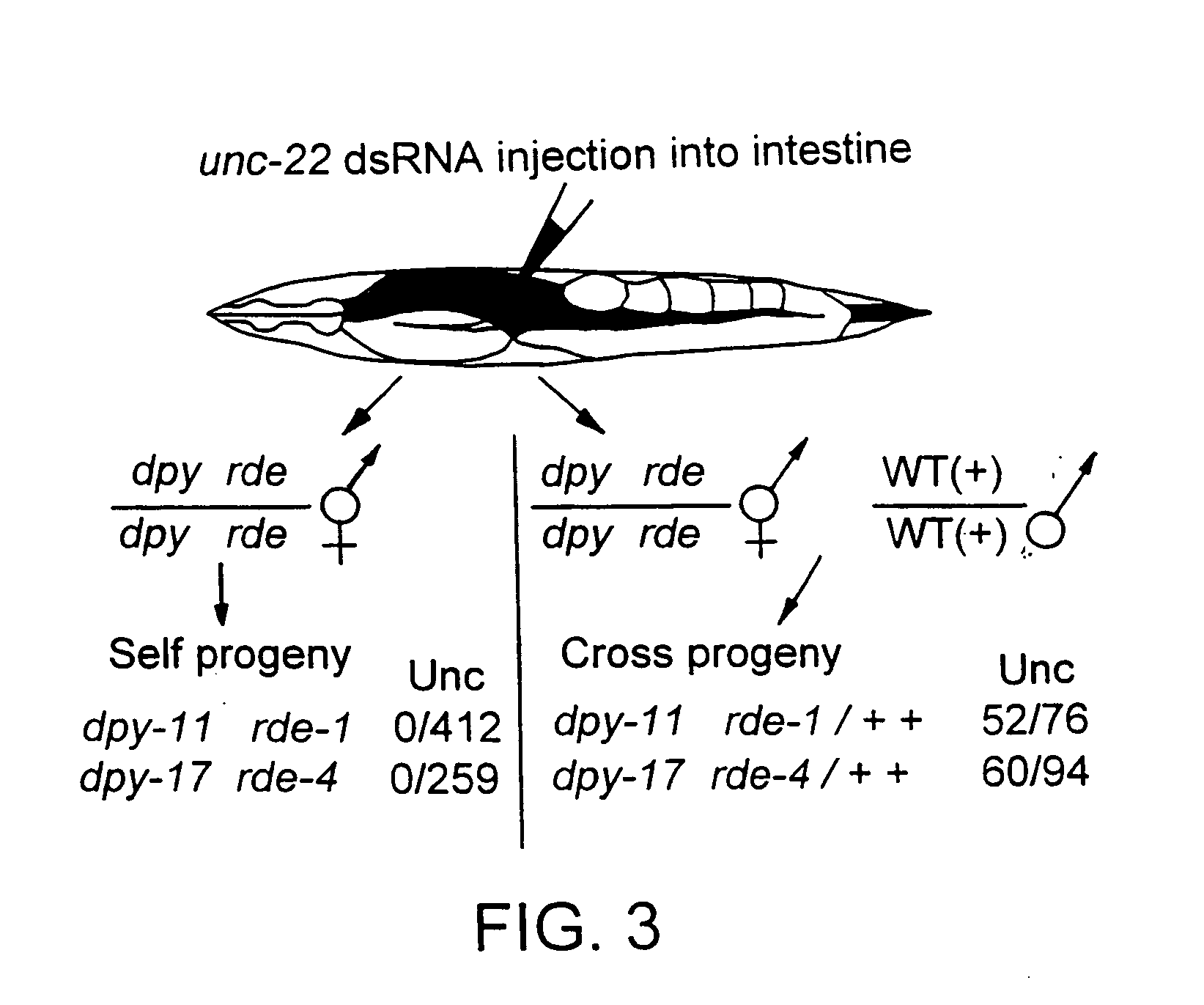
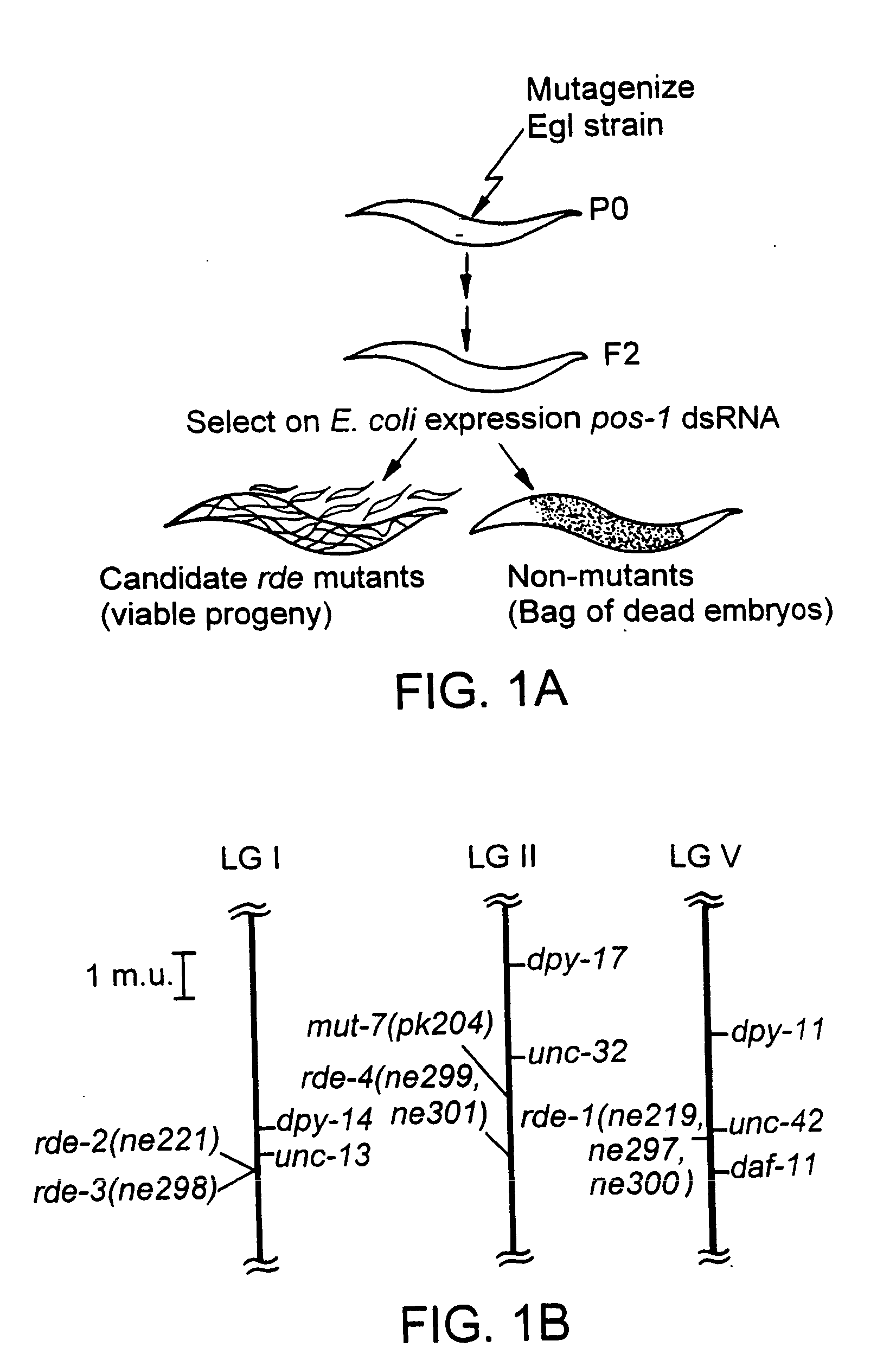
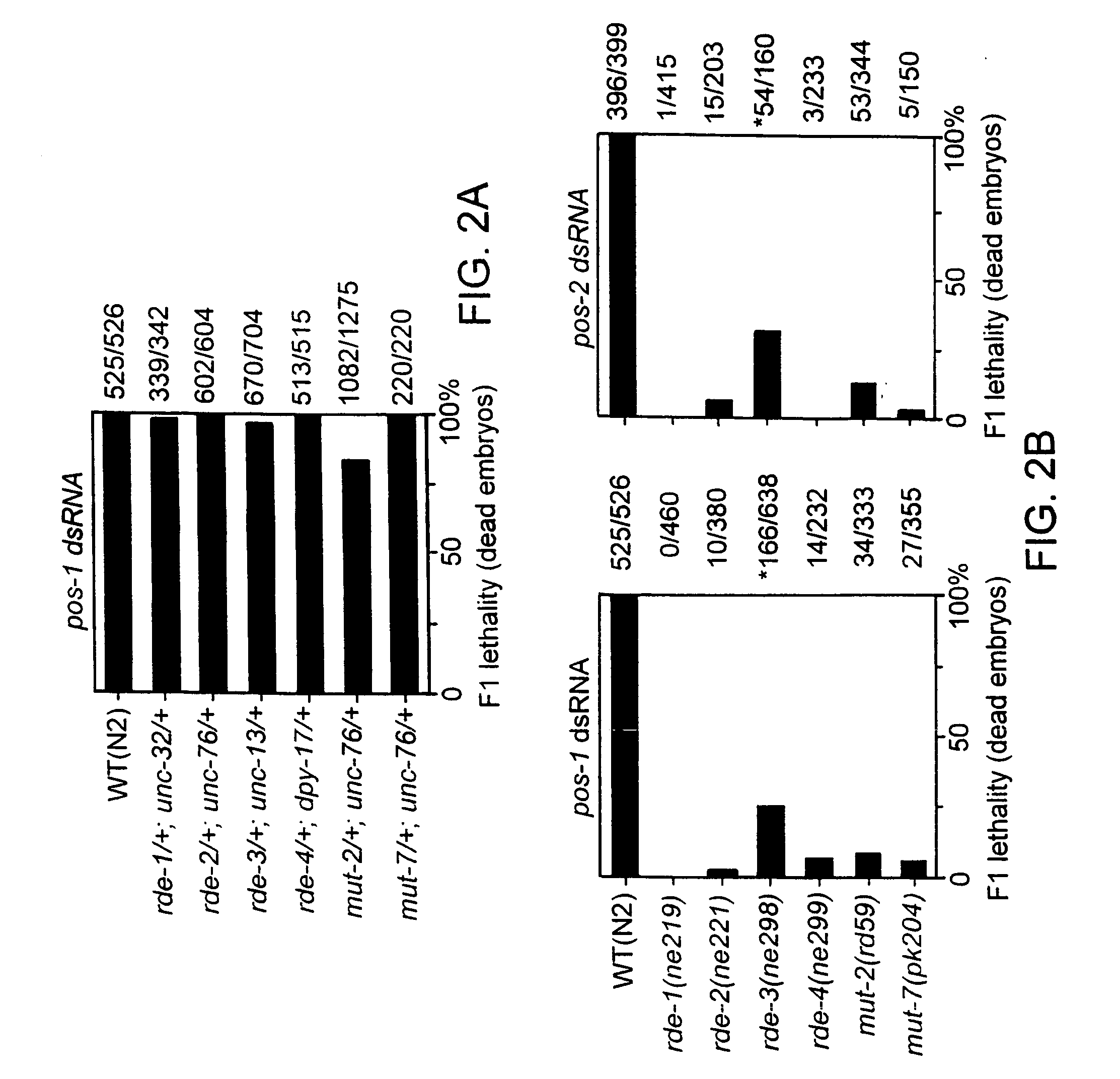
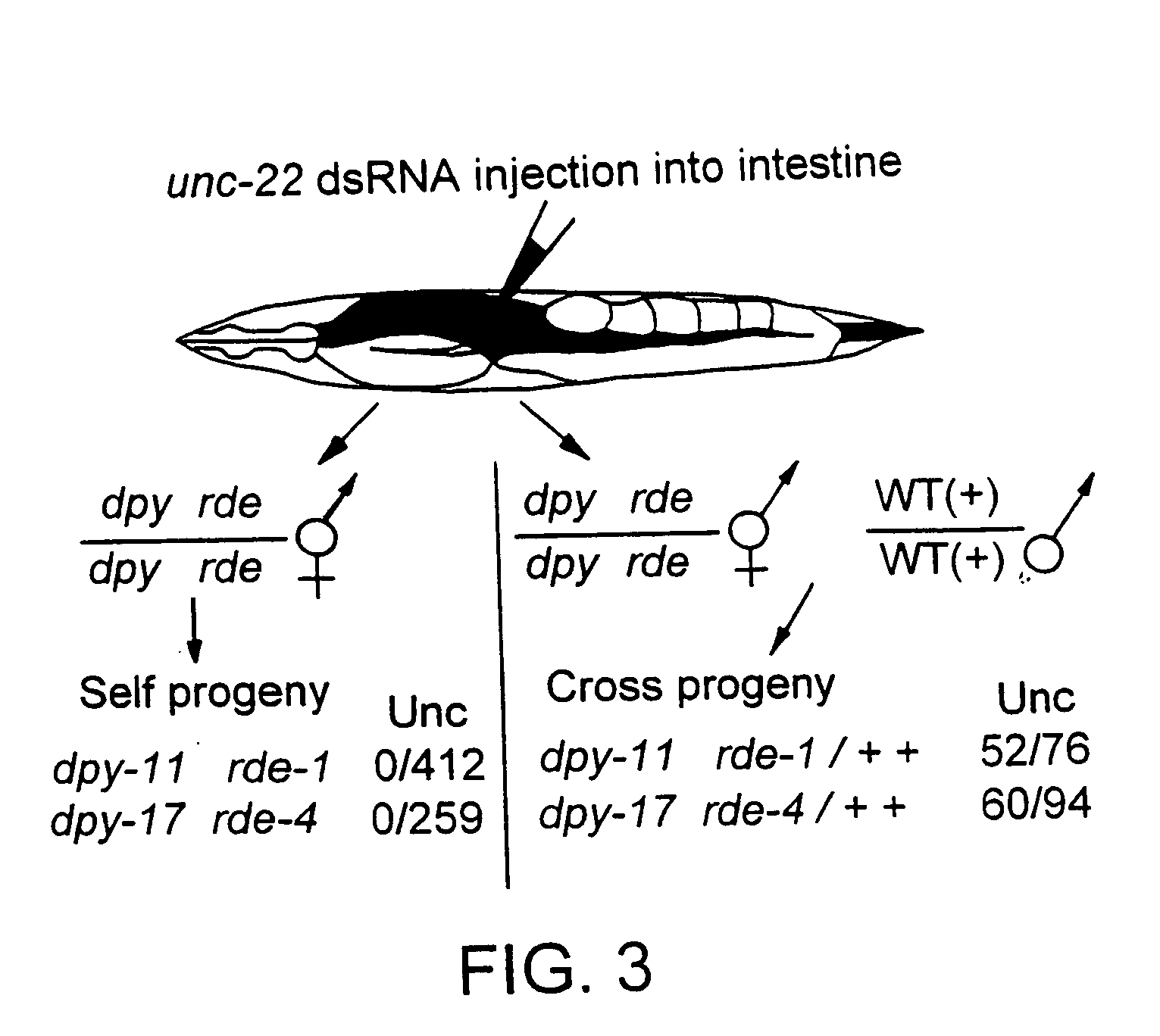
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
41 results about "Gene pathway" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The crucial difference between a gene set and a pathway is that a gene set is an unordered collection of genes whereas a pathway is a complex model that describes a given process, mechanism or phenomenon. Thus, it is very important to understand the difference between a pathway and its corresponding gene set representation.
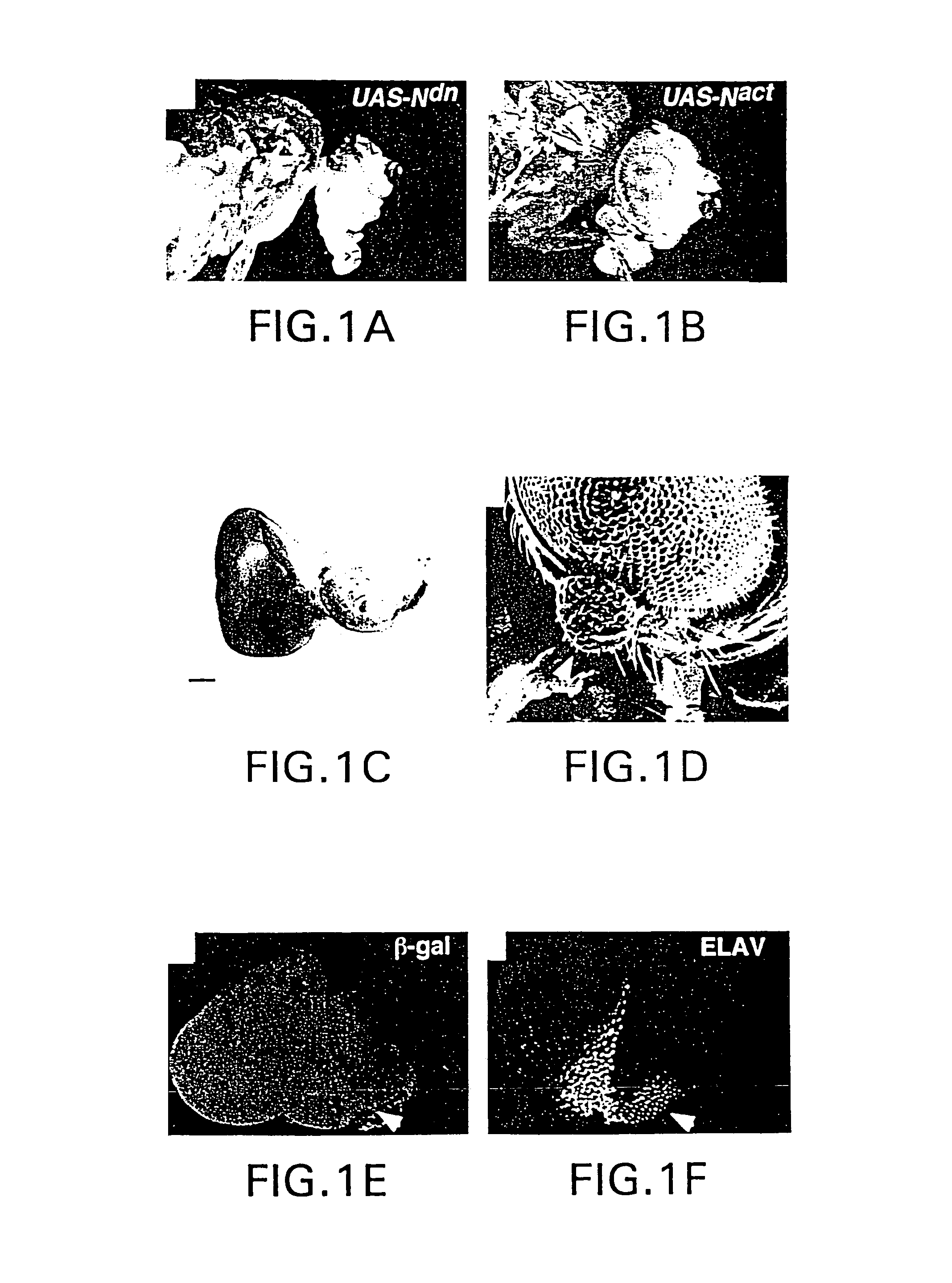
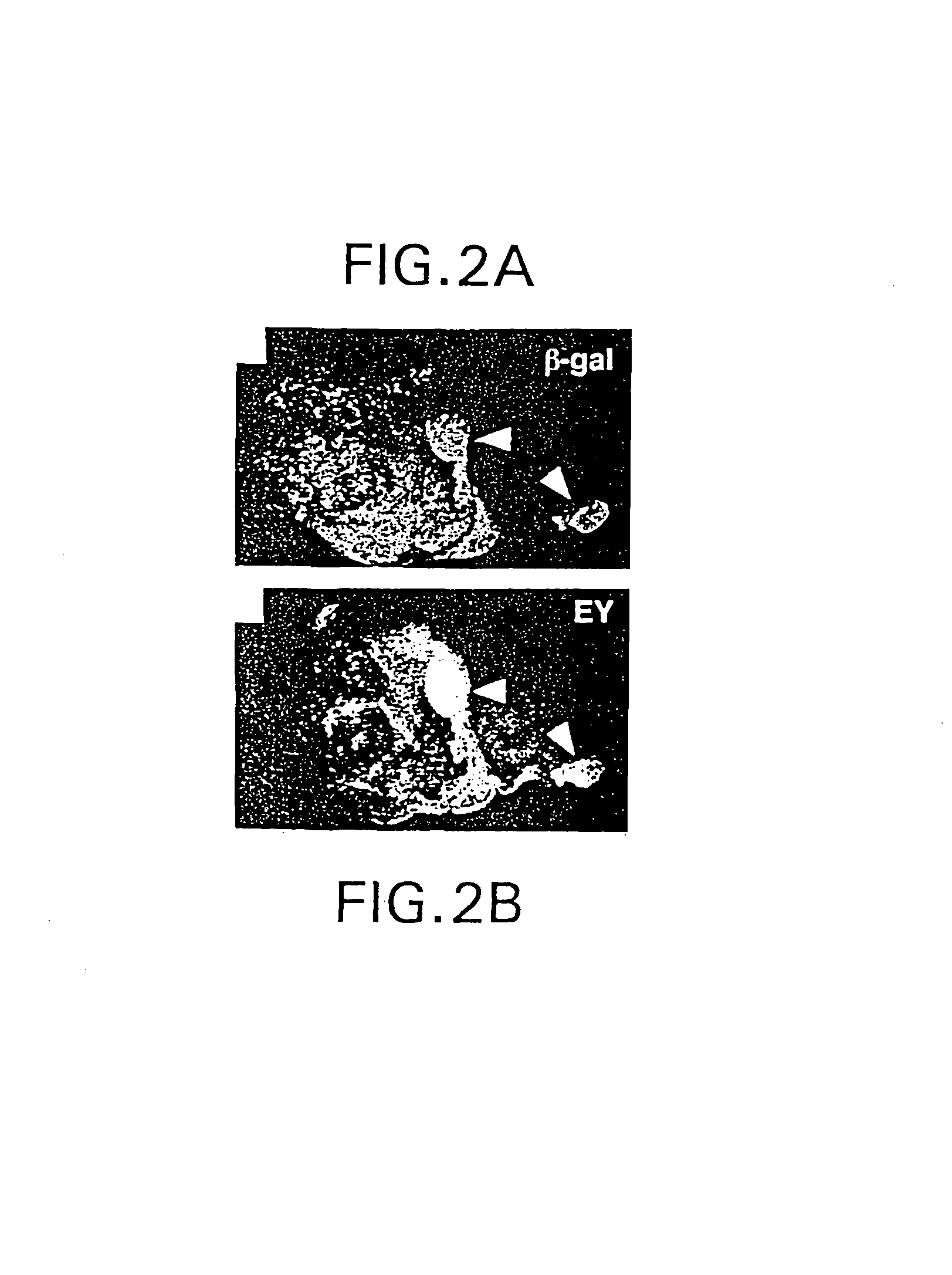
Manipulation of tissue of organ type using the notch pathway
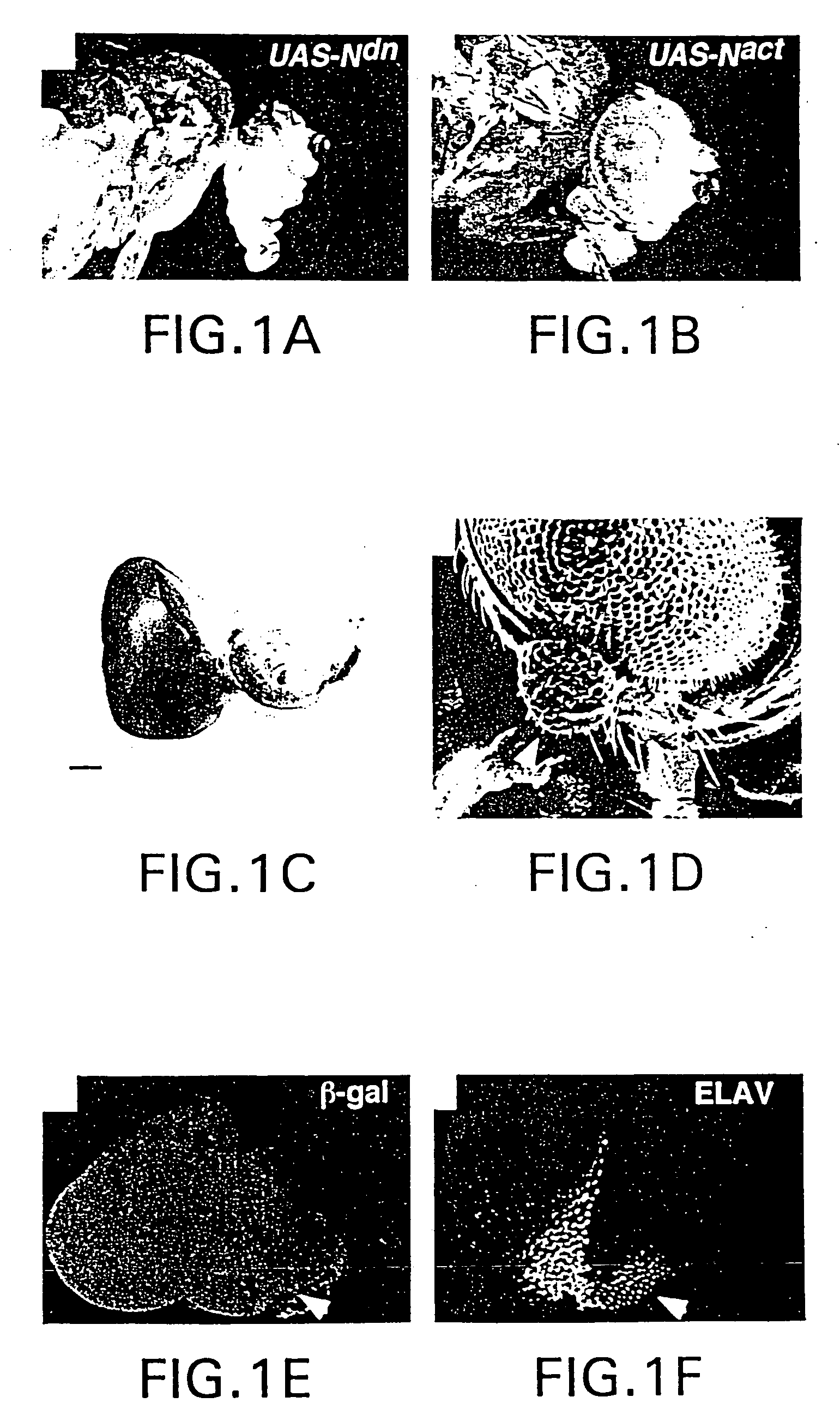
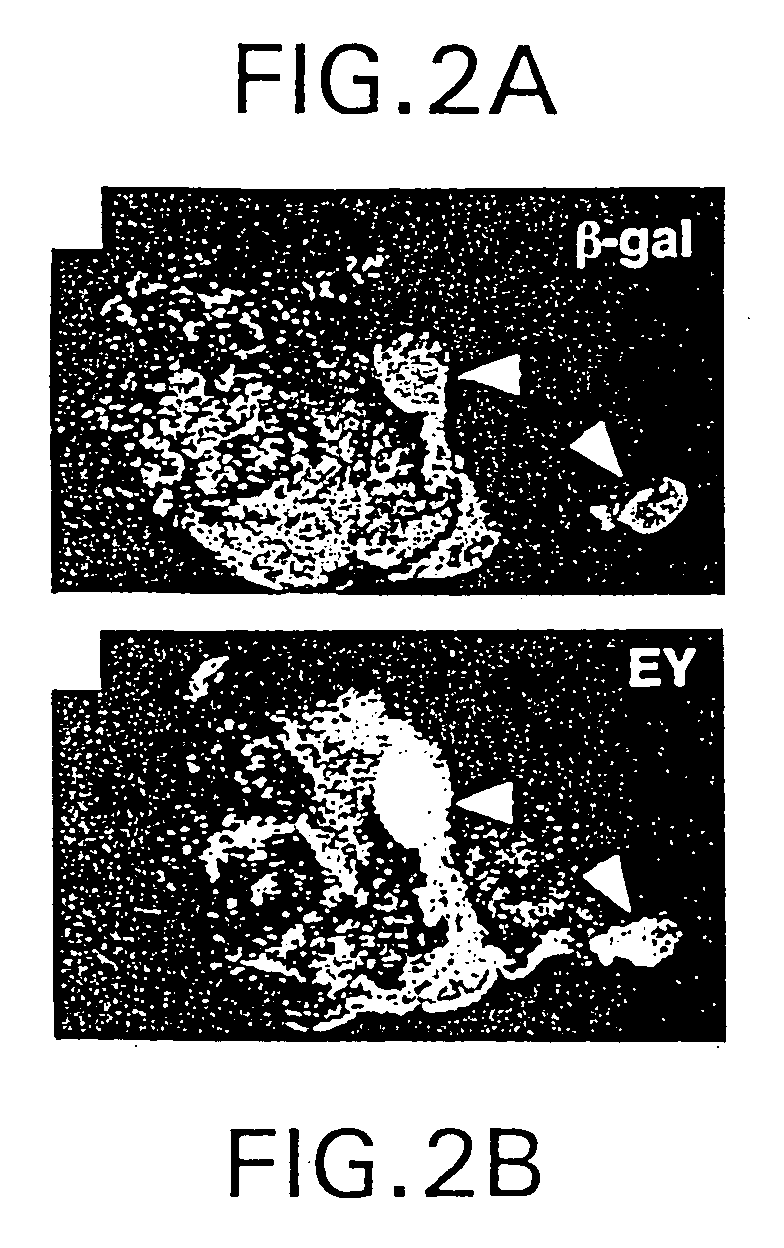
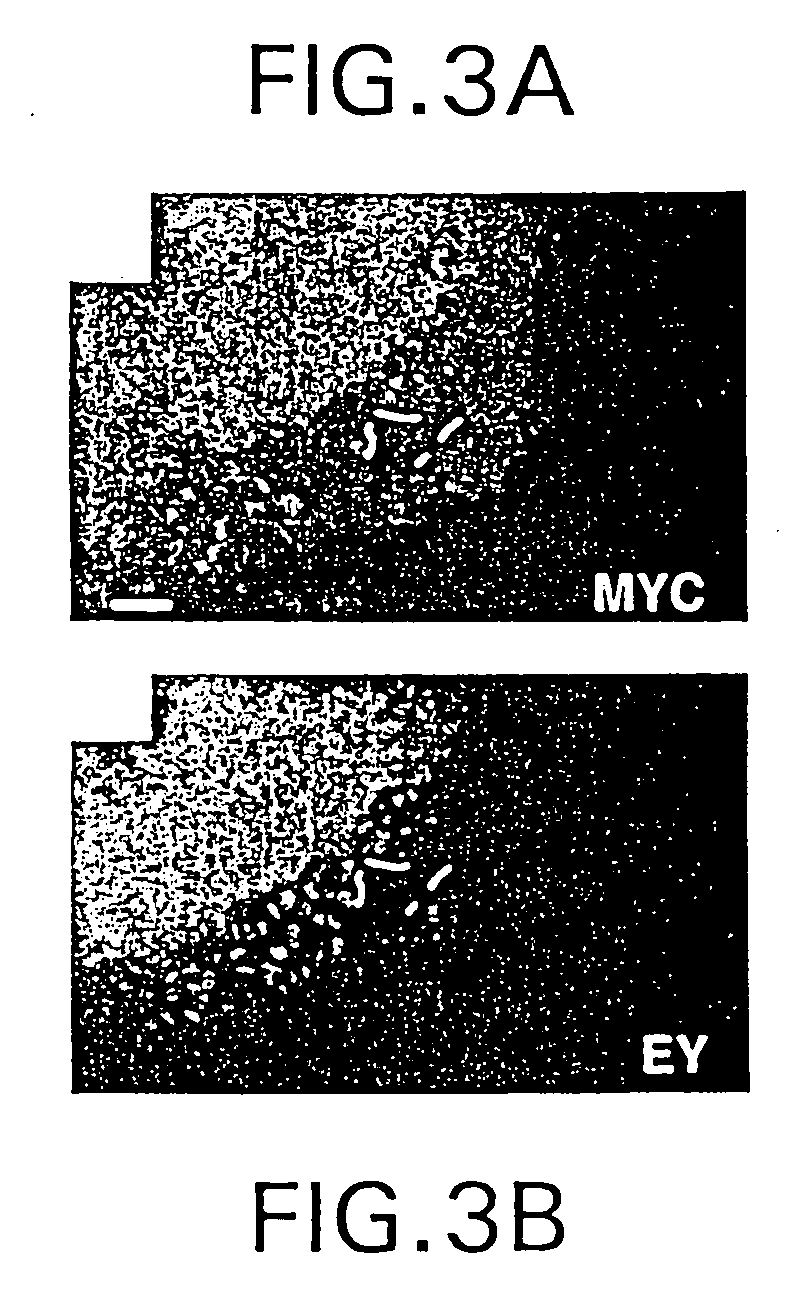
The present invention is directed to methods for altering the fate of a cell, tissue or organ type by altering Notch pathway function in the cell. The invention is further directed to methods for altering the fate of a cell, tissue or organ type by simultaneously changing the activation state of the Notch pathway and one or more cell fate control gene pathways. The invention can be utilized for cells of any differentiation state. The resulting cells may be expanded and used in cell replacement therapy to repopulate lost cell populations and help in the regeneration of diseased and / or injured tissues. The resulting cell populations can also be made recombinant and used for gene therapy or as tissue / organ models for research. The invention is directed to methods for of treating macular degeneration comprising altering Notch pathway function in retinal pigment epithelium cells or retinal neuroepithelium or both tissues. The present invention is also directed to kits utilizing the methods of the invention to generate cells, tissues or organs of altered fates. The invention also provides methods for screening for agonists or antagonists of Notch or cell fate control gene pathway functions.
Owner:UNIVERSITY OF BASEL +1
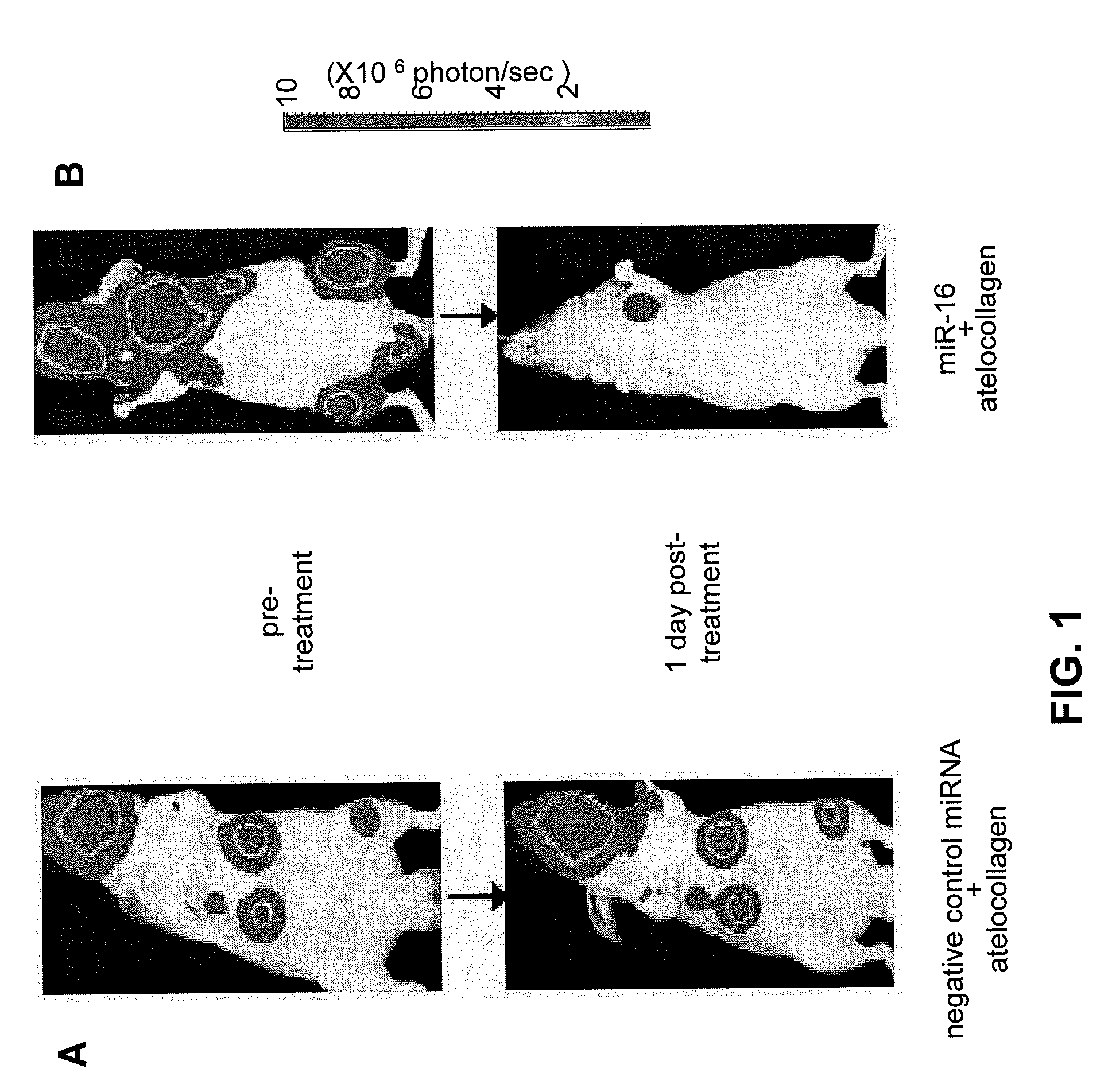
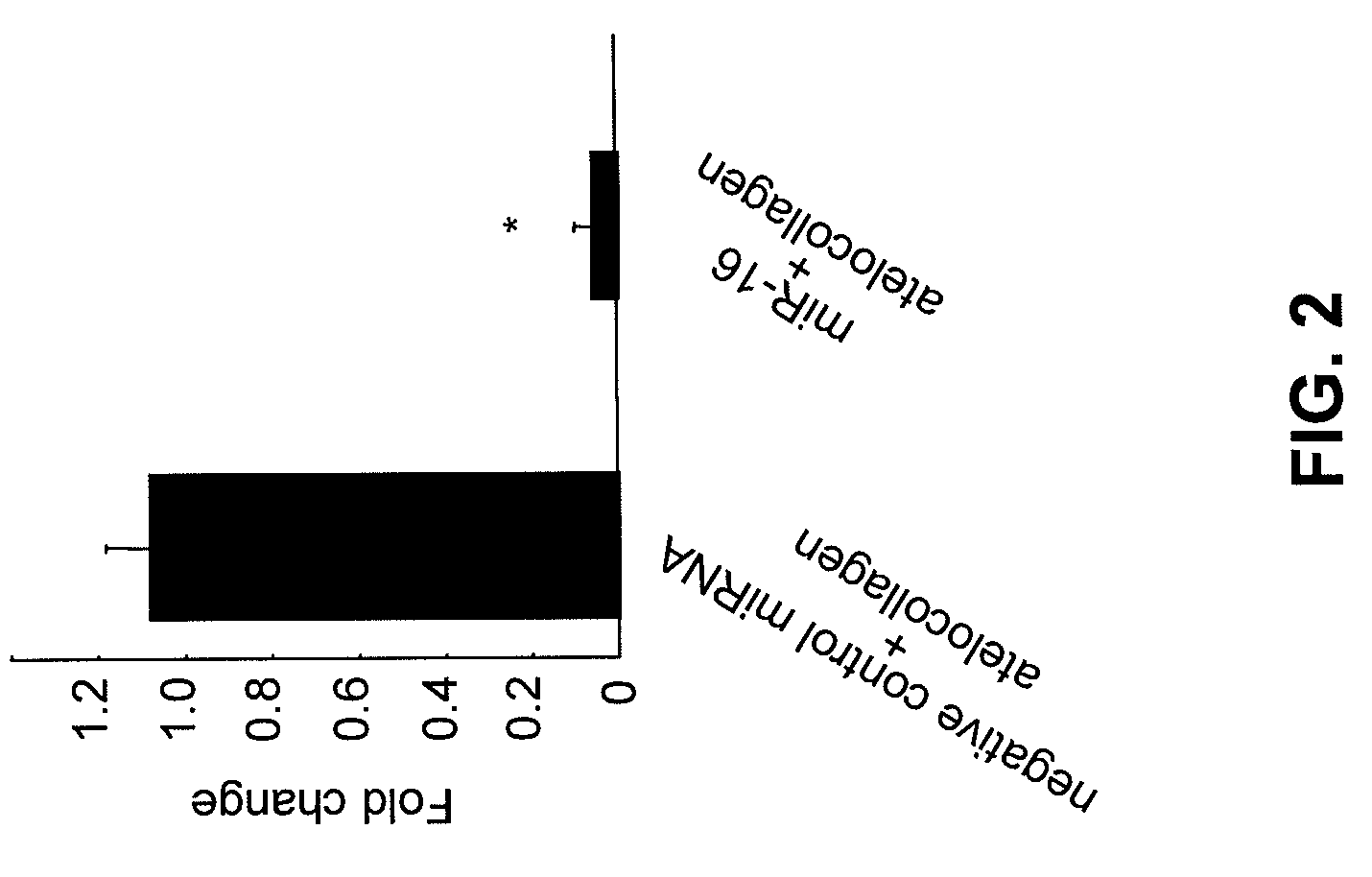
COMPOSITIONS AND METHODS RELATED TO miR-16 AND THERAPY OF PROSTATE CANCER
InactiveUS20090253780A1Growth inhibitionOrganic active ingredientsSpecial deliveryProper treatmentProstate cancer
Owner:TAKESHITA FUMITAKA +2
Mir-20 regulated genes and pathways as targets for therapeutic intervention
InactiveCN101622348AMicrobiological testing/measurementDNA/RNA fragmentationGene pathwayGenetic pathways
Owner:ASURAGEN
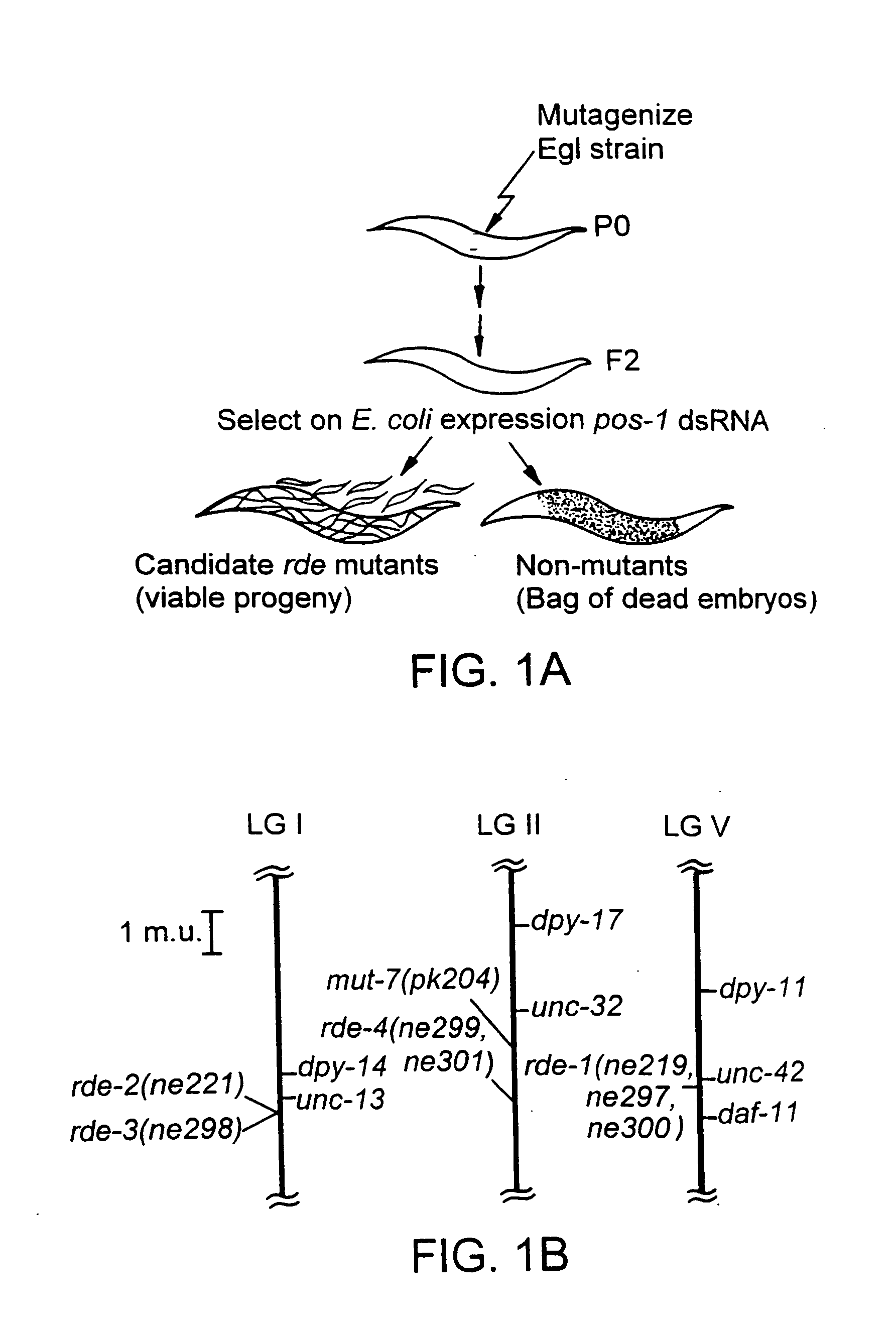
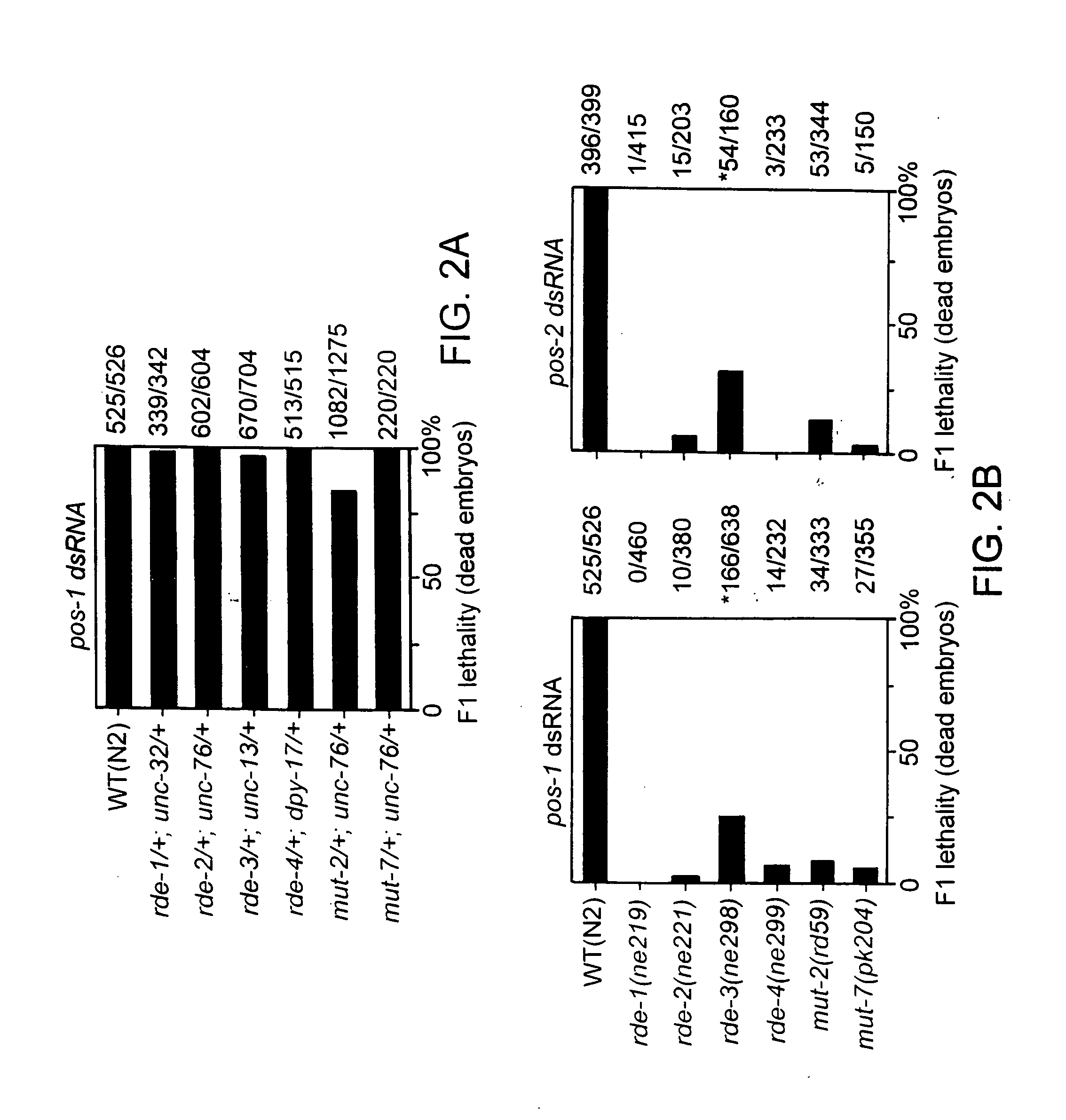
RNA interference pathway genes as tools for targeted genetic interference
InactiveUS20060024798A1Promoting dsRNA-mediated genetic interferenceDecrease and increase RNAi pathway activityPeptide/protein ingredientsMicroencapsulation basedPathway activityGene pathway
Genes involved in double-stranded RNA interference (RNAi pathway genes) are identified and used to investigate the RNAi pathway. The genes and their products are also useful for modulating RNAi pathway activity.
Owner:UNIV OF MASSACHUSETTS
RNA interference pathway genes as tools for targeted genetic interference
InactiveUS20050100913A1Decrease and increase RNAi pathway activityPromoting dsRNA-mediated genetic interferencePeptide/protein ingredientsHydrolasesPathway activityGene pathway
Genes involved in double-stranded RNA interference (RNAi pathway genes) are identified and used to investigate the RNAi pathway. The genes and their products are also useful for modulating RNAi pathway activity.
Owner:UNIV OF MASSACHUSETTS MEDICAL
Mirna regulated genes and pathways as targets for therapeutic intervention
InactiveCN101627121AMicrobiological testing/measurementDNA/RNA fragmentationGene pathwayRegulator gene
Owner:ASURAGEN
miR-20 regulated genes and pathways as targets for therapeutic intervention
InactiveCN101622349AMicrobiological testing/measurementDNA/RNA fragmentationGene pathwayGenetic pathways
Owner:ASURAGEN
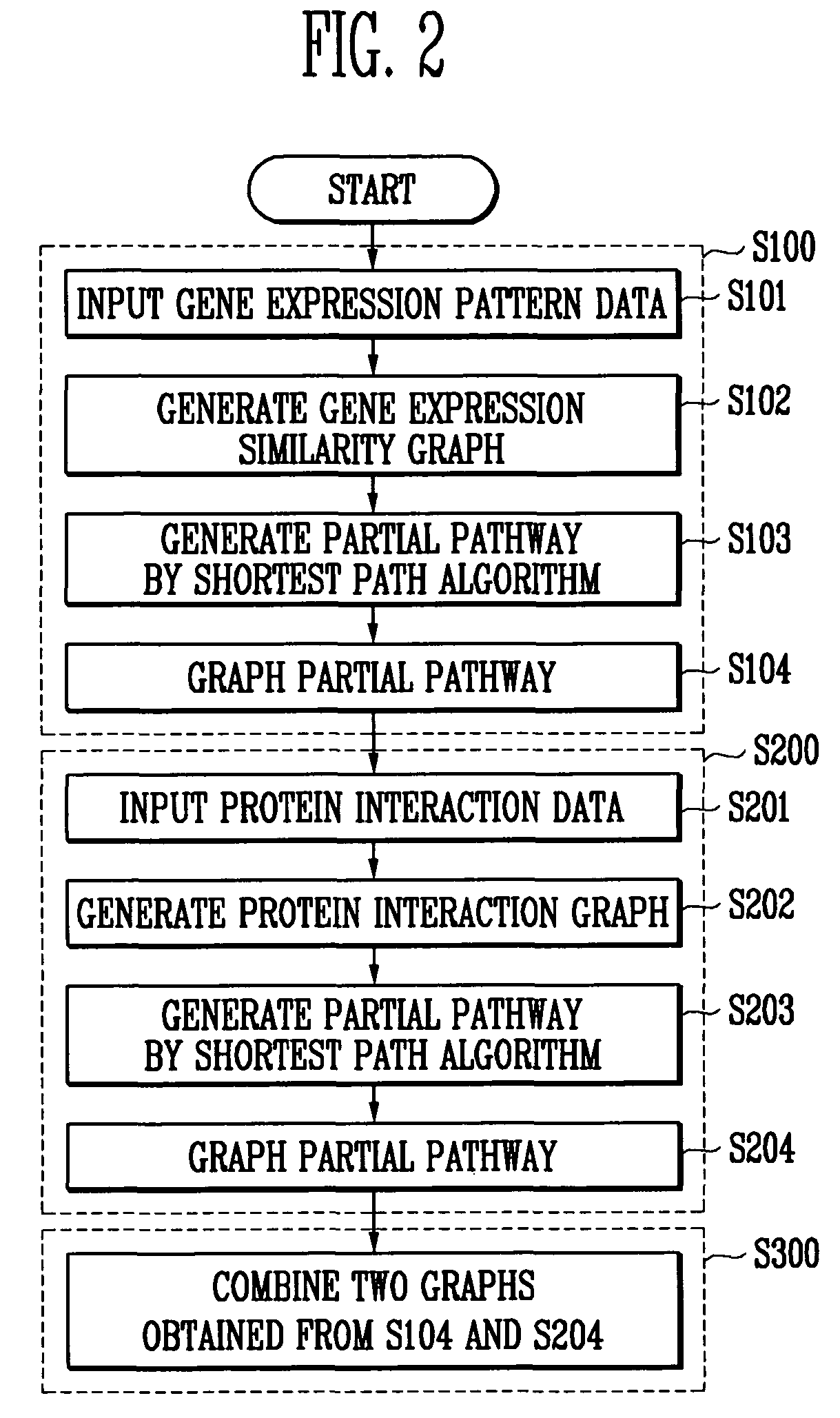
Method and system for predicting gene pathway using gene expression pattern data and protein interaction data
Owner:ELECTRONICS & TELECOMM RES INST
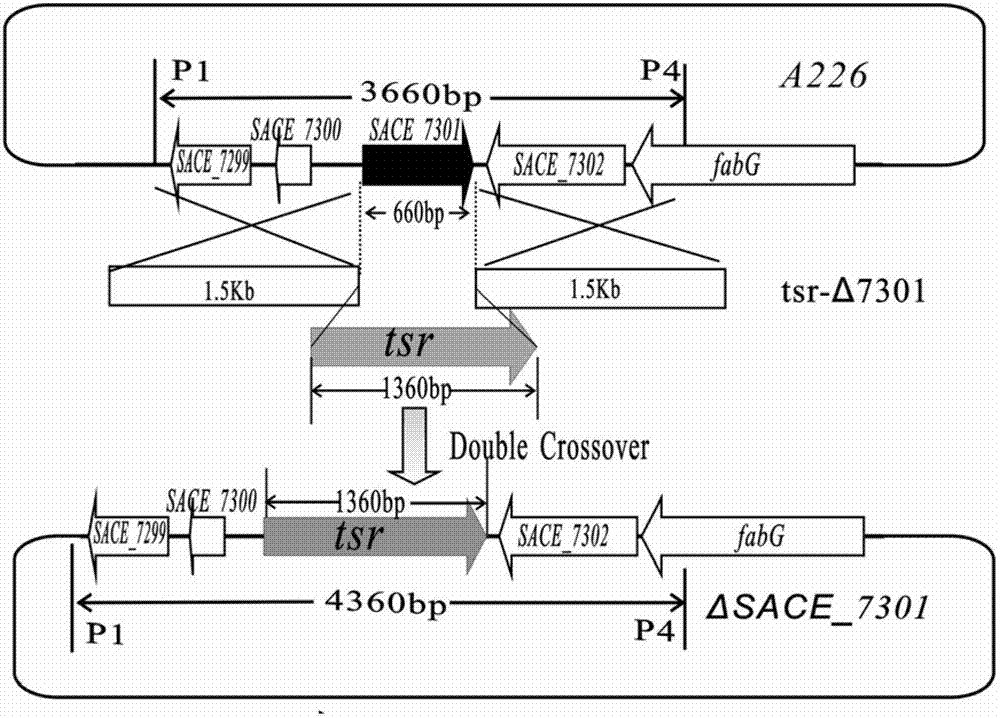
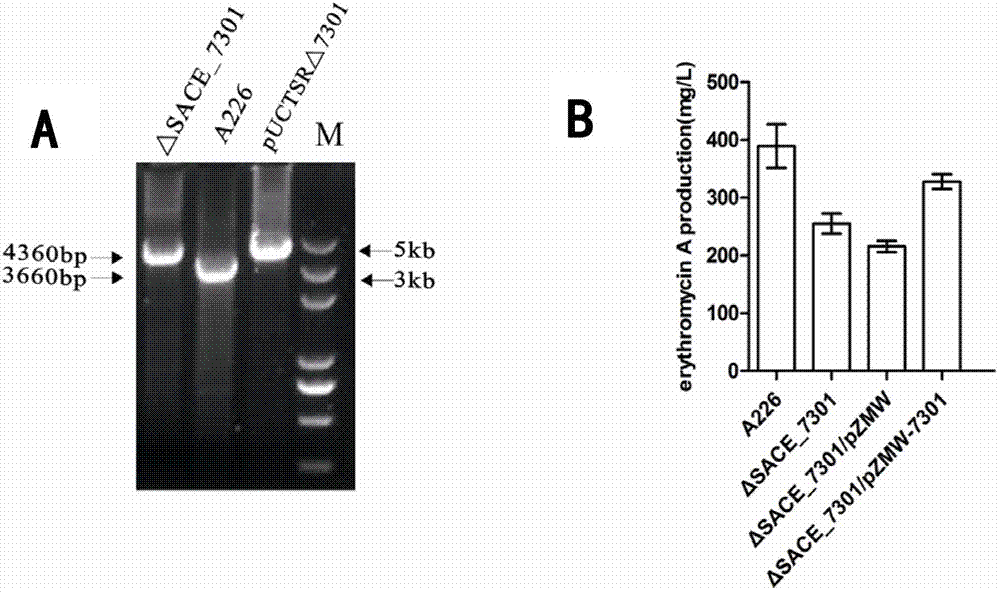
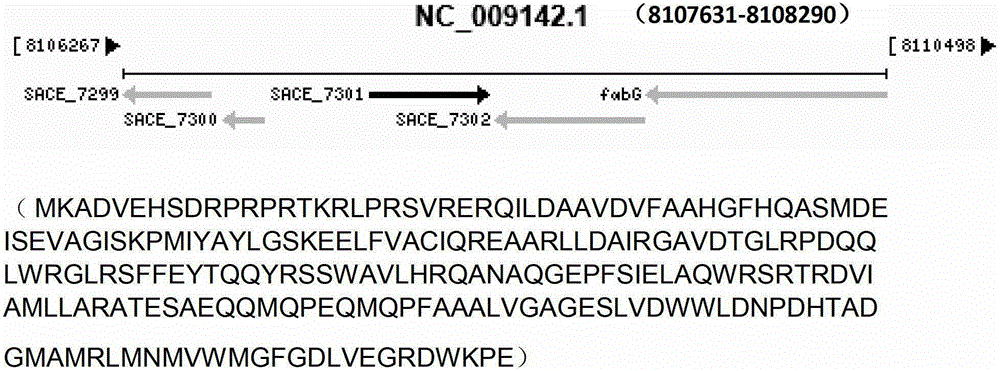
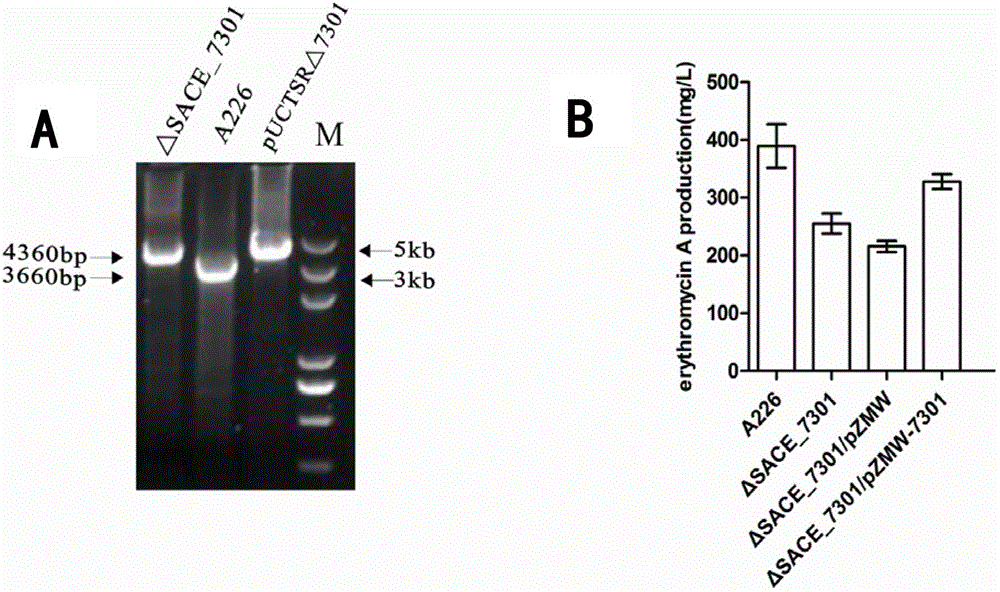
Method for improving erythromycin yield by saccharopolyspora erythraea SACE_7301 gene pathway
ActiveCN103205451AImprove fermentation yieldReduce outputBacteriaMicroorganism based processesBiotechnologyGene pathway
The invention discloses a method for improving erythromycin yield by a saccharopolyspora erythraea SACE_7301 gene pathway. The method is characterized in that gene copy number of the saccharopolyspora erythraea SACE_7301 or gene expression quantity of the SACE_7301 is increased by a gene engineering pathway so as to obtain high-yield engineering strains of saccharopolyspora erythraea erythromycin, and yield of the erythromycin can be improved by fermenting the strains obtained by the technology.
Owner:ANHUI UNIVERSITY
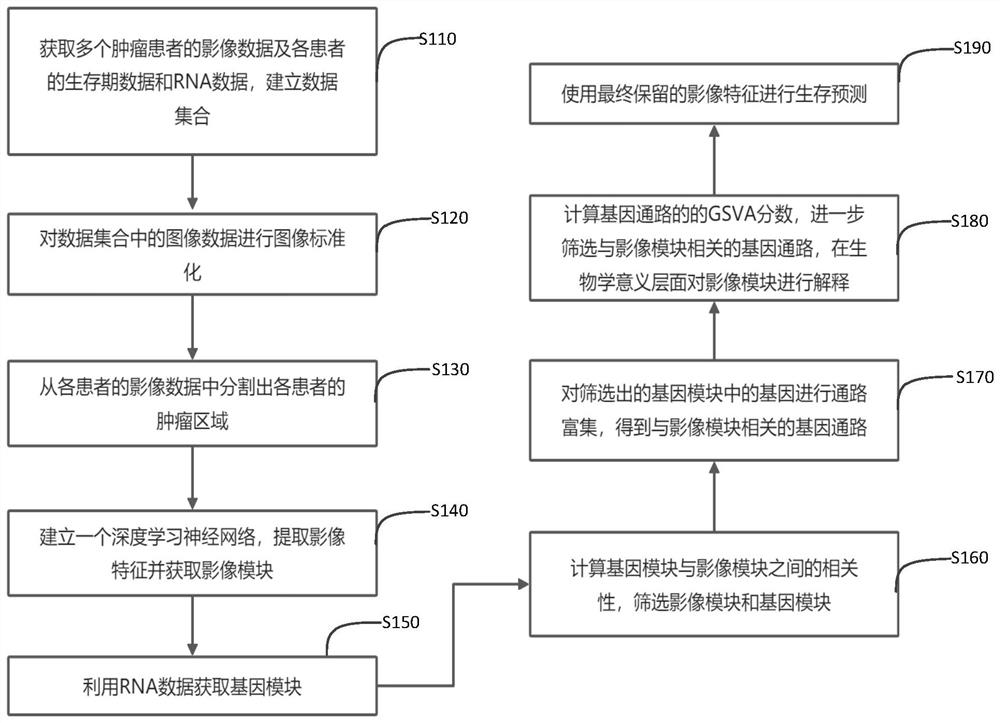
Survival prediction method and system based on image genomics
ActiveCN112907555AImprove generalization abilityImprove interpretabilityImage enhancementImage analysisData setComputational gene
The invention discloses a survival prediction method and system based on image genomics. The method comprises the following steps: acquiring image data of tumor patients and lifetime data and RNA data of each patient, and establishing a data set; segmenting a tumor area of each patient from the image data; inputting the image data of each patient into a neural network to extract image features and cluster the image features to obtain a plurality of image modules; obtaining a gene module of each patient by using the RNA data; performing screening according to the correlation between the gene modules and the image modules, and selecting a plurality of gene modules and image modules which are strongly correlated; performing pathway enrichment on genes in the selected gene module to obtain a gene pathway related to the image module; calculating a gene set variation analysis score of the gene pathway, and retaining the gene pathway having strong correlation with the image module; and carrying out survival prediction by using the retained image features. According to the invention, the biological interpretability in the aspect of survival prediction can be improved, and the generalization ability of deep learning can be improved at the same time.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
Method for recognizing gene pathway based on PAGIS
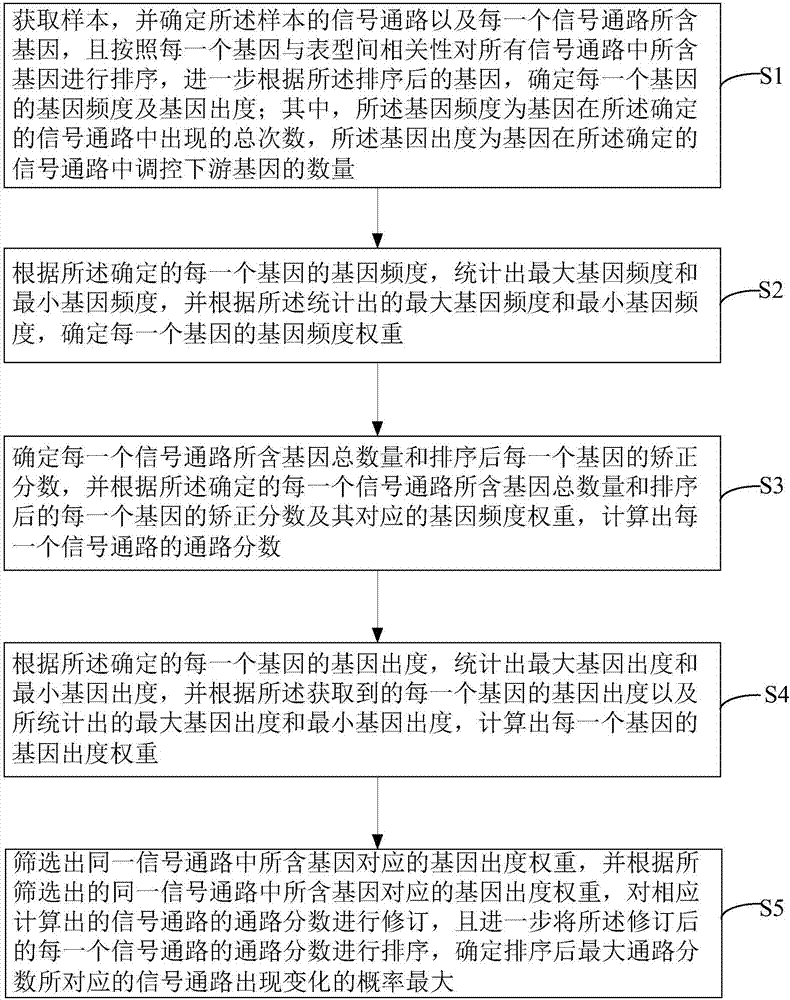
ActiveCN107133492AImprove recognition accuracyBiostatisticsHybridisationHigh probabilitySignalling pathways
An embodiment of the invention discloses a method for recognizing gene pathway based on PAGIS; the method comprises acquiring a sample, determining signal pathways and genes of the sample, and further acquiring gene frequency and gene outdegree of each gene; statistically acquiring maximum gene frequency, minimum gene frequency, maximum gene frequency and minimum gene frequency according to the gene frequency and gene outdegree of each gene so as to obtain gene frequency weight and gene outdegree weight of each gene; calculating comprehensive weight of each gene according to the gene frequency weight and gene outdegree weight of each gene to obtain weight of each signal pathway, sorting the weights of the signal pathways to determine that the signal pathway corresponding to the maximum signal pathway weight has the highest probability to vary. By implementing the embodiment of the invention to recognize pathways in conjunction with gene importance and specificity, and pathway recognition precision is improved.
Owner:WENZHOU UNIVERSITY
miR-126 regulated genes and pathways as targets for therapeutic intervention
InactiveCN101622350AMicrobiological testing/measurementDNA/RNA fragmentationRegulator geneGene pathway
Owner:ASURAGEN
Method for identifying gene pathway based on PADOG
ActiveCN107220526AImprove recognition accuracySequence analysisSpecial data processing applicationsSignalling pathwaysGene pathway
The embodiment of the invention discloses a method for identifying a gene pathway based on PADOG. The method comprises the steps that a sample is obtained, signal pathways and genes of the sample are determined, the genes included in all the signal pathways are sorted, and the gene frequentness and gene outgoing degree of each gene are determined; the gene frequentness weight and correction fraction of each gene are determined, and pathway fraction of each signal pathway is calculated; the gene outgoing degree weight of each gene is determined after sorting; the gene outgoing degree weight of the same signal pathway is screened, pathway fractions of corresponding signal pathways are revised according to the gene outgoing degree weight of the same signal pathway, the revised pathway fractions are sorted, and it is determined that the change probability of the signal pathway corresponding to the maximum pathway fraction after sorting is maximum. By carrying out the method, the pathway identifying precision can be improved by considering the importance of the gene ratio for regulating and controlling a large number of genes on regulation and control of a small number of genes.
Owner:GUANGZHOU UNIVERSITY
Method for identifying gene pathways based on GSA
ActiveCN107203704AImprove recognition accuracyHybridisationSpecial data processing applicationsSignalling pathwaysGene pathway
The embodiment of the invention discloses a method for identifying gene pathways based on GSA. The method includes the steps of obtaining samples, determining the signal pathways and genes of the samples, and sorting the genes contained in all the signal pathways; determining the total number of the genes in each signal pathway and the average value of the positive and negative points of each gene, and calculating the pathway fraction of each signal pathway; obtaining the gene out-degree of each gene, calculating out the maximum and minimum gene out-degree, and working out the gene out-degree weight of each gene; screening the gene out-degree weight in the same signal pathway out, revising the pathway fraction of the corresponding signal pathway according to the gene out-degree weight of the same signal pathway, and sorting the modified pathway fraction to determine that the changing probability of the signal pathway corresponding to the maximum pathway fraction after sorting pathway is maximum. By the method, the importance of regulating a large number of genes compared with only regulating a small number of genes is considered, and therefore the recognition accuracy of the pathways is improved.
Owner:GUANGZHOU UNIVERSITY
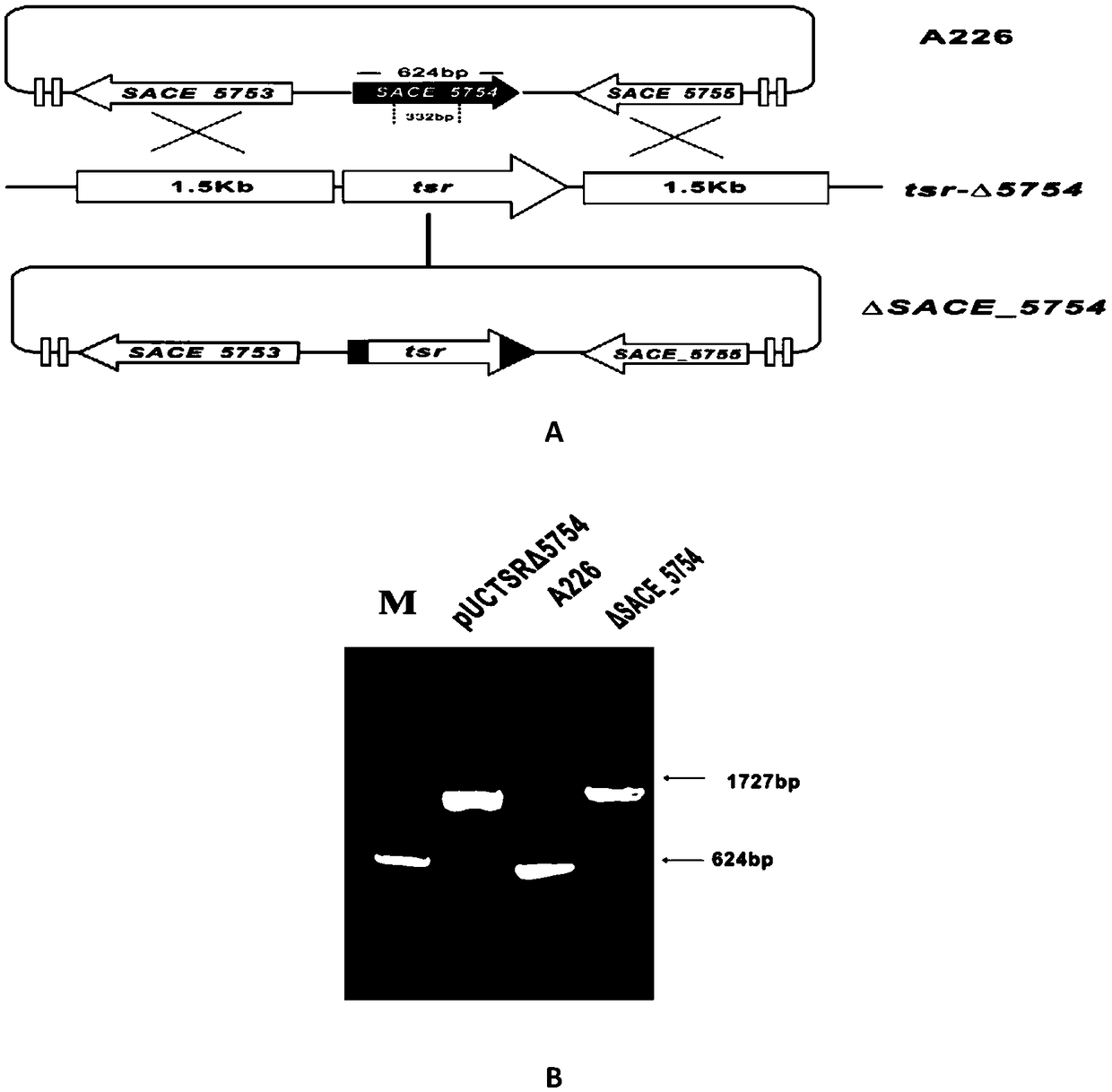
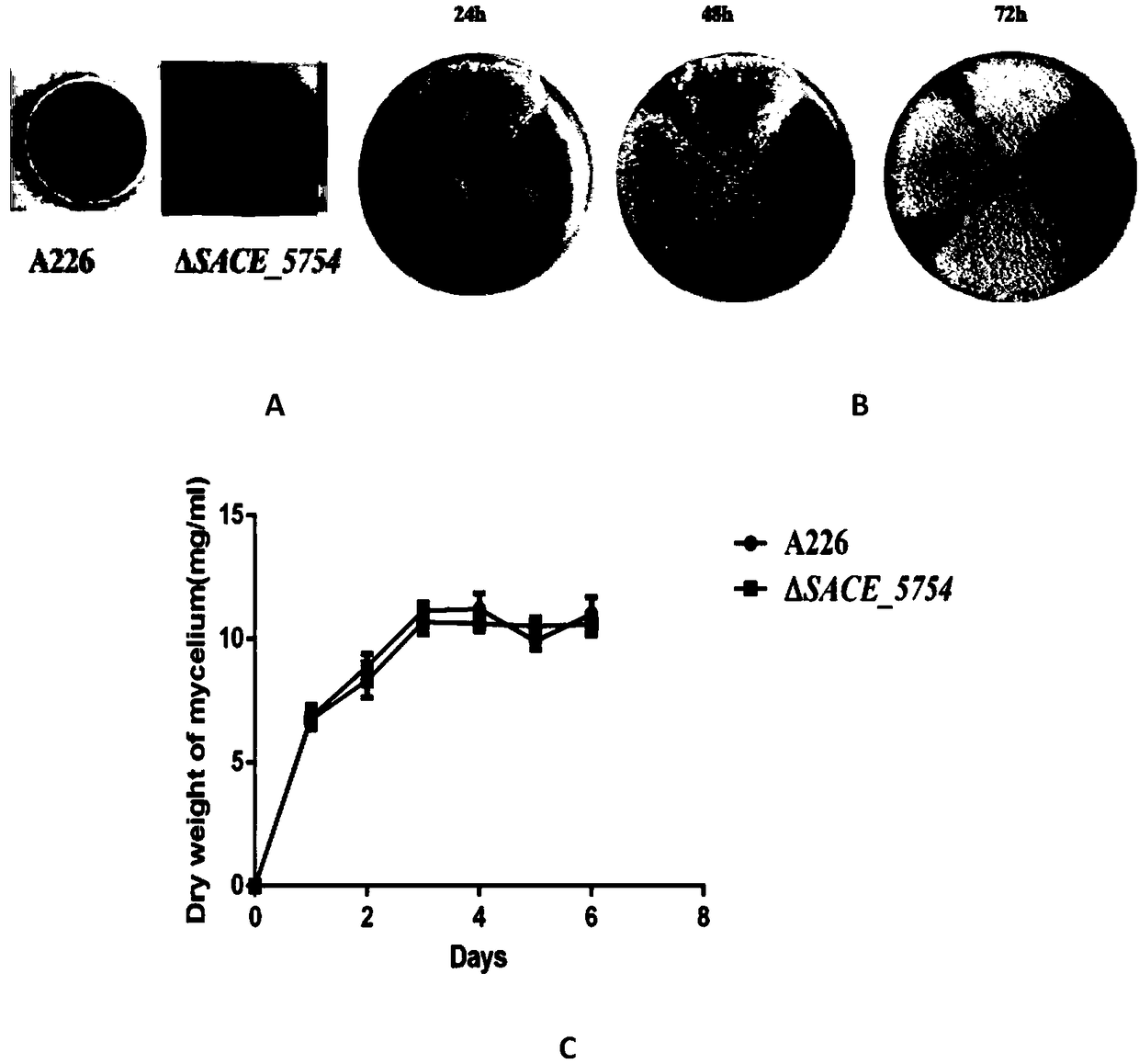
Method for increasing yield of erythromycin by SACE_5754 gene pathway of Saccharopolyspora erythraea
ActiveCN109136253AIncrease productionReduce outputMicroorganism based processesFermentationBiotechnologyGene pathway
The invention discloses a method for increasing yield of erythromycin by a SACE_5754 gene pathway of Saccharopolyspora erythraea, and belongs to the technical field of genetic engineering. According to the method, SACE_5754 is deleted from the Saccharopolyspora erythraea by the genetic engineering pathway, target genes SACE_0388 and SACE_6149 of the Saccharopolyspora erythraea are overexpressed, ahigh-yield erythromycin engineering strain is obtained, and when the obtained strain is applied to fermented production of erythromycin, yield can be substantially increased, and therefore, a new technical support is provided for erythromycin yield increase in industrial production.
Owner:ANHUI UNIVERSITY
Four-gene pathway for wax ester synthesis
ActiveUS8962299B2Reduce the amount of solutionEfficient and cost-effectiveSugar derivativesHydrolasesAcyl Coenzyme A SynthetasesPhylum Cyanobacteria
The invention relates to methods for producing a wax ester in recombinant host cells engineered to express a thioesterase, an acyl-CoA synthetase, an alcohol-forming fatty acyl reductase, and a wax ester synthase. The methods of the invention may take place in photosynthetic microorganisms, and particularly in cyanobacteria. Isolated nucleotide molecules and vectors expressing the thioesterase, acyl-CoA synthetase, alcohol-forming fatty acyl reductase, and wax ester synthase, recombinant host cells expressing the thioesterase, acyl-CoA synthetase, alcohol-forming fatty acyl reductase, and wax ester synthase, and systems for producing a wax ester via a pathway using these four enzymes, are also provided.
Owner:EXXON RES & ENG CO
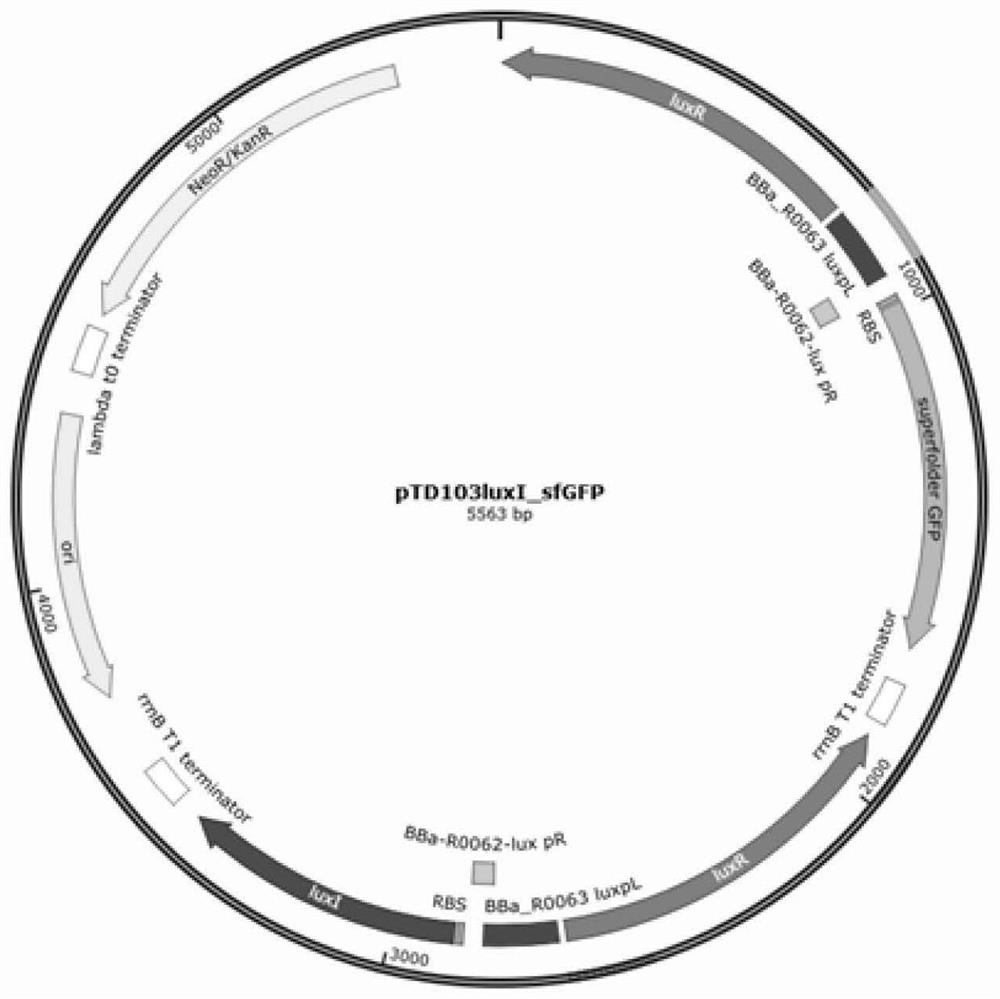
Method for synthesizing 2'-FL in escherichia coli by utilizing microbial quorum sensing and application
InactiveCN112011583AAchieve separationHigh yieldMicroorganism based processesOxidoreductasesBiotechnologyEscherichia coli
The invention provides application of microbial quorum sensing. The microbial quorum sensing is used for self-induction synthesis of 2'-fucosyllactose (2'-FL) in a genome integration mode. The invention also provides a method for carrying out self-induced expression of related proteins in a 2'-FL synthetic pathway by utilizing microbial population induction in escherichia coli and synthesizing the2'-FL. According to the method for synthesizing the 2'-FL, a self-induced gene pathway is constructed by utilizing a promoter and a structural gene corresponding to vibrio fischer quorum sensing; wherein the gene pathway is luxpL-RBS-luxI-TT-luxpL-RBS-luxR-TT-lux pR-RBS-X-TT, and the X is a CDS sequence of a gene to be expressed. According to the method provided by the invention, a microbial quorum sensing mode is integrated into a genome of the escherichia coli, and related genes in the 2'-FL synthetic pathway are expressed, so that the escherichia coli obtains a positive feedback mechanism,and in-vivo synthesis of the 2'-FL can be automatically started when the density reaches a certain threshold value.
Owner:苏州一兮生物技术有限公司
Method of inferring gene pathway activity
InactiveCN108763862AGreat practicabilitySpecial data processing applicationsCorrelation coefficientPrincipal component analysis
The invention provides a method of inferring gene pathway activity. The method comprises: obtaining samples and corresponding pathway networks and expression values of all genes thereof, and using t values of a gene t test and Pearson correlation coefficients as weights of the genes for weighting processing on the expression values of all the genes; obtaining types of interaction among genes in the same pathways and corresponding intensity thereof according to topology structures of the pathway networks of all the samples, and using the intensity of the types of interaction among the genes andthe weighted expression values of all the genes to obtain expression values of interaction among all the genes; and integrating the expression values and the interaction expression values of all thegenes, using a principal-component-analysis (PCA) method for analysis, and further defining all obtained first principal components as activity scores of the pathways. By implementing the method, importance of the genes and importance of interaction among the genes are simultaneously considered for inferring the activity of the pathways, and thus evaluation on biological pathway sample status is realized.
Owner:WENZHOU UNIVERSITY
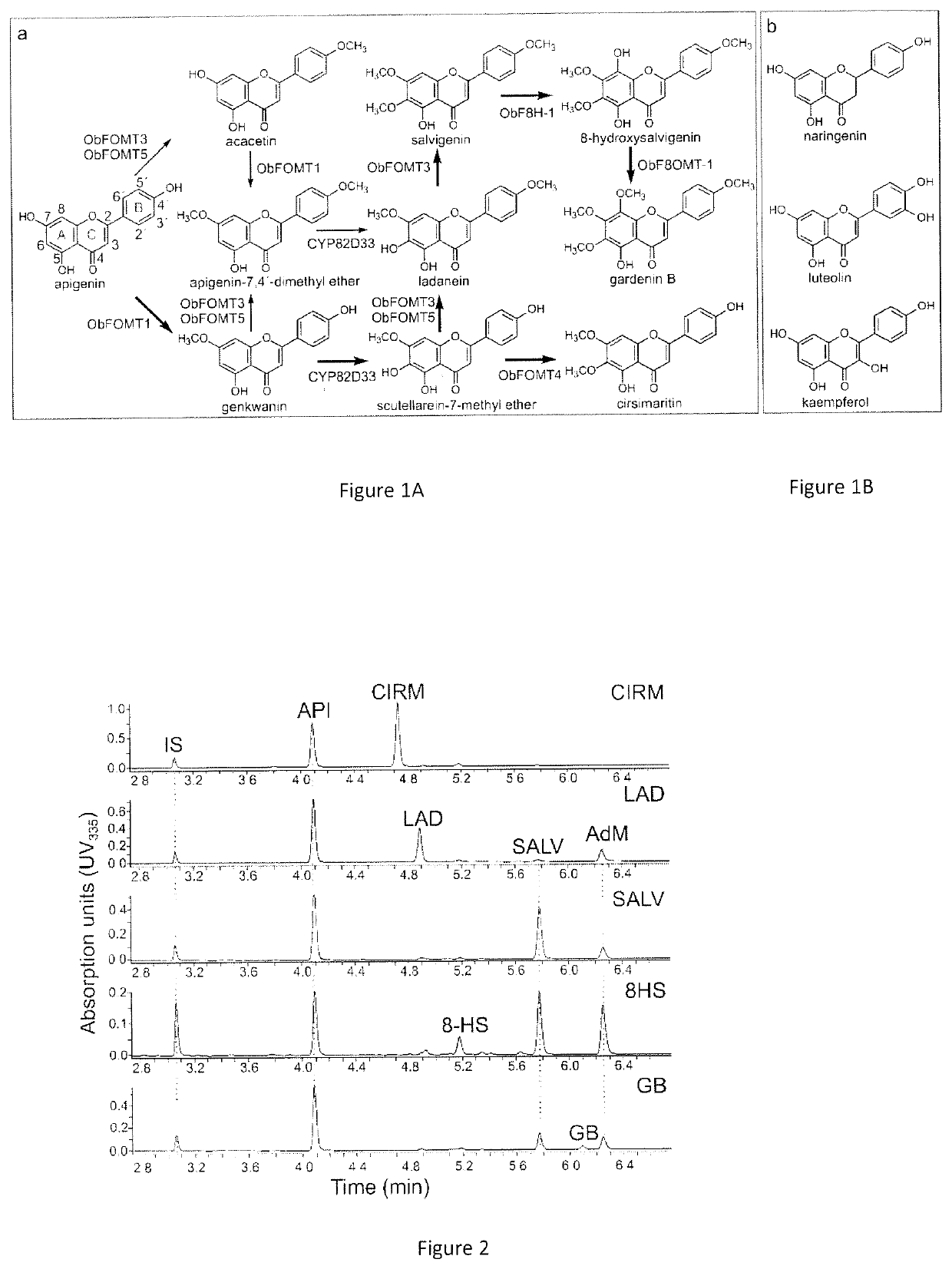
Production of hydroxylated and methoxylated flavonoids in yeast by expression specific enzymes
Recombinant yeast that are genetically modified to contain and express genes or gene pathways that produce hydroxylated and / or methoxylated flavonoids are provided. The genes or gene pathways are derived from plants, for example, from sweet basil (Ocimum basilicum) and the hydroxylated and / or methoxylated flavonoids include, for example, 6-methoxylated naringenin, 6-methoxylated luteolin and 6-methoxylated kaempferol.
Owner:WASHINGTON STATE UNIVERSITY
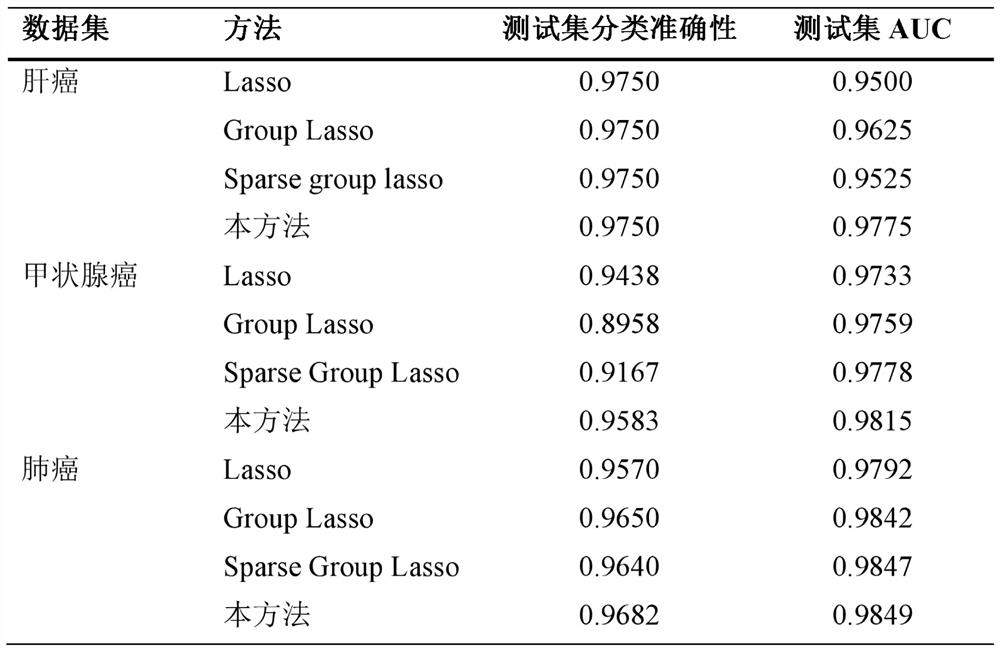
Cancer classification and characteristic gene selection method
ActiveCN113436684AImprove accuracyImprove stabilityBiostatisticsSystems biologyGenetic enhancementGene selection
The invention belongs to the field of biological information, and discloses a cancer classification and characteristic gene selection method, which comprises the following steps of: establishment of a primary learner: establishing T logistic regression models and a spark group lasso regularized loss function solving model corresponding to the T logistic regression models, and outputting a secondary learner training set; establishing a secondary learner: establishing a multi-response regression model and a loss function solving model corresponding to L1 regularization, and outputting a training set prediction result; and a prognosis feature selection model: establishing a prognosis feature selection SGL model. According to the cancer classification and feature gene selection method, the three standards of prediction, stabilization and selection are met, the accuracy and stability of the model on cancer classification prediction are improved through stacking integration, oncogenes and cancer-related genes are accurately selected, and the interpretability of the model is enhanced; gene and gene pathway priori knowledge are fused, and the accuracy of cancer classification and the effectiveness of feature selection are improved.
Owner:NANCHANG UNIV
Manipulation of tissue of organ type using the notch pathway
The present invention is directed to methods for altering the fate of a cell, tissue or organ type by altering Notch pathway function in the cell. The invention is further directed to methods for altering the fate of a cell, tissue or organ type by simultaneously changing the activation state of the Notch pathway and one or more cell fate control gene pathways. The invention can be utilized for cells of any differentiation state. The resulting cells may be expanded and used in cell replacement therapy to repopulate lost cell populations and help in the regeneration of diseased and / or injured tissues. The resulting cell populations can also be made recombinant and used for gene therapy or as tissue / organ models for research. The invention is directed to methods for of treating macular degeneration comprising altering Notch pathway function in retinal pigment epithelium cells or retinal neuroepithelium or both tissues. The present invention is also directed to kits utilizing the methods of the invention to generate cells, tissues or organs of altered fates. The invention also provides methods for screening for agonists or antagonists of Notch or cell fate control gene pathway functions.
Owner:UNIVERSITY OF BASEL +1
System and method for cell-type specific comparative analyses of different genotypes to identify resistance genes
InactiveUS20140296086A1Rapid responseMax resistanceMicrobiological testing/measurementLibrary screeningGlycinePericycle
Glycine max L. Merr. (soybean) resistance to Heterodera glycines Ichinohe is classified into two cytologically-defined responses, the G. max [Peking]- and G. max[PI 88788]-types. Microarray analyses comparing these cytologically and developmentally distinct resistant reactions reveal differences in gene expression in pericycle and surrounding cells even before infection. Gene pathway analyses compare the two genotypes (1) before, (2) at various times during, (3) constitutively throughout the resistant reaction and (4) at all time points prior to and during the resistant reaction. The amplified levels of transcriptional activity of defense genes may explain the rapid and potent reaction in G. Max[Peking / PI 548402] as compared to G. max[PI 88788].
Owner:MISSISSIPPI STATE UNIVERSITY
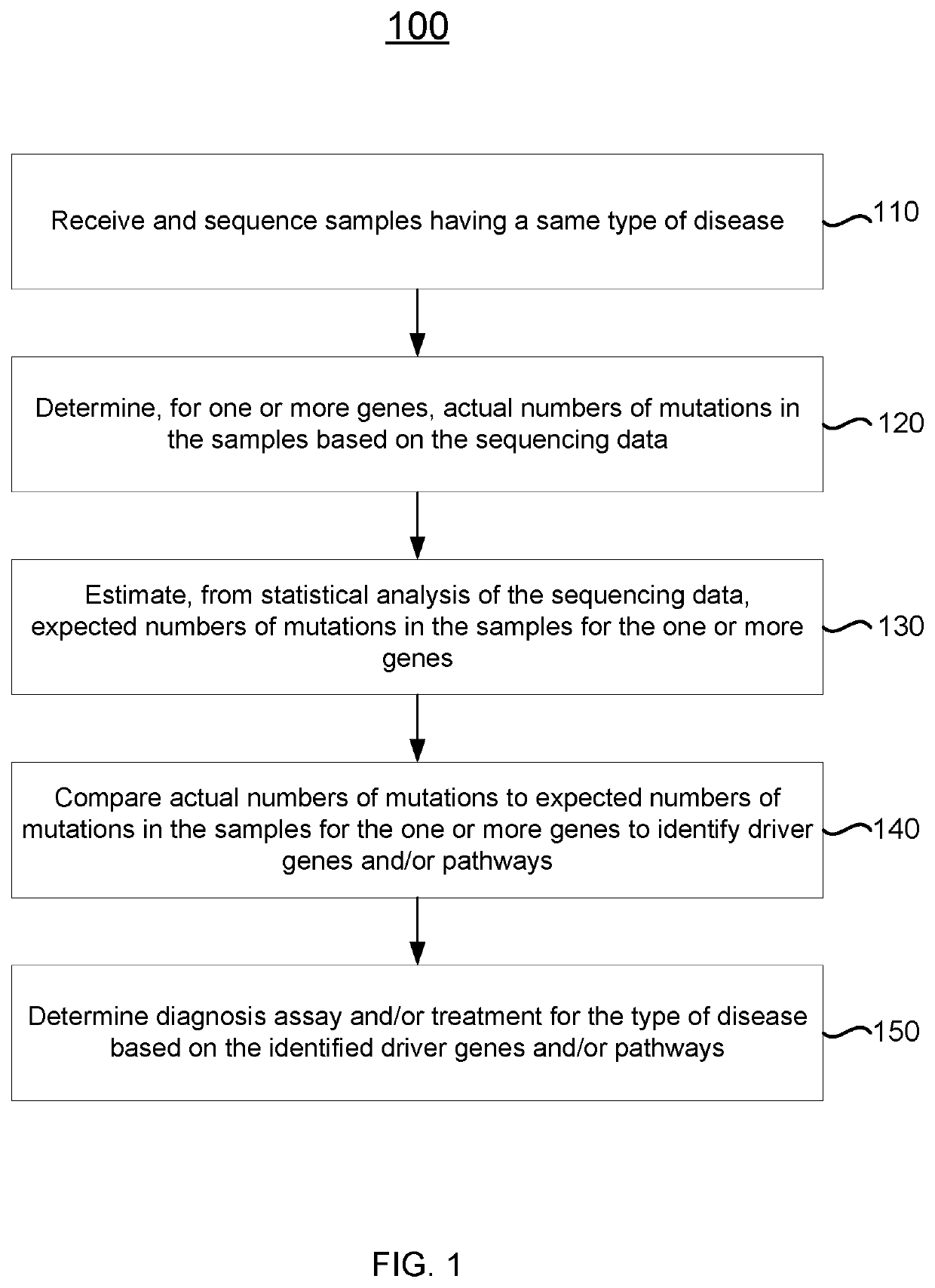
Detecting cancer driver genes and pathways
Described herein are methods, systems, and apparatuses for detecting significantly mutated genes / pathways in a cancer cohort. A driver gene detection technique taking into account the heterogeneous mutational context in a cancer cohort is disclosed. A statistical model of a gene-specific mutation rate distribution (e.g., using an optimized gene specific mean estimation and / or a gene-specific dispersion estimation) is used to model a sample / gene-specific background mutation rate. The statistical model may then be used to detect gene / pathway enrichment and distinguish tumor suppressors and oncogenes based on the spatial distribution of non-silent mutations, loss-of-function mutations, and / or gain-of-function mutations.
Owner:ROCHE SEQUENCING SOLUTIONS INC
Production of hydroxylated and methoxylated flavonoids in yeast by expression specific enzymes
Owner:WASHINGTON STATE UNIVERSITY
High-throughput screening method based on biosensor and application of high-throughput screening method in multi-gene metabolic pathway optimization
PendingCN113061642AMetabolic pathway optimizationAvoiding Fluorescence Quenching ProblemsVectorsMicrobiological testing/measurementBiotechnologyHigh-Throughput Screening Methods
The invention discloses a high-throughput screening method based on a biosensor and application of the high-throughput screening method in multi-gene metabolic pathway optimization, and belongs to the technical field of high-throughput screening. According to the invention, an efficient high-throughput screening method is constructed by combining two biosensors of specific response glycollic acid, high-throughput screening is carried out on a multi-gene pathway library of metabolite micromolecule glycollic acid, and optimization of glycollic acid multi-gene metabolic pathways is realized in a short time; high-yield strains are selected based on plate resistance, so that the screening flux can be greatly expanded, and the high-yield strains are simply and rapidly enriched; the concentration of glycollic acid in the fermentation liquor is rapidly detected by combining a 96-well plate fluorescence determination method; according to the method, repeated elimination of plasmids can be realized by combining a glycollic acid sensor and a temperature-sensitive replicon, and iterative high-throughput screening is facilitated; and through the improvement, the screening sensitivity of the target high-yield strain is effectively improved, and the enrichment efficiency of the high-yield strain is improved.
Owner:JIANGNAN UNIV
Improving erythromycin production through the sace_7301 gene pathway of Saccharopolyspora erythromycetes
ActiveCN103205451BImprove fermentation yieldReduce outputBacteriaMicroorganism based processesBiotechnologyGene pathway
The invention discloses a method for improving erythromycin yield by a saccharopolyspora erythraea SACE_7301 gene pathway. The method is characterized in that gene copy number of the saccharopolyspora erythraea SACE_7301 or gene expression quantity of the SACE_7301 is increased by a gene engineering pathway so as to obtain high-yield engineering strains of saccharopolyspora erythraea erythromycin, and yield of the erythromycin can be improved by fermenting the strains obtained by the technology.
Owner:ANHUI UNIVERSITY
An anti-tumor traditional Chinese medicine compound extract that inhibits the overexpression of ras proto-oncogene
ActiveCN105267619BExtract works wellLow toxicityAntineoplastic agentsPlant ingredientsPancreas CancersBiological activation
The invention relates to a traditional Chinese medicine compound extract, and a preparation method and an application thereof. The traditional Chinese medicine compound extract is composed of ginseng and Radix Ophiopogonis. Researches show that about 90% of pancreas cancer, 50% of colorectal carcinoma, 30-40% of lung adenocarcinoma and 5-40% of leukemia are caused by ras gene over-expression, so an ras gene becomes a universally accepted target for screening antitumor drugs. Compared with traditional Chinese medicine compound extract water decoction treatment liquids, the traditional Chinese medicine compound extract obtained through the method in the invention has the advantages of low toxicity and good ras gene pathway excessive activation effect. The invention also provides a new use of the traditional Chinese medicine compound extract, an application of the extract in inhibition of ras proto-oncogene over-expression, and an application of the extract in antitumor medicines for inhibiting ras proto-oncogene mutated over-expression.
Owner:LANZHOU UNIVERSITY
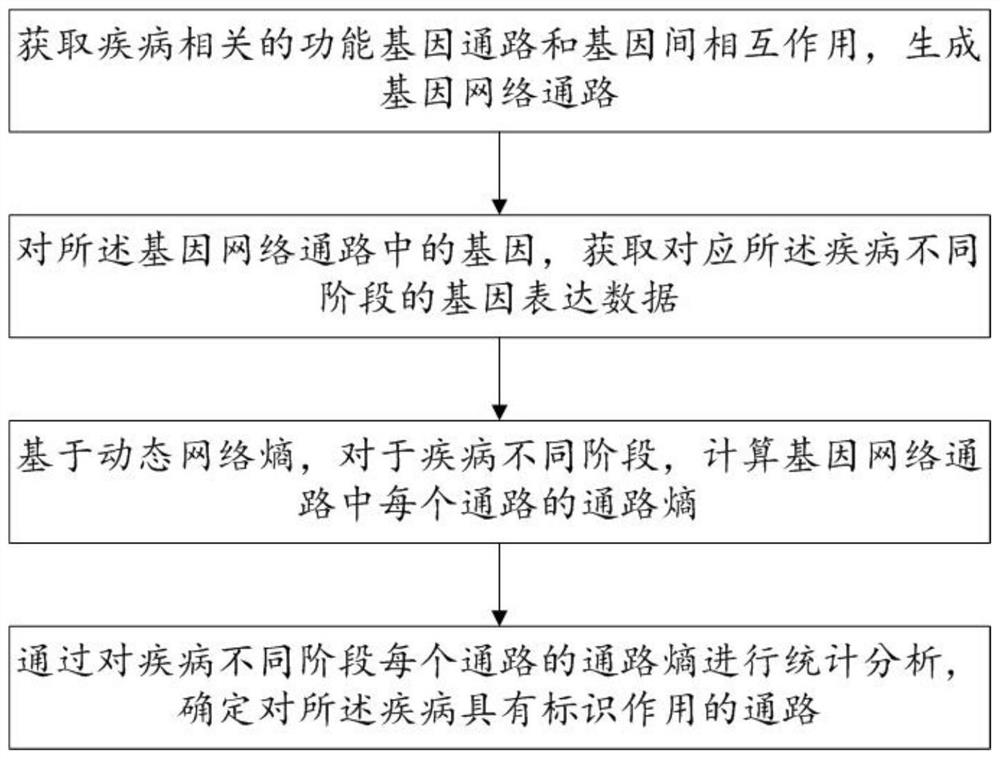
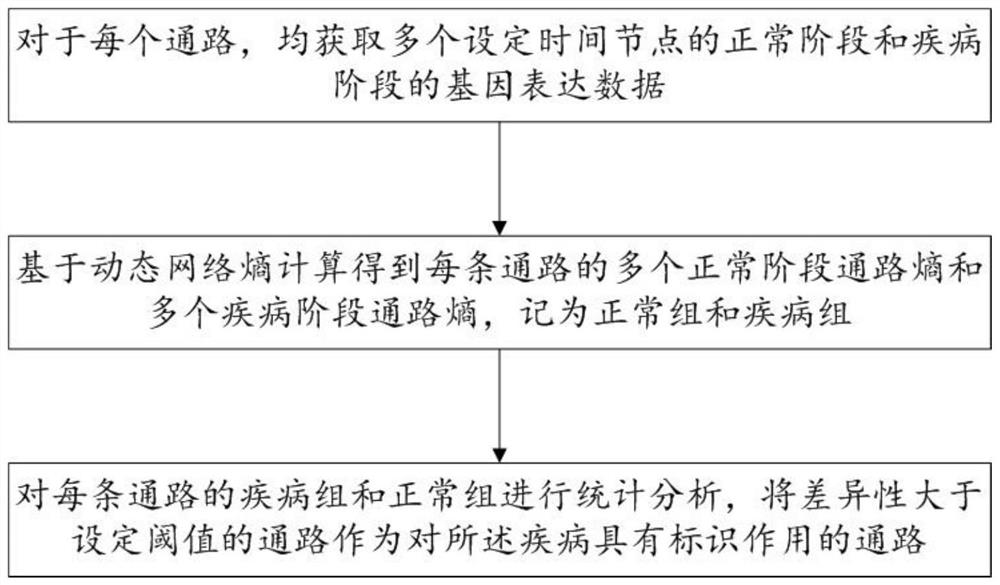
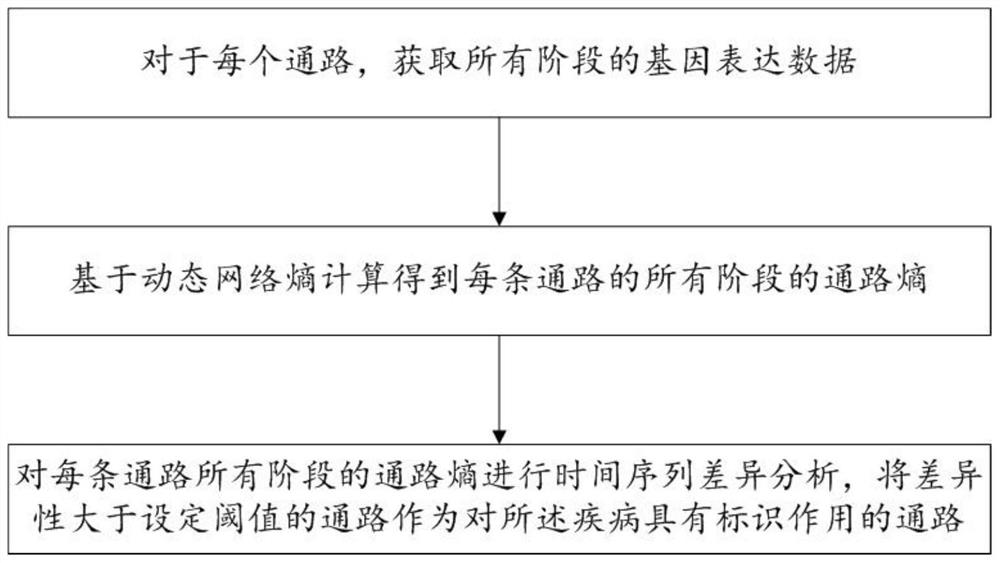
Biomarker identification method and system based on dynamic network entropy
PendingCN113889180AInteraction data sources are accurate and reliableEasy accessBiostatisticsCharacter and pattern recognitionDiseaseStatistical analysis
The invention discloses a biomarker identification method and system based on dynamic network entropy, and the method comprises the following steps: obtaining a functional gene pathway related to a disease and interaction between genes, and generating a gene network pathway; acquiring gene expression data corresponding to different stages of the disease for the genes in the gene network pathway; based on the dynamic network entropy, for different stages of the disease, calculating the pathway entropy of each pathway in the gene network pathways; and performing statistical analysis on the channel entropy of each channel in different stages of the disease to determine a channel with an identification effect on the disease. According to the method, the gene network pathway is constructed, information measurement is carried out on pathways in different stages of the disease by adopting entropy, and the gene pathway biomarkers related to the disease progress can be accurately identified by analyzing the change of the entropy.
Owner:SHANDONG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com