5S818, CSF1PO, D8S1179, D7S820, TH01, vWA, D13S17, D16S539, D18S51 and D21S11, and thesex gene locus is Amelogenin. The sequences of the amplification primers are separately shown as SEQ ID NO: 1-SEQ ID NO: 28. The 13 CODIS STR gene loci are independent of one another without a chaineffect, and can be matched with the gene loci of an existing commercial kit and a database of the Ministry of Public Security, so that the adaptability of an analytical result is achieved; meanwhile,a single DNA molecule can be amplified and detected; on the premise of ensuring the efficiency of the system, the detection rate of degraded / broken DNA templates can be increased; the reagent and kitcan be widely applied to the fields of individual recognition by legal medical experts, paternity test and other group genetics analysis.
Owner:SUZHOU GENO TRUTH BIOTECHNOLOGY CO LTD +1
Primer group and kit for simultaneously amplifying 13 human STR gene loci and application of primer group and kit
PendingCN112852973ARapid expansionImprove amplification specificityMicrobiological testing/measurementDNA/RNA fragmentationCore geneMolecular genetics technique
The invention provides a primer group and a kit for simultaneously amplifying 13 human STR gene loci and application of the primer group and the kit, and belongs to the technical field of molecular genetics. In the invention, the 13 STR gene loci comprise 12 autosomal STR gene loci and one sex identification STR gene locus. According to the method, 13 STR gene loci with fragments smaller than 320 bp can be amplified in one reaction at the same time, 8 core gene loci and 5 preferable gene loci specified by the Ministry of Public Security are included, meanwhile, a sex identification gene locus is further included, and the problem that detection of trace and degraded large-fragment gene loci of a detected material is lost can be effectively prevented. The 13 STR gene loci are combined with mainstream kits in the market, and more gene locus information can be obtained for trace and degraded DNA, so that the individual recognition capability is effectively improved.
Owner:百特元生物科技(北京)有限公司
Group-of-soybean quantitative trait QTL loci and screening method thereof
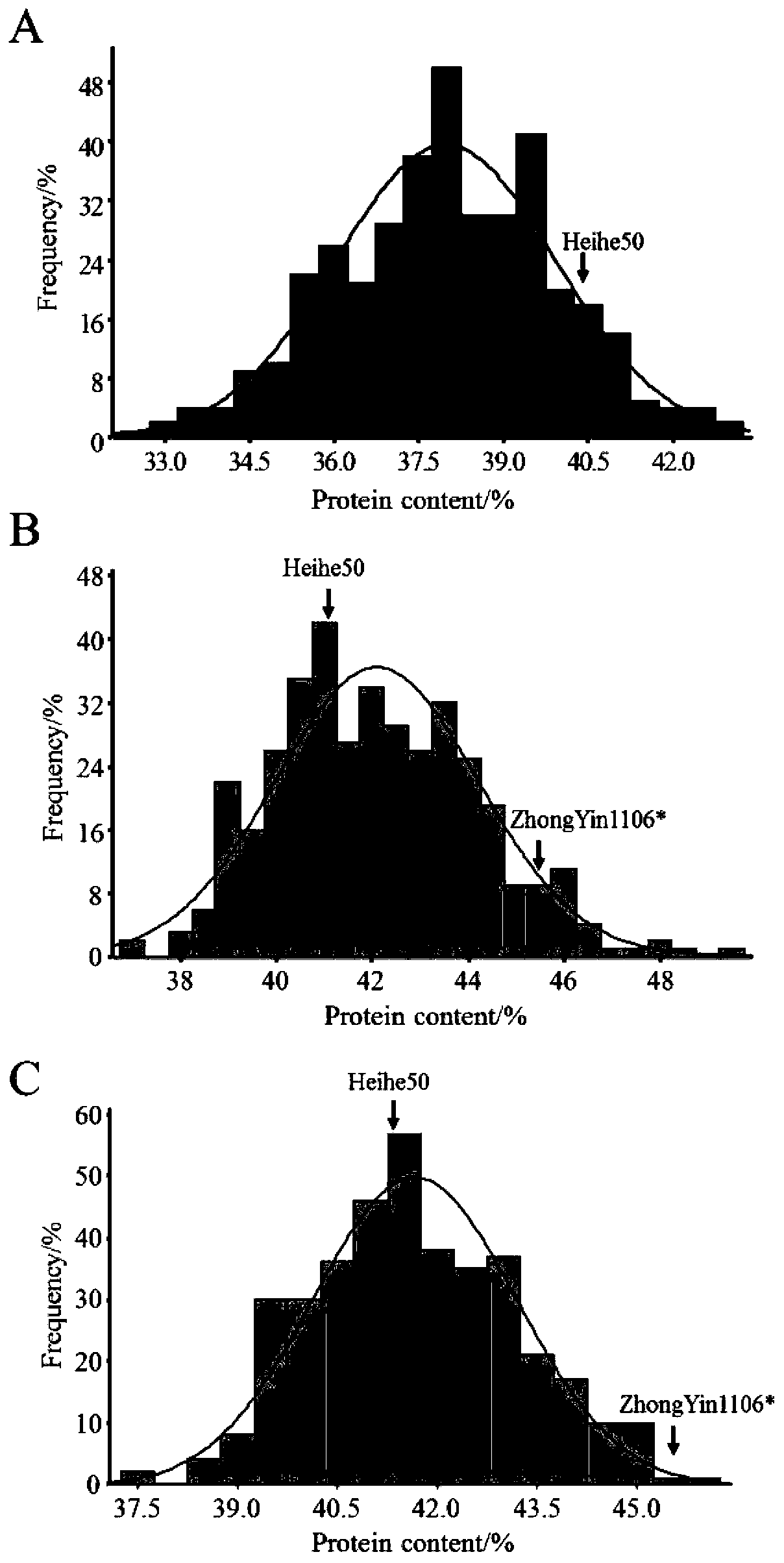
The invention discloses group-of-soybean quantitative trait QTL loci and a screening method thereof, and belongs to the field of molecular genetics. The method comprises the following steps: determining a target character; screening parents and constructing a genetic population; obtaining phenotypic data of the target character; extracting genome DNA of the parent and the genetic population; constructing a mixing pool, further screening molecular markers linked with target traits between parents and the mixing pool, and selecting the molecular markers with p less than 0.0001 to further identify genotypes in the genetic population; performing qTL localization analysis. By utilizing the method, two QTL loci for controlling the content of soybean protein and QTL loci for controlling the branching number of soybeans are screened out. The soybean quantitative trait QTL locus screening method is established, a foundation is laid for gene initial positioning, fine positioning, cloning and newvariety breeding of soybean related traits, and meanwhile available markers are provided for soybean molecular marker-assisted selection.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Primer group and kit for simultaneously amplifying 34 human STR gene loci and application of primer group and kit
PendingCN112852972AImprove individual recognitionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationCore geneY chromosome deletions
The invention provides a primer group and a kit for simultaneously amplifying 34 human STR gene loci and application of the primer group and the kit, and belongs to the technical field of molecular genetics. In the invention, the 34 STR gene loci comprise 29 autosomal STR gene loci, one Y chromosome STR gene locus and 4 sex identification STR gene loci. According to the invention, 34 STR gene loci can be simultaneously amplified in one reaction, 20 core gene loci and 10 preferable gene loci specified by Ministry of Public Security are included, all loci of mainstream kits in the current market are included, and meanwhile, 5 gender identification gene loci are also included; and the risk of gender identification errors caused by Y chromosome deletion can be effectively prevented. 34 STR gene loci are combined, so that the method has the characteristics of high individual recognition capability and high non-father exclusion rate.
Owner:百特元生物科技(北京)有限公司
SNP (Single Nucleotide Polymorphism) marker for influencing wool length of alpine merino sheep and application of SNP marker
ActiveCN114752681AImprove accuracyThe detection process is fastFood processingMicrobiological testing/measurementNucleotideMolecular genetics
The invention belongs to the technical field of molecular genetics, and particularly relates to an SNP (Single Nucleotide Polymorphism) marker related to influence on the wool length of alpine merino and application of the SNP marker, and the SNP molecular marker is located at the 6296384 nucleotide site T / C mutation on the No.3 chromosome of the international sheep reference genome Oarv4.0 version. The invention further relates to a specific primer pair for detecting the SNP molecular marker by using a PCR technology, a kit containing the primer pair and a nucleotide polymorphism detection method, and compared with a traditional detection method, the technology has the advantages of high accuracy, high detection speed, low cost, easiness in result judgment and the like. SNP locus detection is used for carrying out early selection of the wool length of the high-mountain merino sheep, shortening the breeding period and accelerating the breeding process, a technology for selecting the wool length of the high-mountain merino sheep in the early stage is established, the breeding time of excellent characters of the wool length of the high-mountain merino sheep is shortened, the breeding cost is reduced, and the application value is very high.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
Primer group and kit for simultaneously amplifying 37 Y-STR gene loci of human and application thereof
PendingCN112852970ARapid expansionImprove amplification specificityMicrobiological testing/measurementDNA/RNA fragmentationForensic databaseForensic dna
The invention provides a primer group and a kit for simultaneously amplifying 37 Y-STR gene loci of human and application thereof, and belongs to the technical field of molecular genetics. In the invention, the 37 Y-STR gene loci comprise 36 Y chromosome STR gene loci and 1 Y-indel gene locus; 37 gene loci can be amplified in one reaction at the same time, the compatibility of current public security DNA database comparison is fully met, the amplification time is shortened, the identification efficiency and the detection material adaptability of the kit are improved, and the multifunctional STR identity identification requirements of current forensic identity identification, forensic DNA database construction and judicial genetic relationship identification can be met.
Owner:百特元生物科技(北京)有限公司
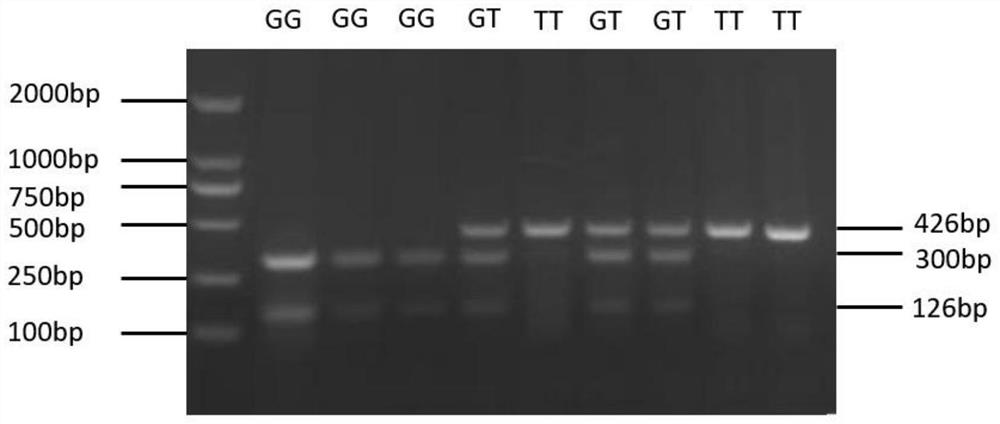
Method for identifying chicken abdominal fat mass by using chicken CEBP zeta gene and application of method
ActiveCN112375811ASpeed up the breeding processHigh precisionMicrobiological testing/measurementBiotechnologyEnzyme digestion
The invention discloses a method for identifying chicken abdominal fat mass by using a chicken CEBP zeta gene and application of the method, belongs to the technical field of animal molecular genetics, and aims to more accurately and conveniently identify low-fat broiler chickens and accelerate the breeding process of the broiler chickens. Primers are designed according to upstream 123bp and downstream 302bp sequences of the chicken CEBP zeta gene SNP (single nucleotide polymorphism) locus g.764T>G, the obtained primers are used for performing PCR (polymerase chain reaction) amplification on chicken genome DNA, an amplification product is subjected to enzyme digestion by restriction endonuclease, an enzyme digestion product is subjected to agarose gel electrophoresis separation, and a marker genotype of chicken abdominal fat content is obtained. The method is simple to operate, low in cost and high in accuracy, can be used for automatic detection, is an effective molecular marker breeding means, and provides a scientific basis for marker-assisted selection of chickens, and with adoption of the method, acceleration of the broiler chicken breeding process and cultivation of low-fat broiler chickens meeting market requirements are facilitated, so that relatively good economic benefits are obtained.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Multiplex amplification system and kit for 23 short tandem repeats
ActiveCN103834732BImprove performanceIncrease independenceMicrobiological testing/measurementMultiplexGenome human
Owner:SUZHOU MICROREAD GENETICS
SNP (Single Nucleotide Polymorphism) molecular marker for identifying age-round shearing amount of alpine merino and application of SNP molecular marker
ActiveCN114657266AImproving Breeding AccuracyThe detection process is fastFood processingMicrobiological testing/measurementNucleotideMolecular genetics technique
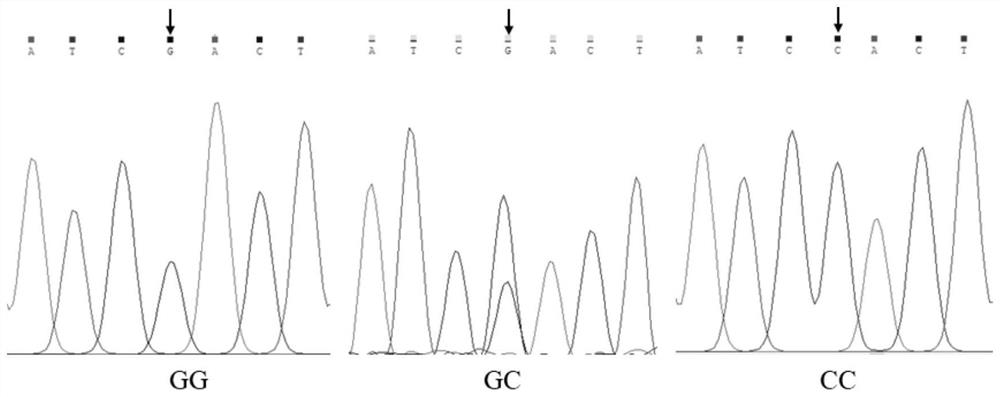
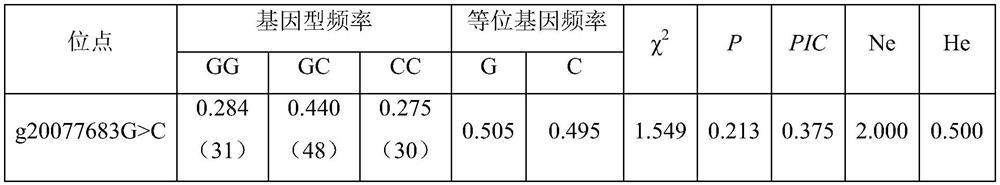
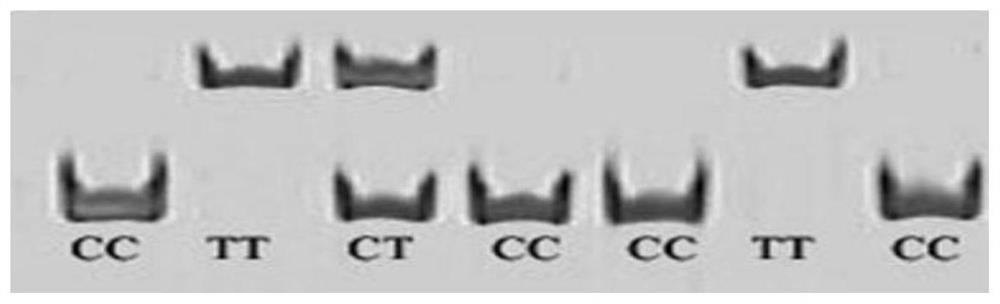
The invention belongs to the technical field of molecular genetics, and particularly relates to an SNP (Single Nucleotide Polymorphism) molecular marker for identifying the age-round shearing amount of alpine merino and application of the SNP molecular marker. The SNP molecular marker is located at the 20077683th nucleotide site G / C mutation on the fifth chromosome of the international sheep reference genome Oarv4.0 version. The invention further relates to a specific primer pair for detecting the SNP molecular marker by using a PCR technology, a kit containing the primer pair and a nucleotide polymorphism detection method, and compared with a traditional detection method, the technology has the advantages of high accuracy, high detection speed, low cost, easiness in result judgment and the like. SNP locus detection is used for carrying out early selection of the annual shearing amount of the alpine merino sheep, shortening the breeding period and accelerating the breeding process, an early selection technology of the annual shearing amount of the alpine merino sheep is established, the breeding time of the excellent character of the annual shearing amount of the alpine merino sheep is shortened, the breeding cost is reduced, and the application value is very high.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
Gene locus closely associated with level of milk polyunsaturated fatty acid and application thereof
InactiveCN103233002APromote healthy growthMicrobiological testing/measurementDNA/RNA fragmentationNutritionNucleotide
The invention relates to a gene locus closely associated with the level of milk polyunsaturated fatty acid and application thereof, belonging to the technical field of genetics. The invention provides a gene locus closely associated with the level of milk polyunsaturated fatty acid, namely a fatty acid desaturase 1 gene (FADS1) locus, i.e. an FADS1 locus having rs174556 mononucleotide polymorphism. The mononucleotide polymorphism rs174556 base C of the locus is mutated into T, thus being closely associated with the level of milk polyunsaturated fatty acid. The levels of arachidonic acid and eicosapentaenoic acid in milk of a wet nurse carrying FADS1 gene rs174556T / T minor allele homozygote are obviously degraded, and the locus has mononucleotide polymorphism determining the level of milk polyunsaturated fatty acid. The FADS1 locus and a coding protein thereof are of great importance in elucidating a polyunsaturated fatty acid metabolic mechanism, establishing individual nutrition intervention and establishing appropriate dietary recommended intakes of polyunsaturated fatty acid based on different genetic backgrounds.
Owner:JILIN UNIV
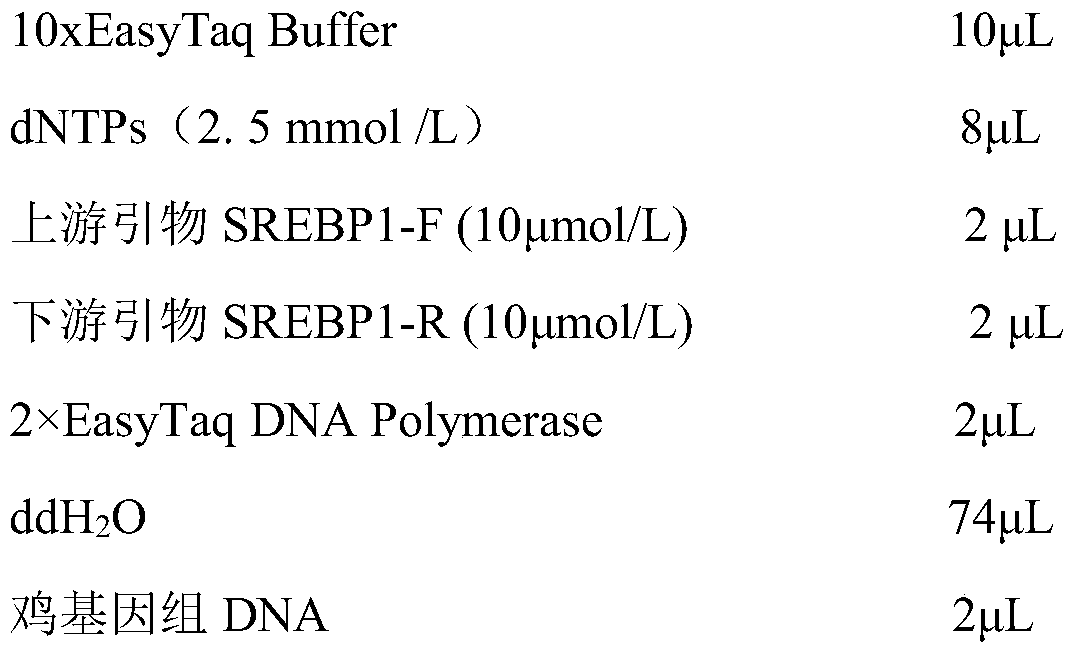
A molecular marker method and application for simultaneously predicting abdominal fat traits, leg bone traits and reproductive performance of broilers
ActiveCN111560440BSpeed up the breeding processHigh precisionMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionGenomic DNA
The invention discloses a molecular marking method for simultaneously predicting abdominal fat characters, leg skeleton characters and reproductive performance of broilers and an application, and belongs to the technical field of animal molecular genetics. The invention discloses a method for identifying low-fat broilers more accurately and conveniently and accelerating broiler breeding process. According to the invention, primers are designed according to the sequences from the 208 bp at the upstream to the 72 bp at the downstream of the SNP locus g.3656C > T of the chicken SREBP1 gene exon 3to obtain an amplification primer, then carrying out PCR amplification on the chicken genomic DNA according to the obtained primer, and carrying out enzyme digestion on the amplification product by using restriction endonuclease to obtain the chicken marker genotype. The method is simple to operate, low in cost and high in accuracy, can be used for automatic detection, is an effective molecular marker breeding means, and can be used for accelerating the breeding process of chickens and cultivating low-fat broilers with healthy leg bones and good reproductive performance, which meet market requirements, so that good economic benefits are obtained.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular markers and screening methods of ppaiv2 gene locus in pear
ActiveCN106701911BSpeed up the selection processQuick filterMicrobiological testing/measurementDNA/RNA fragmentationPEARGermplasm
The invention relates to the field of molecular genetics, discloses a molecular marker of a pear PpAIV2 gene locus and further discloses a screening method of the molecular marker of the pear PpAIV2 gene locus. A genome simple sequence repeat (SSR) locus adjacent to pear soluble acid invertase gene PpAIV2 is predicated and a locus specific primer is designed; one SSR allelic variation locus which is tightly interlocked with the PpAIV2 is obtained through amplification in germplasms with different genetic backgrounds and F1-generation population genomes; the SSR allelic variation locus is named as P.zaas-1. The pear soluble acid invertase gene PpAIV2 is a key gene for accumulating pear fruit sucrose; the P.zaas-1 has potential identification and screening effects on materials with remarkable differences on accumulation of the fruit sucrose.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
SNP (Single Nucleotide Polymorphism) marker for identifying weaning weight of alpine merino and application of SNP marker
ActiveCN114657265AImproving Breeding AccuracyImprove accuracyFood processingMicrobiological testing/measurementWeaningNucleotide
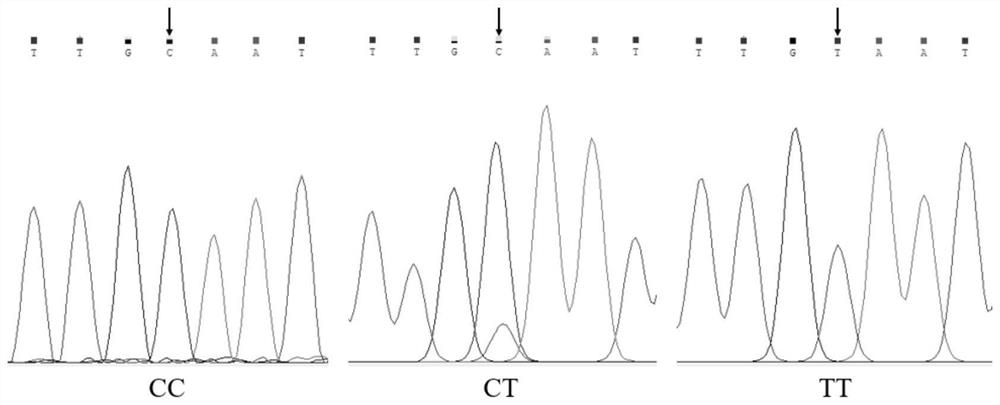
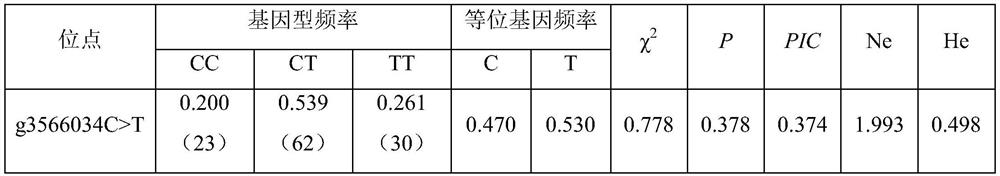
The invention belongs to the technical field of molecular genetics, and particularly relates to an SNP (Single Nucleotide Polymorphism) marker for identifying the weaning weight of alpine merino and application of the SNP marker, and the SNP molecular marker is located at the 3566034th nucleotide site C / T mutation on the 23rd chromosome of the international sheep reference genome Oarv4.0 version. The invention further relates to a specific primer pair for detecting the SNP molecular marker by using a PCR technology, a kit containing the primer pair and a nucleotide polymorphism detection method, and compared with a traditional detection method, the technology has the advantages of high accuracy, high detection speed, low cost, easiness in result judgment and the like. SNP locus detection is used for carrying out early selection of the weaning weight of the high-mountain merino sheep, shortening the breeding period and accelerating the breeding process, a technology for early selection of the weaning weight of the high-mountain merino sheep is established, the breeding time of excellent characters of the weaning weight of the high-mountain merino sheep is shortened, the breeding cost is reduced, and the application value is very high.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
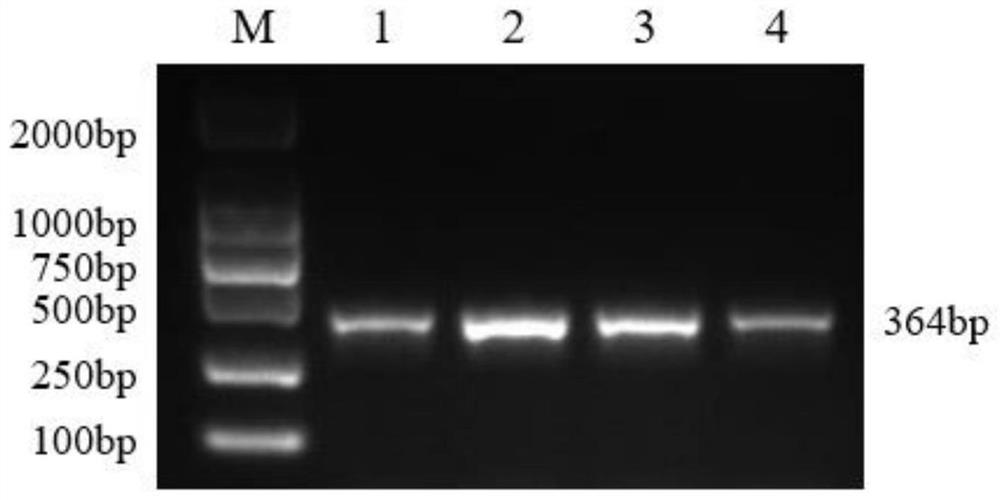
A kind of rice blast resistance locus pi2/9 functional gene molecular marker and its application
ActiveCN107916300BLow costImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyResistant genes
The invention provides a functional gene molecular marker of a rice blast resistance locus Pi2 / 9 and application thereof. The molecular marker of a specific band type for rice blast resistance genes Pi2, Piz-t, Pi9 and Pigm is amplified from rice genome DNA by virtue of a primer pair SEQ ID NO.1-2. The functional gene specific molecular marker of the rice blast resistance locus Pi2 / 9 has importantapplication value, and utilization efficiency of the gene in germplasm resource screening, molecular marker-assisted selective breeding, gene pyramiding breeding and transgenic breeding can be increased by utilizing the marker.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
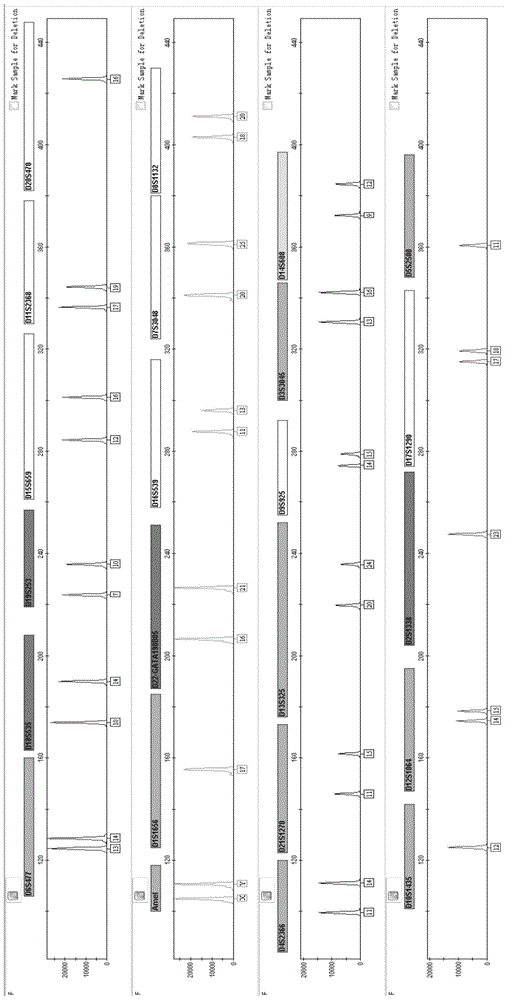
A fluorescently labeled x-indel site compound amplification system and its application
ActiveCN104131067BHigh sensitivityHigh individual recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceInsertion deletionMultiplexing
The invention provides a fluorescently-labeled X-InDel locus composite amplification system. By means of the system, eighteen insertion-deletion genetic polymorphism loci on an X chromosome can be compositely amplified, wherein the eighteen loci are respectively labeled with three fluoresceins FAM, HEX and TAMRA. The composite amplification system is high in sensitivity and individual identification rate, is high in polymorphism, is good in stability and repeatability, is accurate in typing results and can satisfy a practical requirement. A kit can be manufactured on the basis of the composite amplification system, can be used for paternity test, dyad paternity test, grandparent and grandchild test, sibling test and individual identification. New technologies can be provided for the fields such as anthropology, medical genetics and the like.
Owner:ACADEMY OF FORENSIC SCIENCE
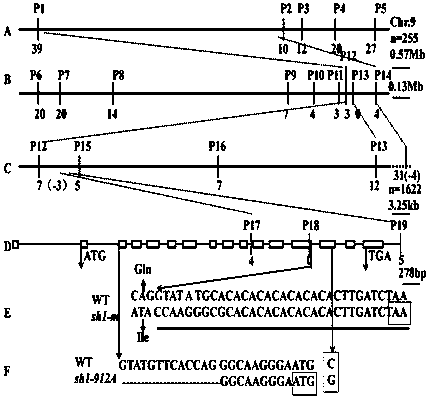
A kind of corn sucrose synthase mutant gene, encoded protein and application thereof
The invention belongs to the technical field of biological genetics, and in particular, relates to a corn sucrose synthase mutation gene, a protein encoded by the gene, and an application of the gene. The corn sucrose synthase mutant sh1-m comprises a nucleotide sequence shown in SEQ ID No:1. The protein encoded by the corn sucrose synthase mutant sh1-m comprises an amino acid sequence shown in SEQ ID No:2. The sh1-m mutant is discovered in an improved Zheng58 selfing line material (Gai-Z58), a gene sequence structure is verified, and the corn sucrose synthase mutant sh1-m is confirmed to be an allele of Sh1 gene locus, can significantly improve the endosperm amylose content (16.5%), and has excellent application value.
Owner:MAIZE RES INST SHANDONG ACAD OF AGRI SCI
A kind of rflp method and its application of quick detection yellow cattle flii gene SNP
ActiveCN105349673BSimple polymorphism detectionPolymorphic precisionMicrobiological testing/measurementAnimal scienceNucleotide
Owner:NORTHWEST A & F UNIV
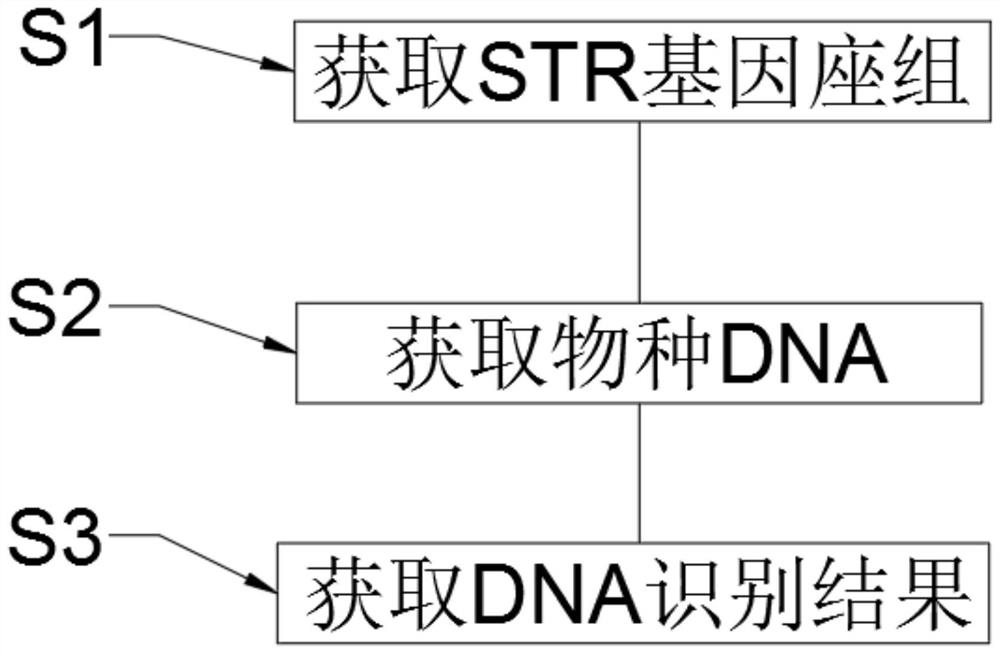
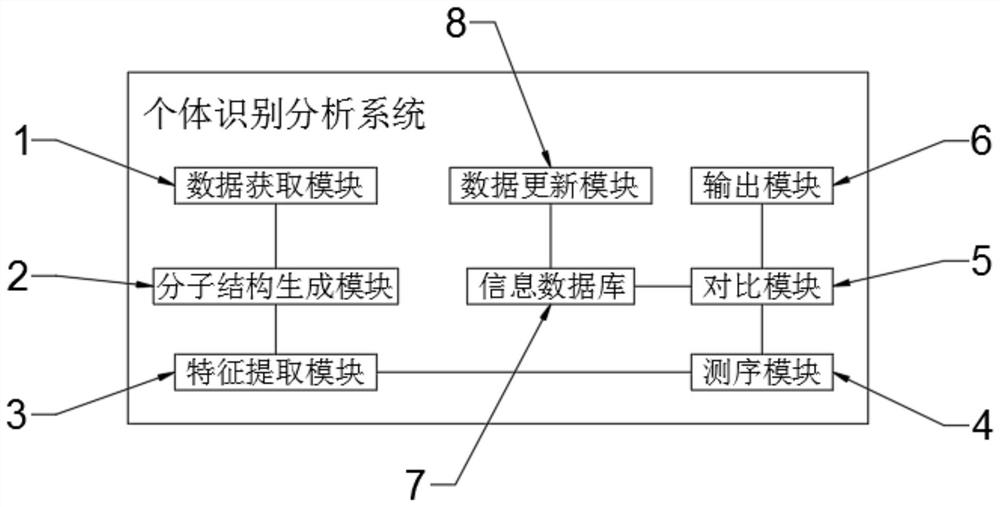
Cross-species individual identification method and individual identification analysis system
PendingCN113736773AAchieving identifiabilityAchieving kinship identificationMicrobiological testing/measurementProteomicsCore geneCommon species
The invention belongs to the technical field of genetics determination, and particularly relates to a cross-species individual identification method and an individual identification analysis system. The cross-species individual identification method comprises the following steps of 1, obtaining an STR gene locus group; and 101) taking out core gene sequences in whole-genome sequences of ten common species including human, pig, cattle, horse, goat, dog, cat, sheep, rabbit and yak. Compared with the prior art, the technology provided by the invention has the most important advantages that cross-species identification of ten common species types is realized compared with individual identification and genetic identification of the existing single species, so that the ten common species types share one STR site, and the related work of maintaining a corresponding DNA analysis system for each single species is greatly simplified; and a plurality of corresponding species only need to share one DNA analysis system, and the individual identification and genetic identification of the ten species can be realized by utilizing the STR gene locus set and an individual identification algorithm provided by the invention.
Owner:深圳百人科技有限公司
Molecular marker method for simultaneously predicting abdominal fat characters, leg skeleton characters and reproductive performance of broiler chickens and application
ActiveCN111560440ASpeed up the breeding processHigh precisionMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionGenomic DNA
The invention discloses a molecular marking method for simultaneously predicting abdominal fat characters, leg skeleton characters and reproductive performance of broilers and an application, and belongs to the technical field of animal molecular genetics. The invention discloses a method for identifying low-fat broilers more accurately and conveniently and accelerating broiler breeding process. According to the invention, primers are designed according to the sequences from the 208 bp at the upstream to the 72 bp at the downstream of the SNP locus g.3656C > T of the chicken SREBP1 gene exon 3to obtain an amplification primer, then carrying out PCR amplification on the chicken genomic DNA according to the obtained primer, and carrying out enzyme digestion on the amplification product by using restriction endonuclease to obtain the chicken marker genotype. The method is simple to operate, low in cost and high in accuracy, can be used for automatic detection, is an effective molecular marker breeding means, and can be used for accelerating the breeding process of chickens and cultivating low-fat broilers with healthy leg bones and good reproductive performance, which meet market requirements, so that good economic benefits are obtained.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
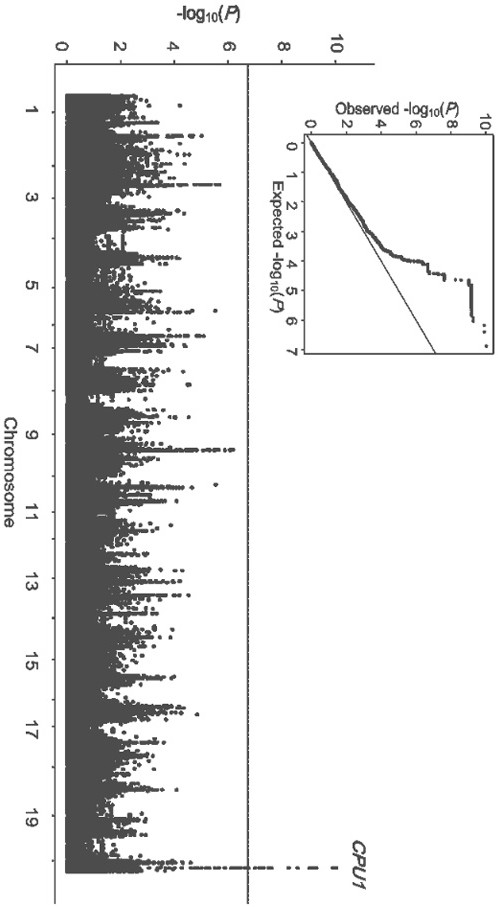
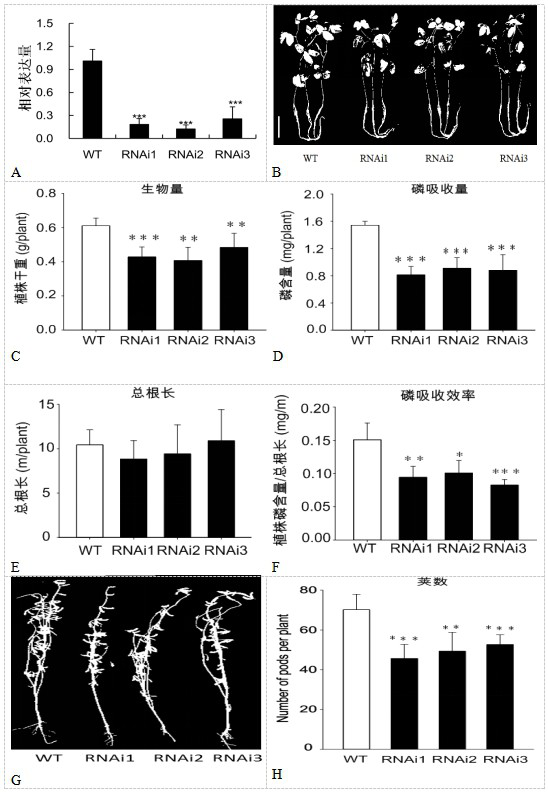
SEC12-like protein gene CPU1 and application thereof in improving soybean phosphorus efficiency
PendingCN113930431AExcellent allele resourcePlant peptidesFermentationBiotechnologyWhole Genome Association Analysis
The invention discloses an SEC12-like protein gene CPU1 and application thereof in improving soybean phosphorus efficiency, and belongs to the field of biotechnology. Through a forward genetics method of whole genome association analysis, a major genetic locus influencing soybean phosphorus absorption efficiency is identified, and a candidate gene CPU1 therein is identified. The gene CPU1 has natural variation in soybeans, and comprises two alleles, namely a phosphorus low-efficiency allele CPU1-H1 and a phosphorus high-efficiency allele CPU1-H2. The research based on the whole transformed plant of the soybean shows that the phosphorus absorption efficiency of the soybean is remarkably reduced by inhibiting the expression of the allele CPU1-H2, and the biomass and yield of the transgenic plant are finally reduced. According to the invention, a new scientific insight is provided for genetic basic research of natural variation of crops including soybeans, and meanwhile, an excellent allele resource is provided for phosphorus high-efficiency molecular breeding.
Owner:FUJIAN AGRI & FORESTRY UNIV
Molecular marking method for peanut flowering habit related gene locus and application of molecular marking method
ActiveCN111471789AClear identification methodFine identification methodMicrobiological testing/measurementClimate change adaptationBiotechnologyMolecular breeding
The invention belongs to the technical fields of molecular genetics and molecular breeding of crops and provides a molecular marking method for a peanut flowering habit related locus and application of the molecular marking method. A molecular marker comprises InDel markers PID12-21 and PID12-29 which are located on a twelfth chromosome and an InDel marker PID2-9 located on a 02-th chromosome. Formonogenic-inheritance hybrid colonies, flowering habits only need to consider a genotype of flowering habit related loci of the twelfth chromosome; for digenic-inheritance hybrid colonies, the flowering habits need to comprehensively consider genotypes of the flowering habit related loci of the 02-th and the twelfth chromosomes; and regardless of the hybrid colonies of monogenic- / digenic-inheritance modes, by only identifying a genotype of an amplified target strap, a perpetual blooming type and an alternate blooming type of peanuts can be distinguished, types of flowering habits of filial generations of the peanuts are screened rapidly, and flowering habit related plant type breeding, yield breeding and the like are assisted.
Owner:QINGDAO AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com