Biomarker identification method and system based on dynamic network entropy
A biomarker and dynamic network technology, applied in biological systems, biostatistics, bioinformatics, etc., can solve problems such as inability to obtain dynamic features, and achieve high accuracy, accurate and reliable sources.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
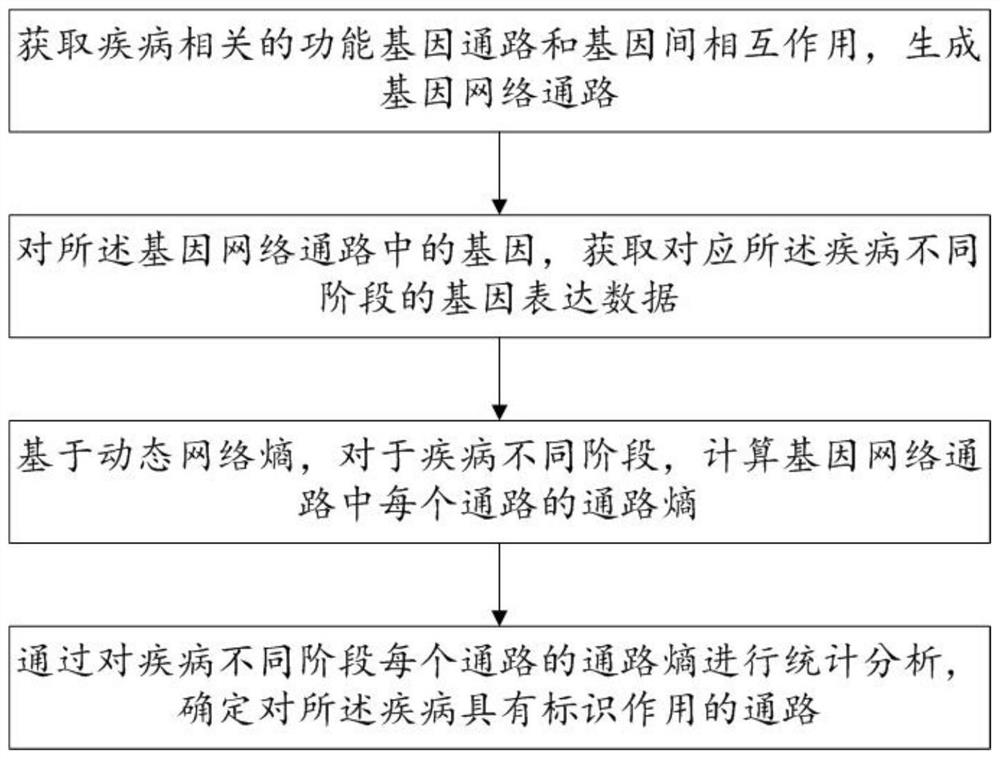
[0059] This embodiment discloses a biomarker identification method based on dynamic network entropy, such as figure 1 shown, including:
[0060] S1: Obtain disease-related functional gene pathways and gene interactions, and generate gene network pathways;
[0061] S2: Obtain gene expression data corresponding to different stages of the disease for the genes in the gene network pathway;
[0062] S3: Based on the dynamic network entropy, for different stages of the disease, calculate the pathway entropy of each pathway in the gene network pathway;
[0063] S4: Through statistical analysis of the pathway entropy of each pathway in different stages of the disease, determine the pathways that have a marker effect on the disease.
[0064] Each step is described in detail below.
[0065] Step S1 specifically includes:
[0066] Step S101: Obtain disease-related functional gene pathways, such as: pathway 1 (gene a, gene b, ...), pathway 2 (gene a, gene m, ...); what needs to be exp...
Embodiment 2
[0107] The first embodiment above provides a method for identifying biomarkers based on dynamic network entropy, which realizes the identification of gene pathways that can identify diseases. As a specific application, this embodiment provides a diabetes diagnosis system based on biomarker identification based on dynamic network entropy.
[0108] The system specifically includes:
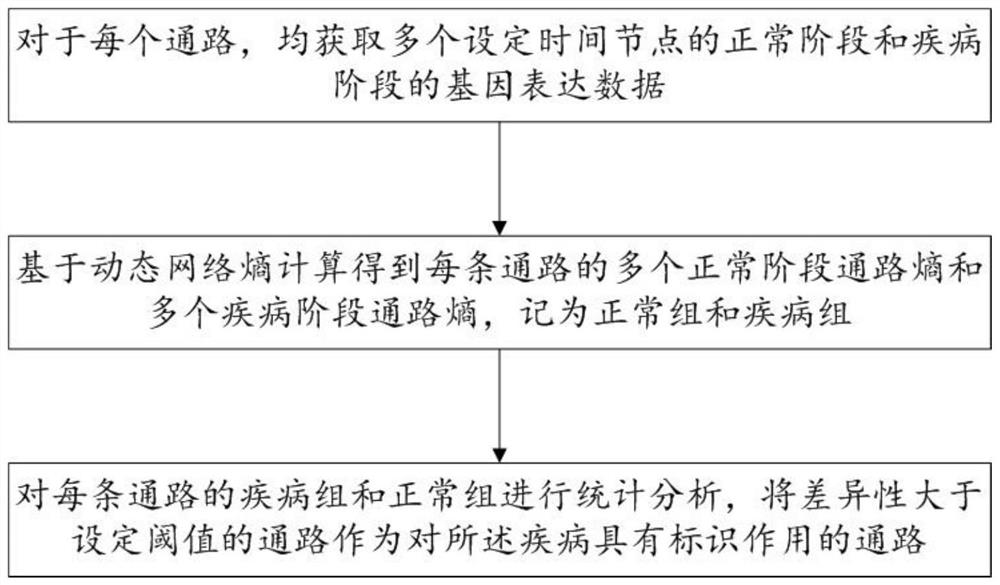
[0109] The gene pathway screening module is used to screen the pathways that have a marker effect on diabetes according to the biomarker identification method described in Example 1, and record them as candidate pathways; wherein, diabetes is divided into two stages: normal and disease;
[0110] The diagnostic model training module is used to obtain the gene expression data of the normal group and the disease group corresponding to the candidate pathway as the initial data set; based on the initial data set, the support vector machine model is trained to obtain the diagnostic model;
[0111] Specif...
Embodiment 3
[0115] The first embodiment above provides a method for identifying biomarkers based on dynamic network entropy, which realizes the identification of gene pathways that can identify diseases. As a specific application, this embodiment provides a liver cancer diagnosis system based on biomarker identification based on dynamic network entropy.
[0116] The system specifically includes:
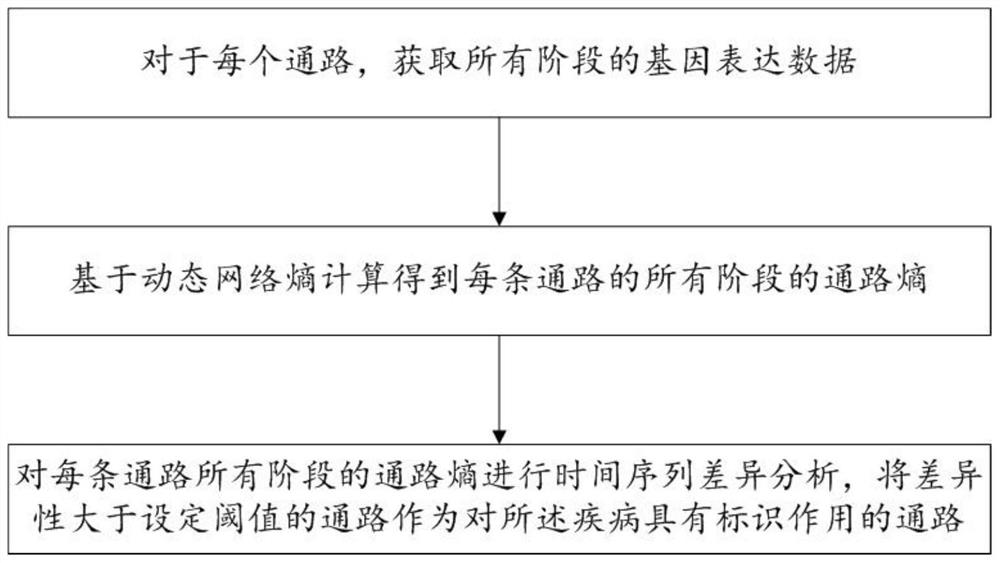
[0117] The gene pathway screening module is used to screen the pathways that have a marker effect on liver cancer according to the biomarker identification method described in Example 1, and record them as candidate pathways; wherein, liver cancer is divided into normal and multiple disease stages;
[0118] The diagnostic model training module is used to obtain the gene expression data of all stages corresponding to the candidate pathway as an initial data set; train the support vector machine model according to the initial data sets of each two adjacent stages to obtain a multi-classification d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com