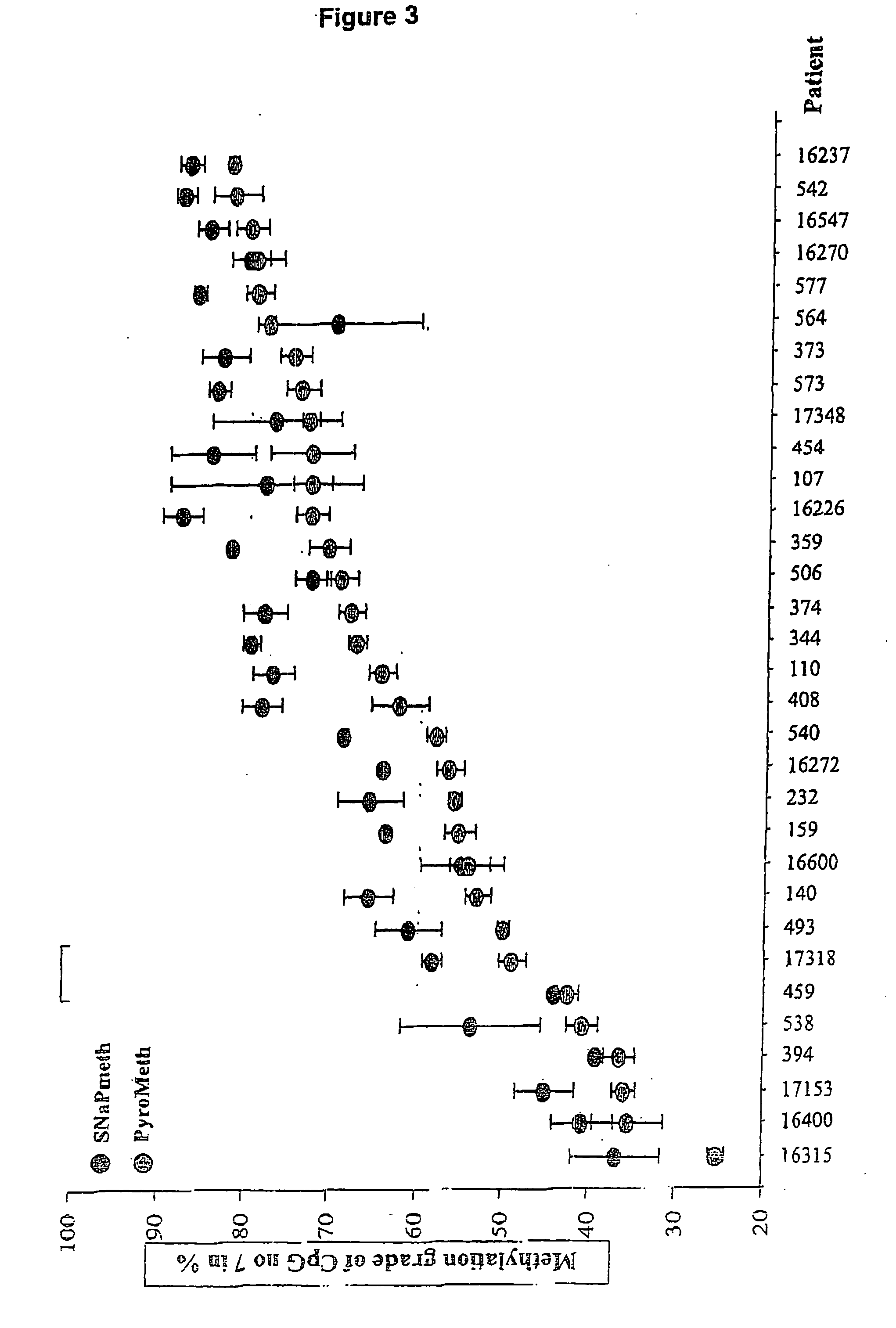
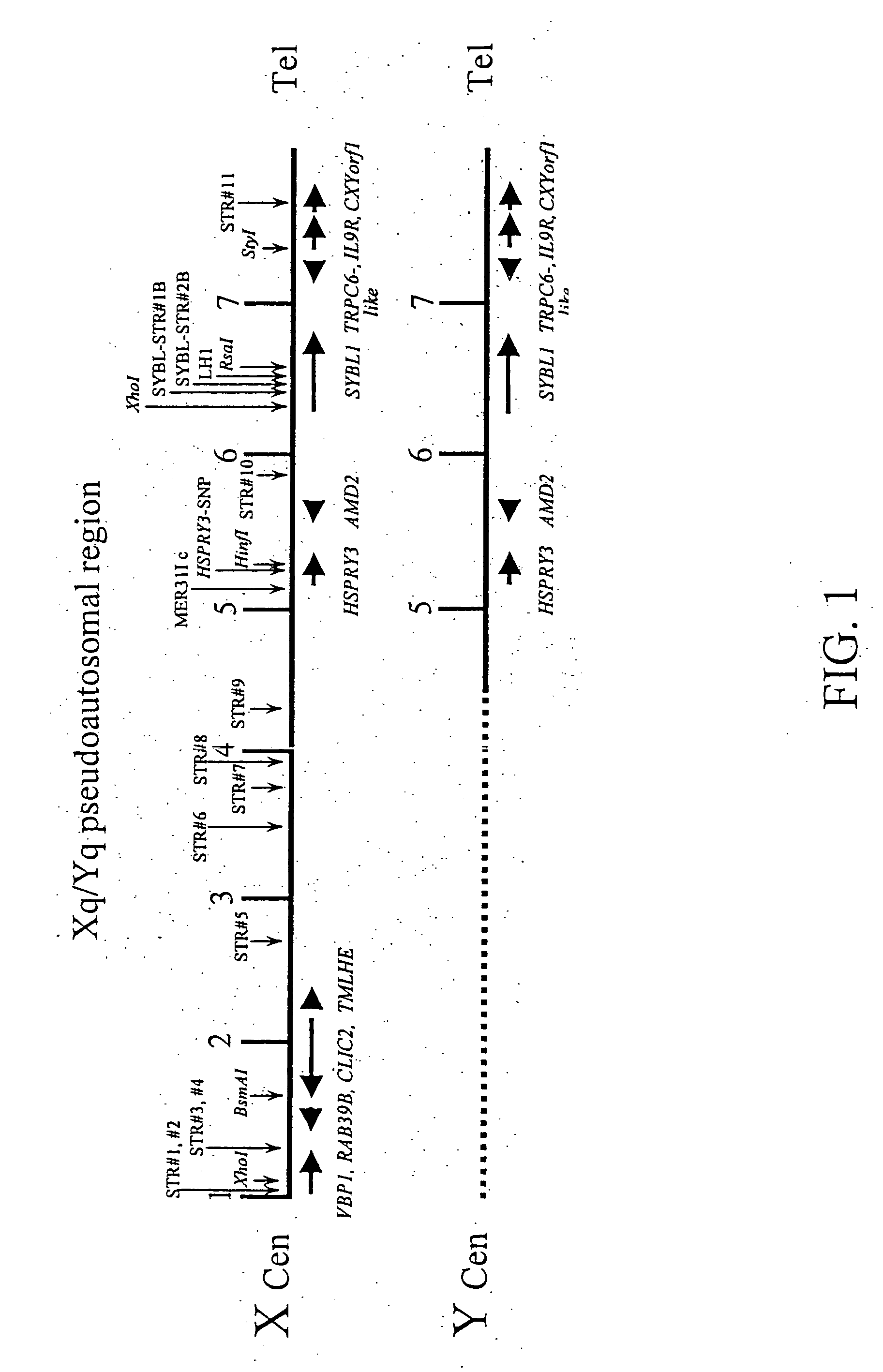
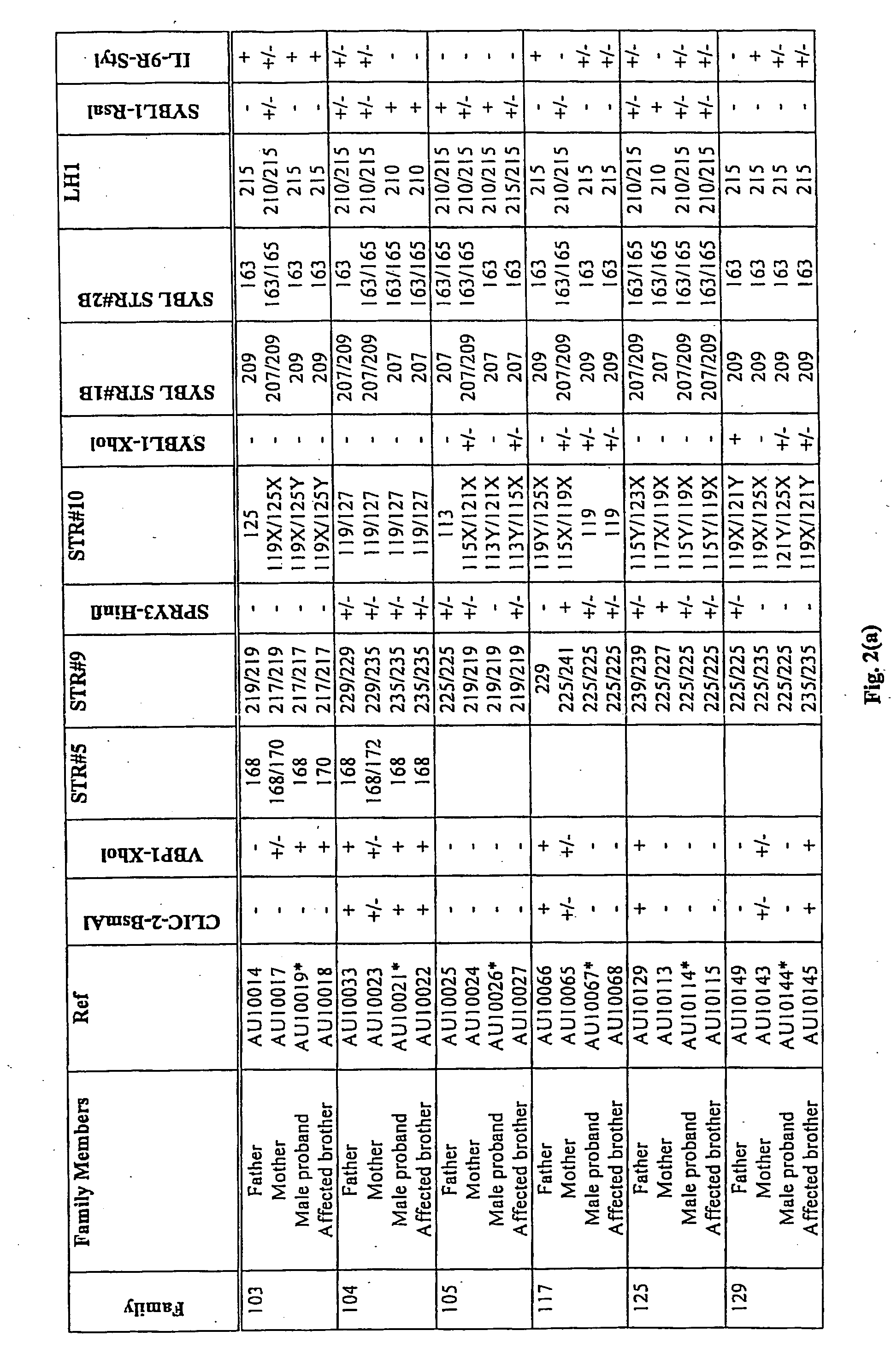
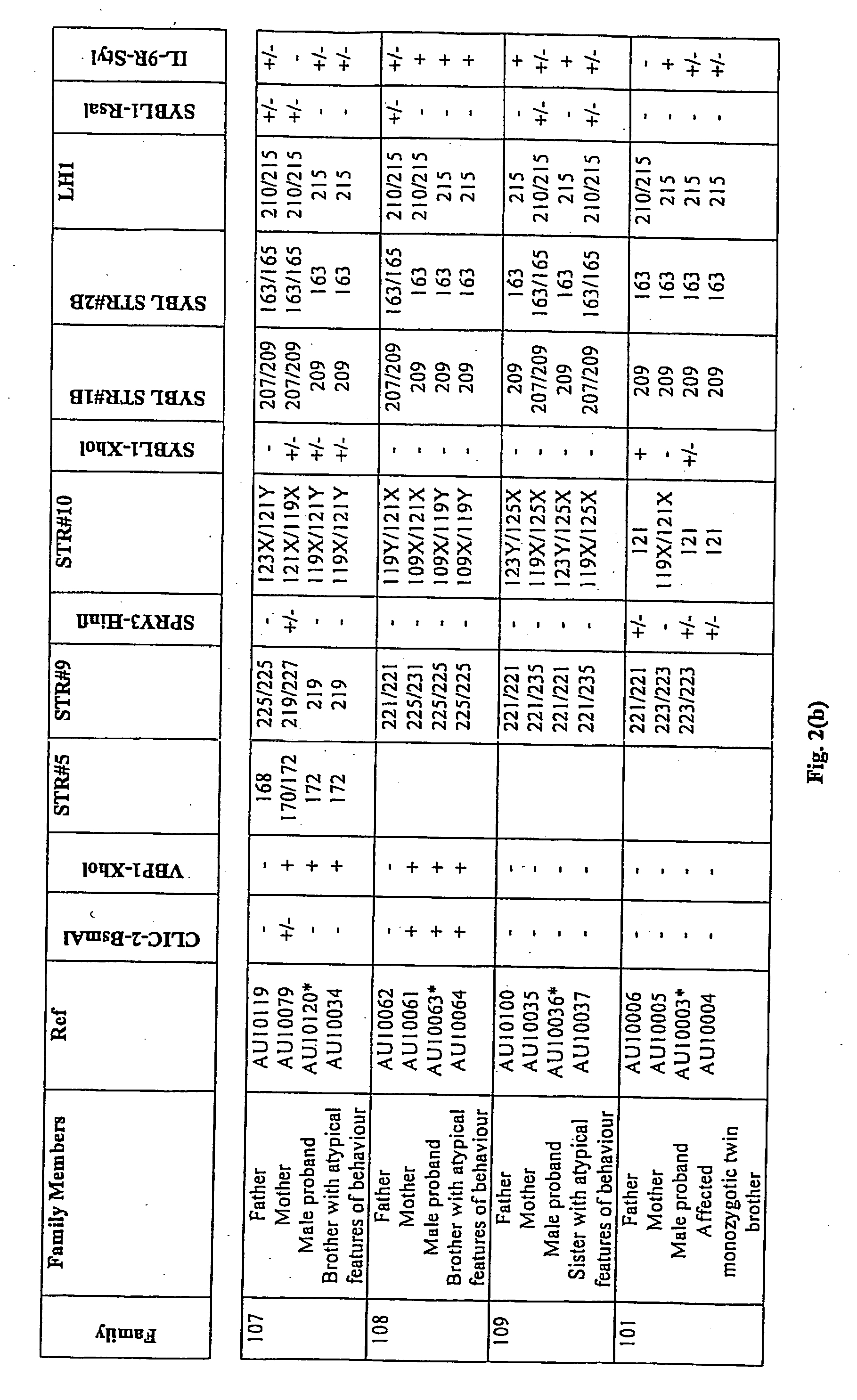
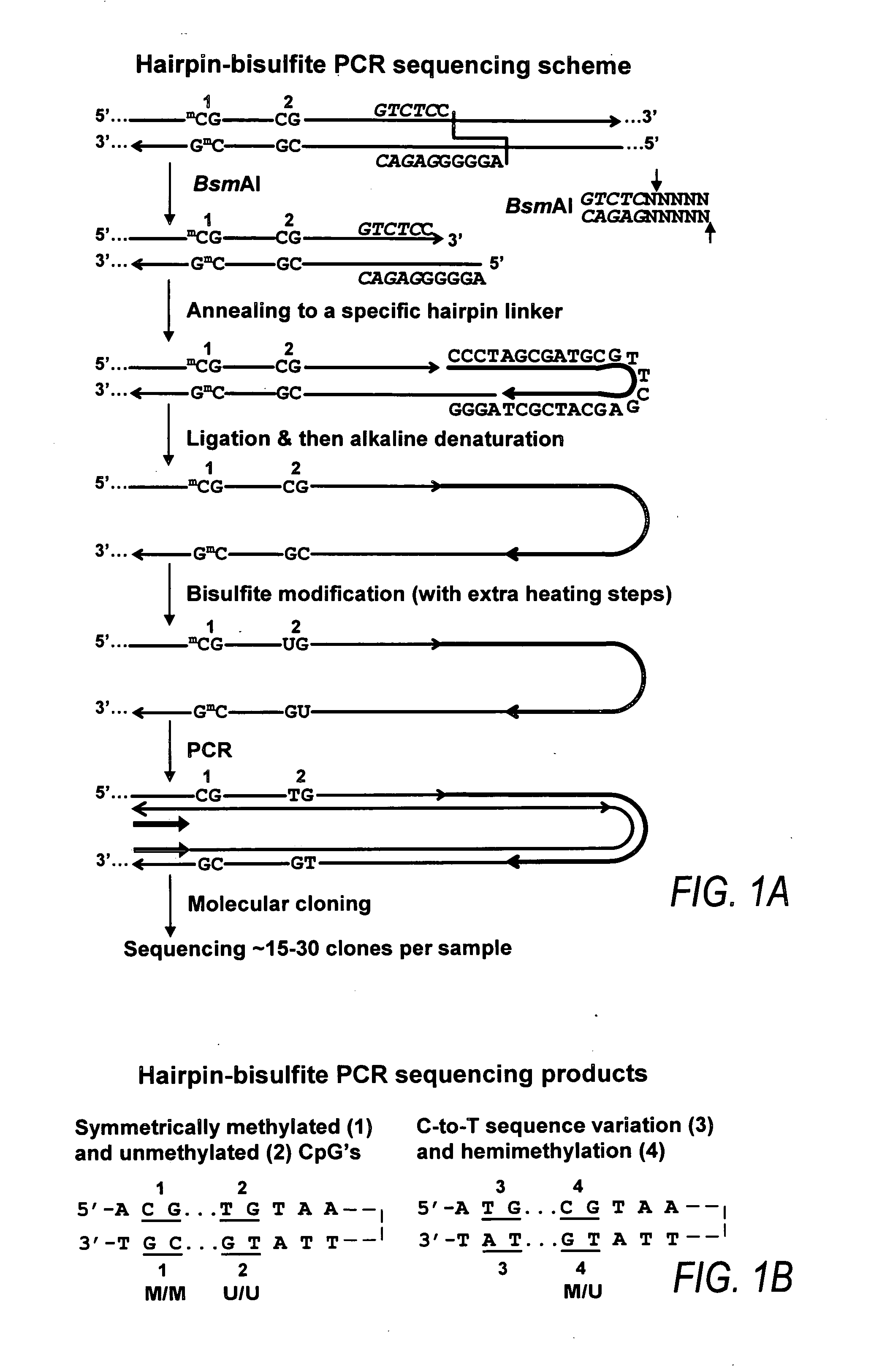
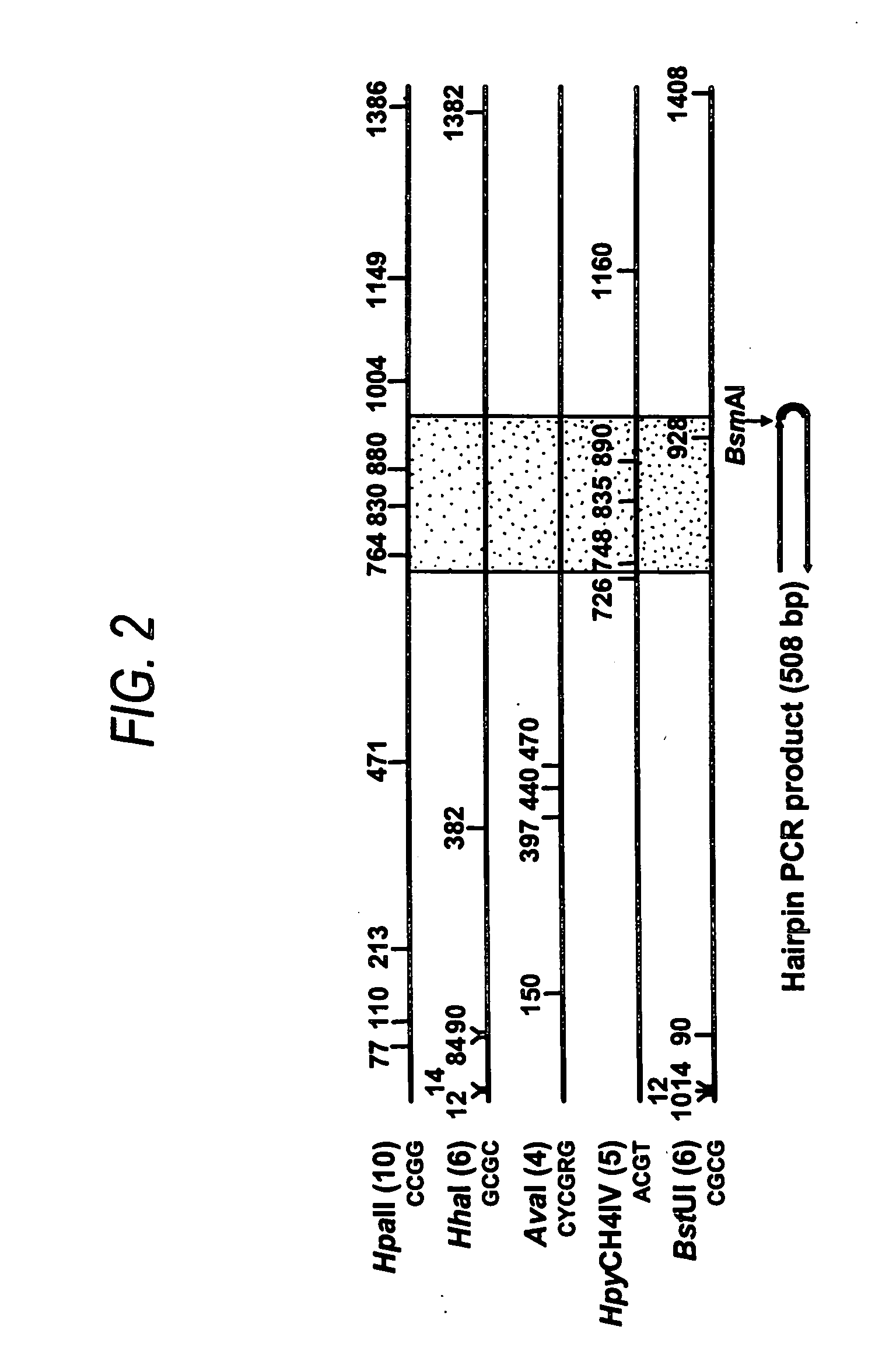
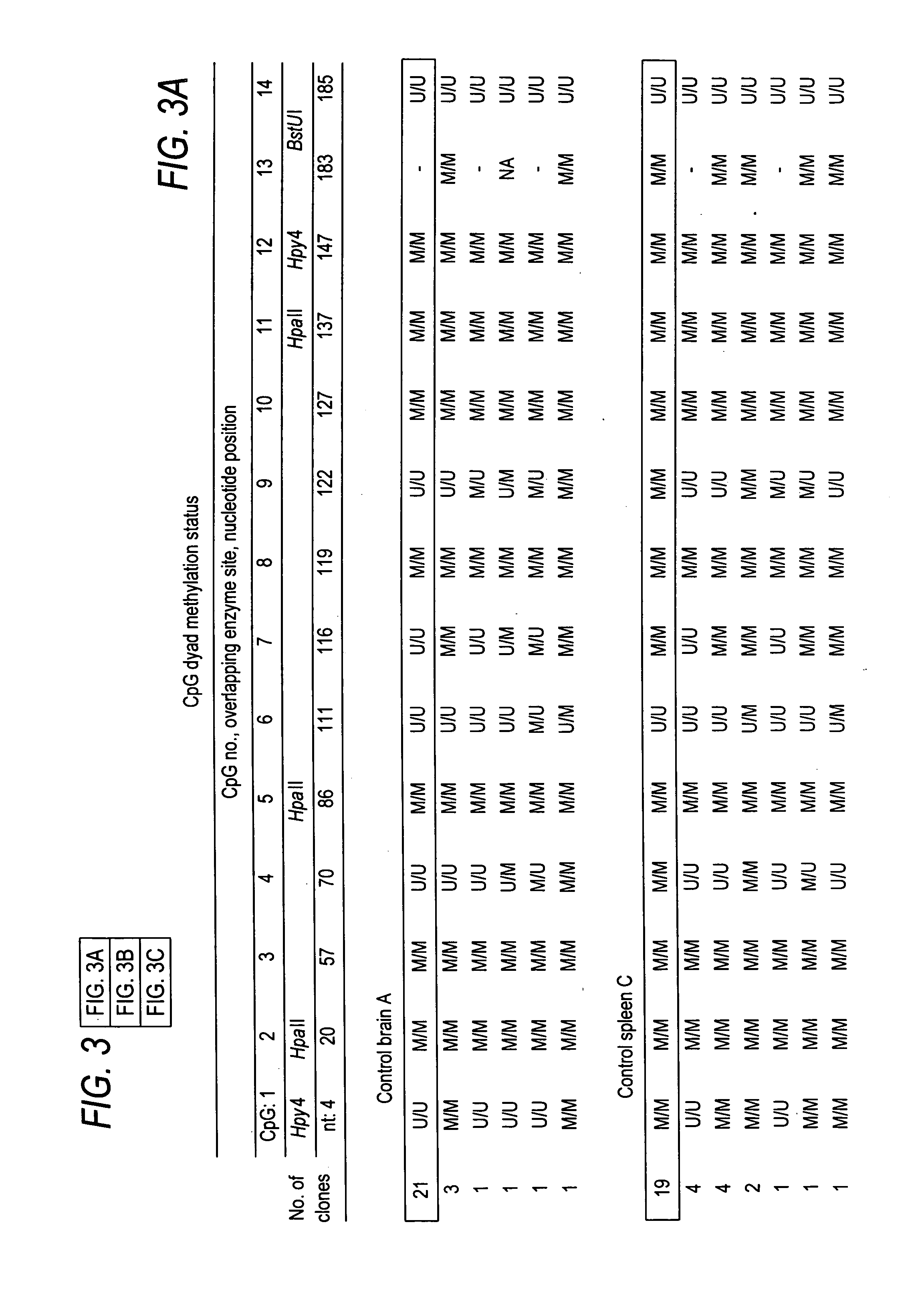
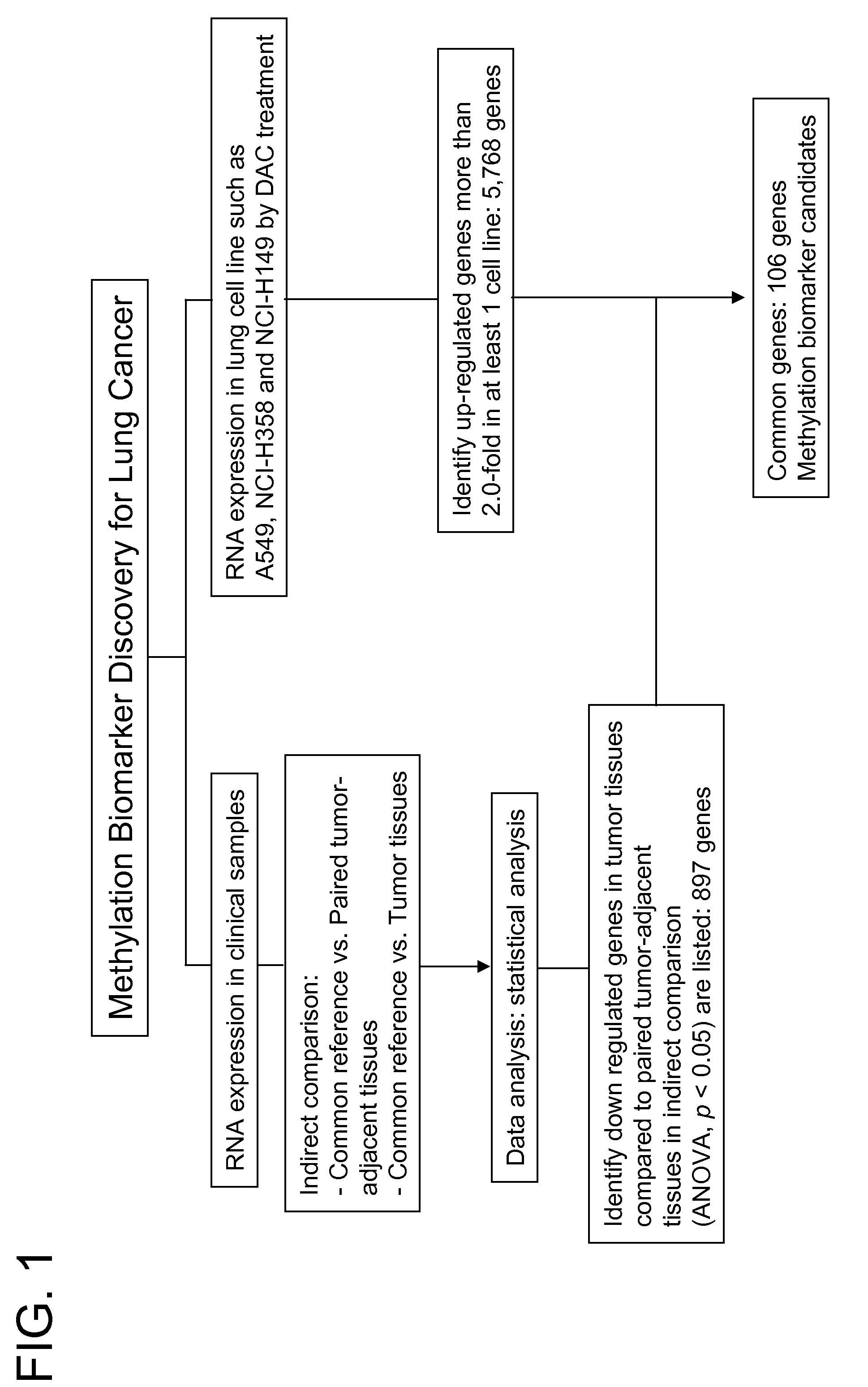
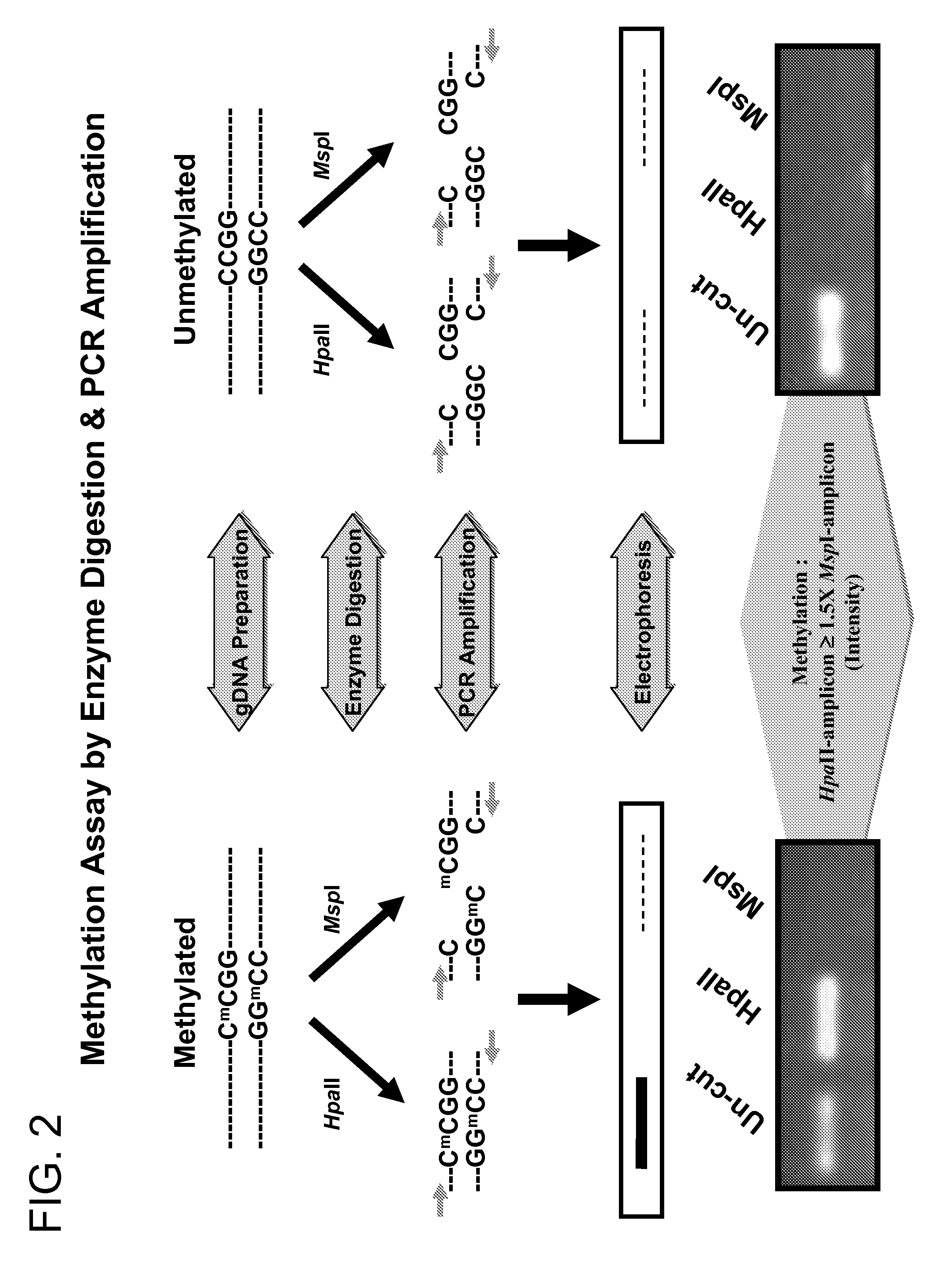
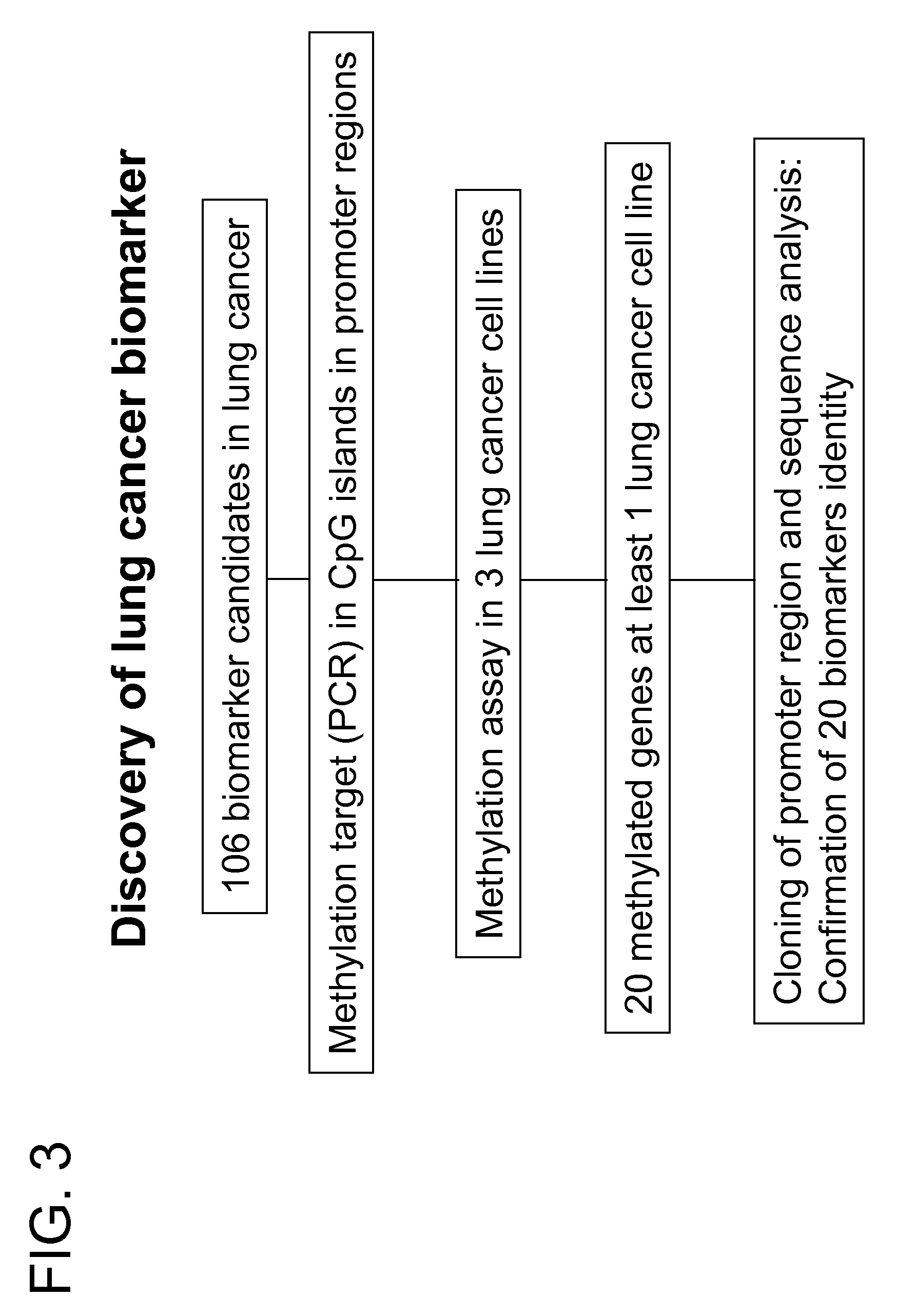
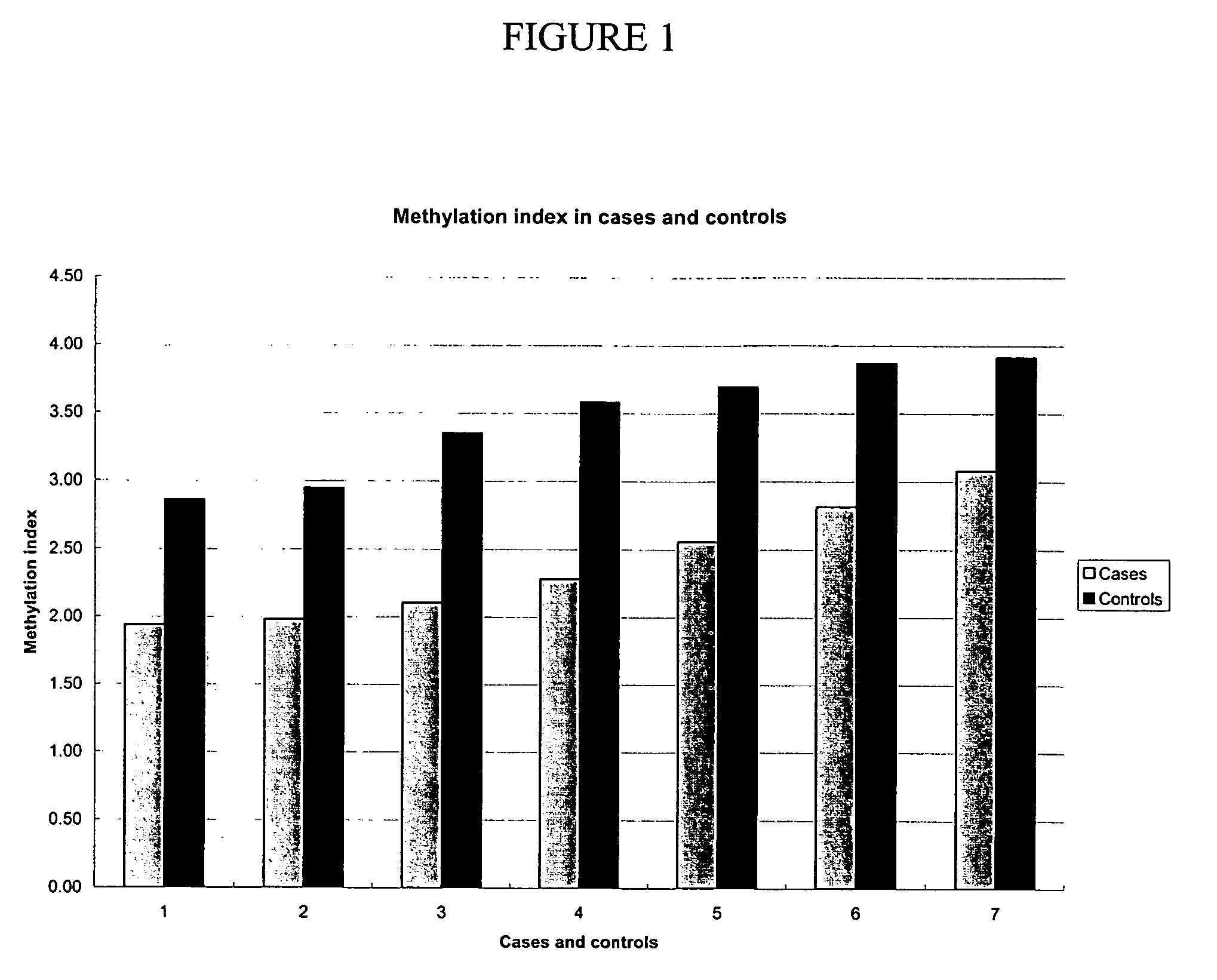
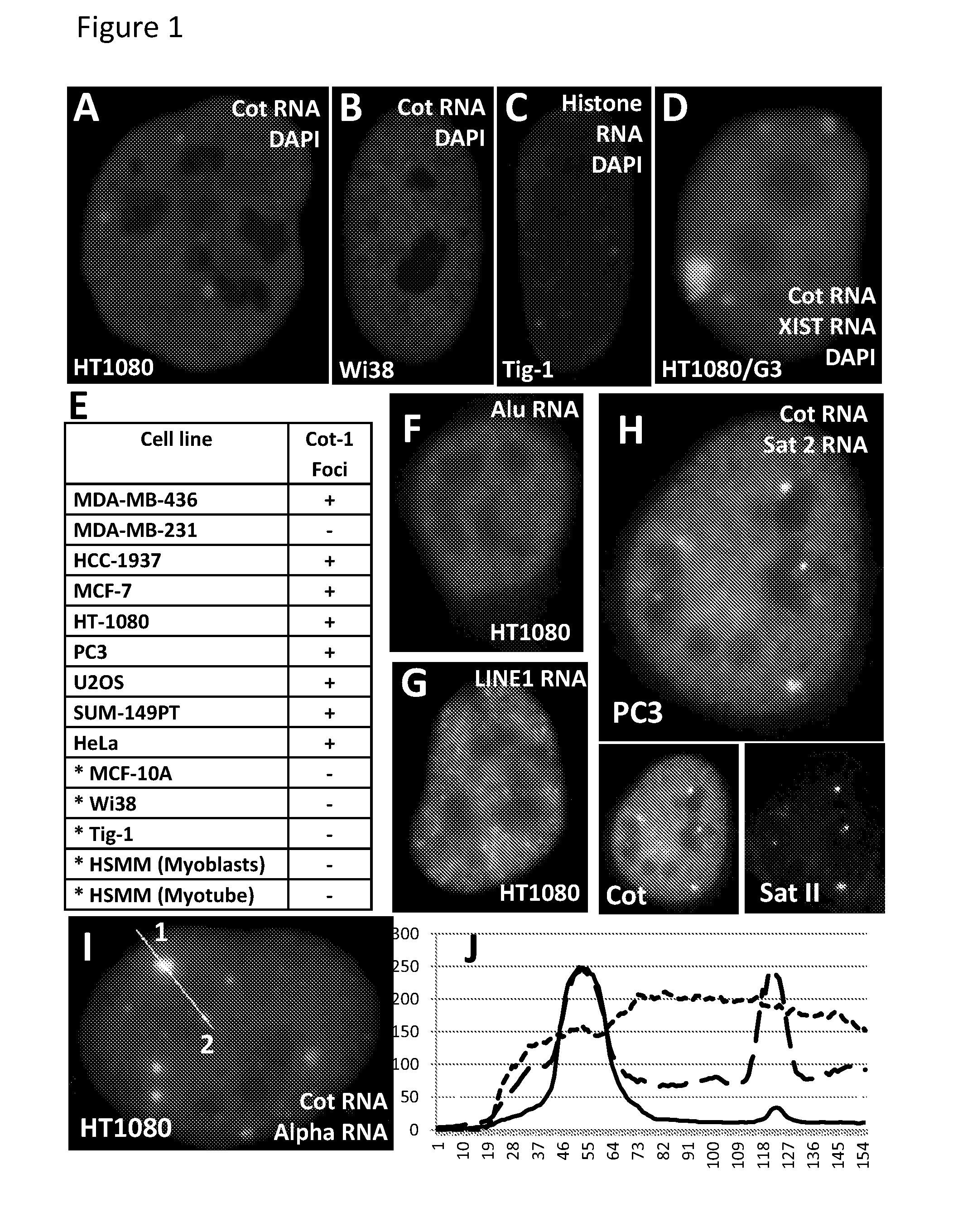
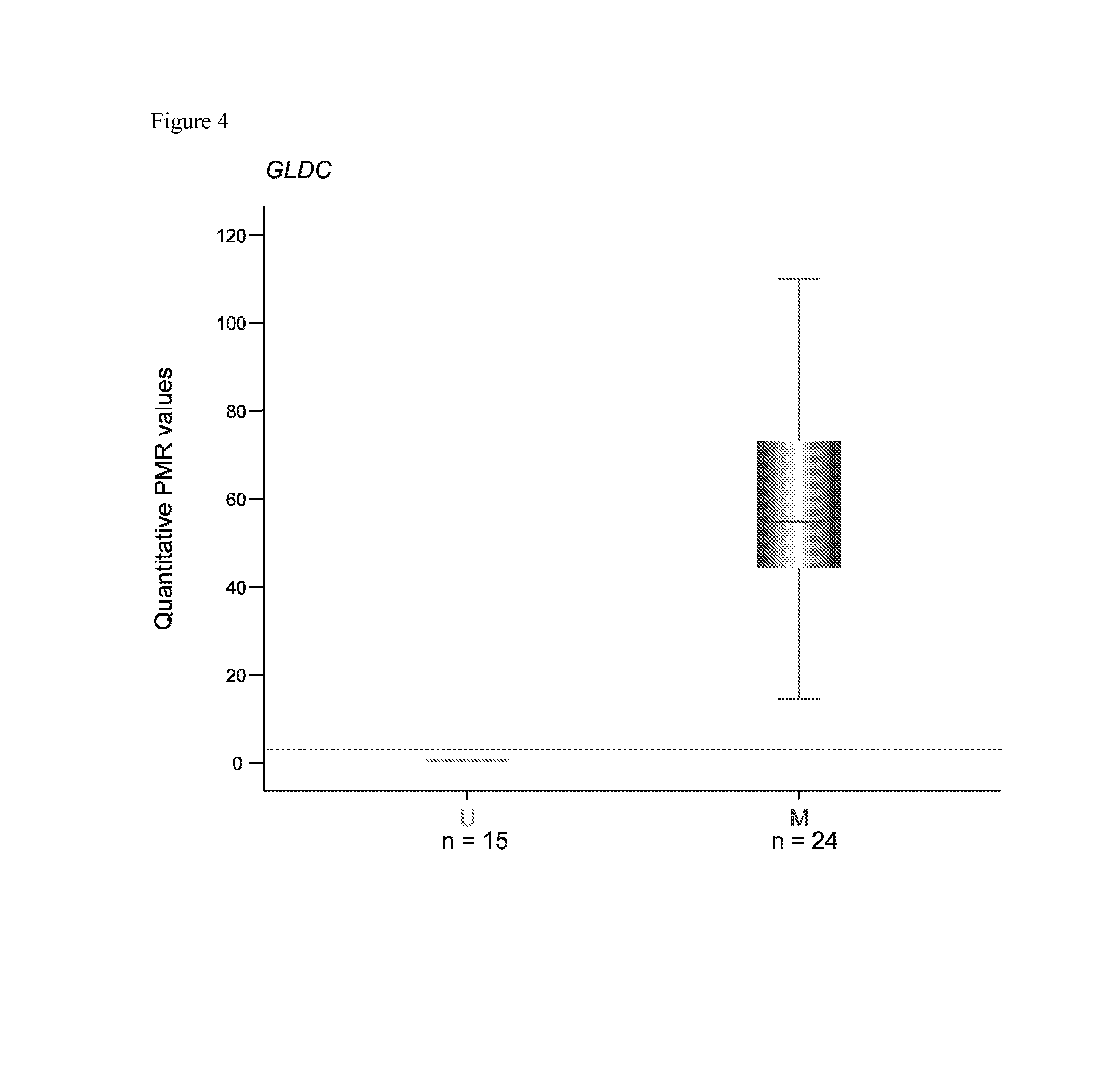
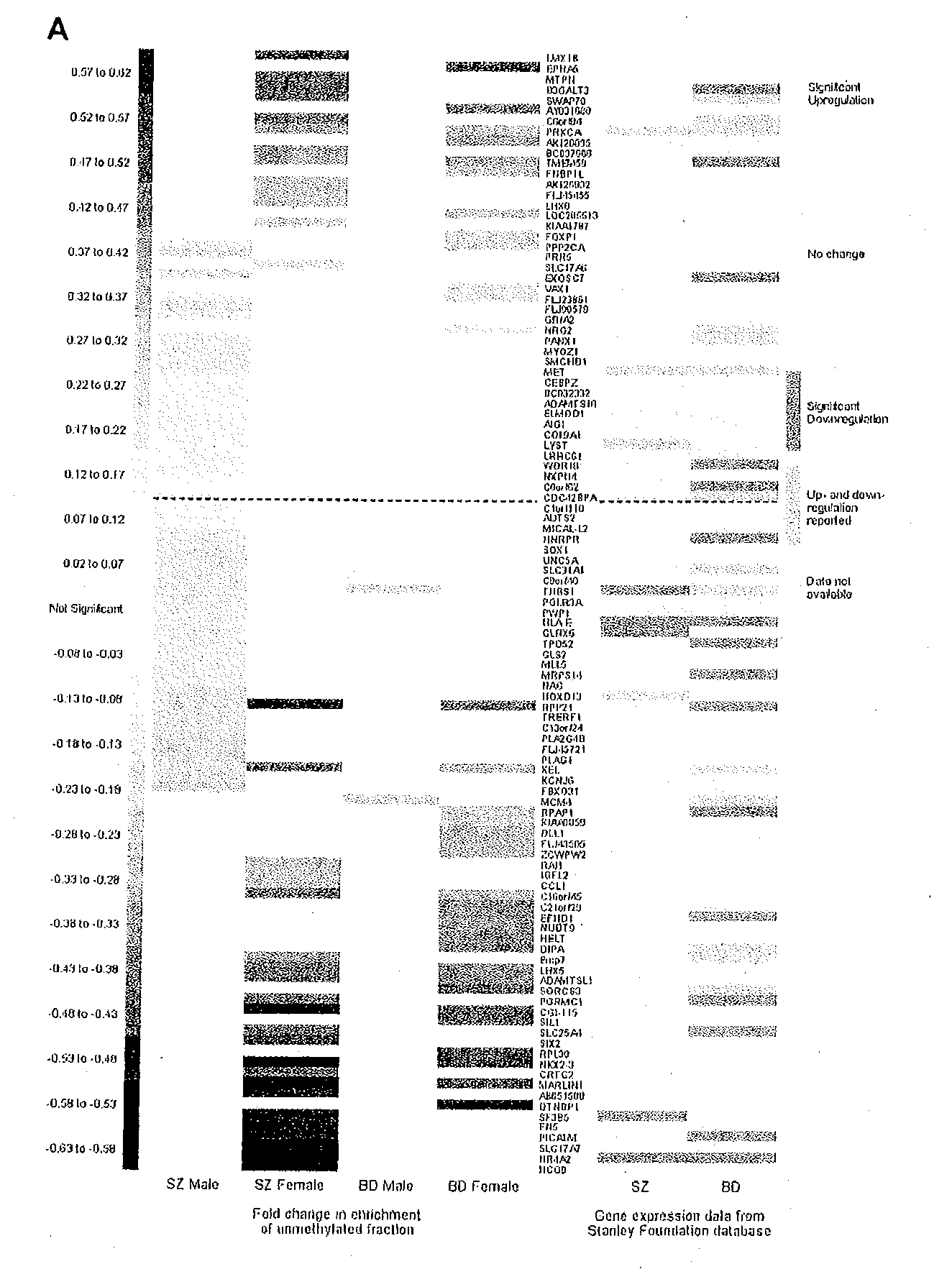
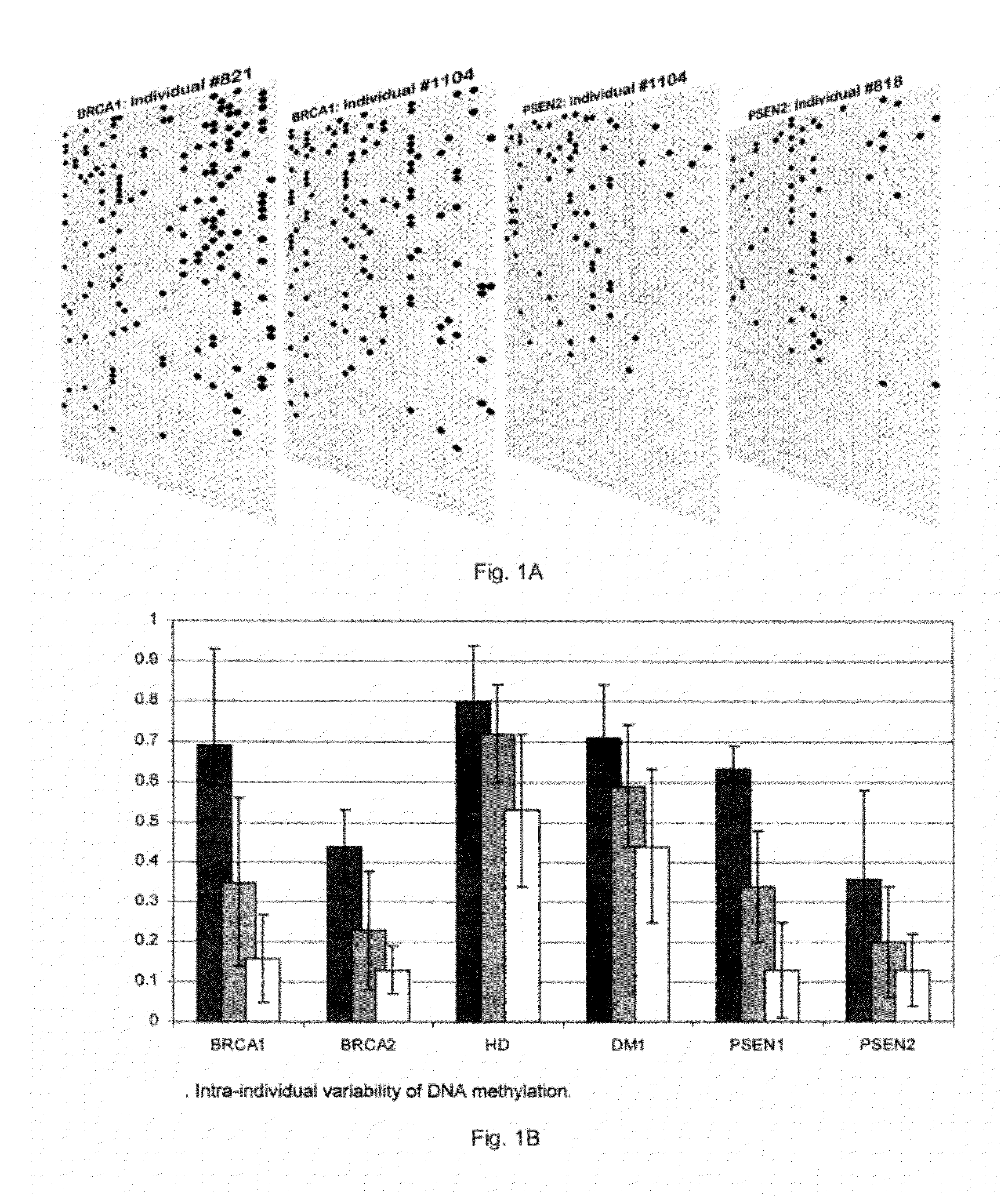
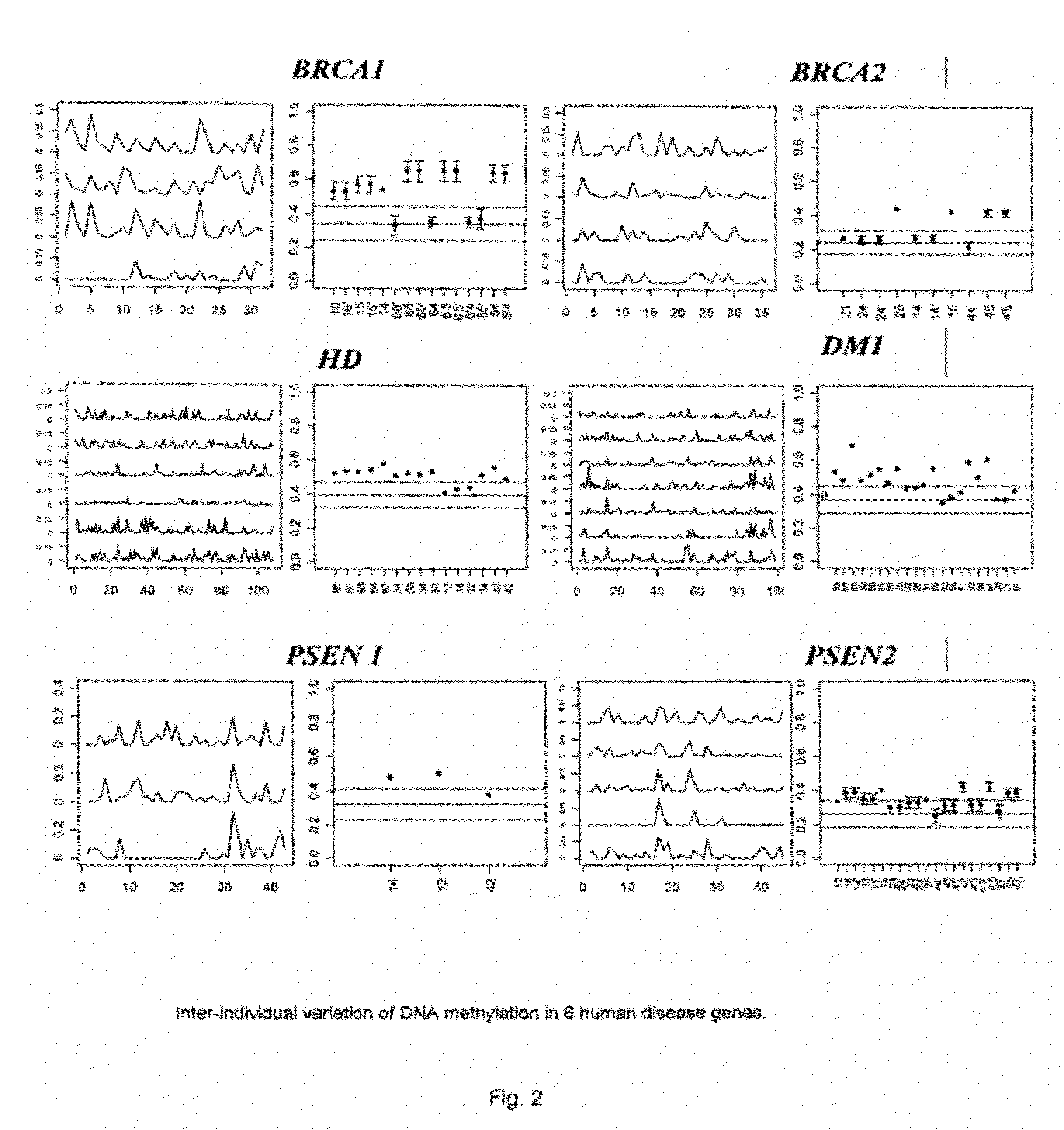
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
61 results about "Epigenetic biomarkers" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
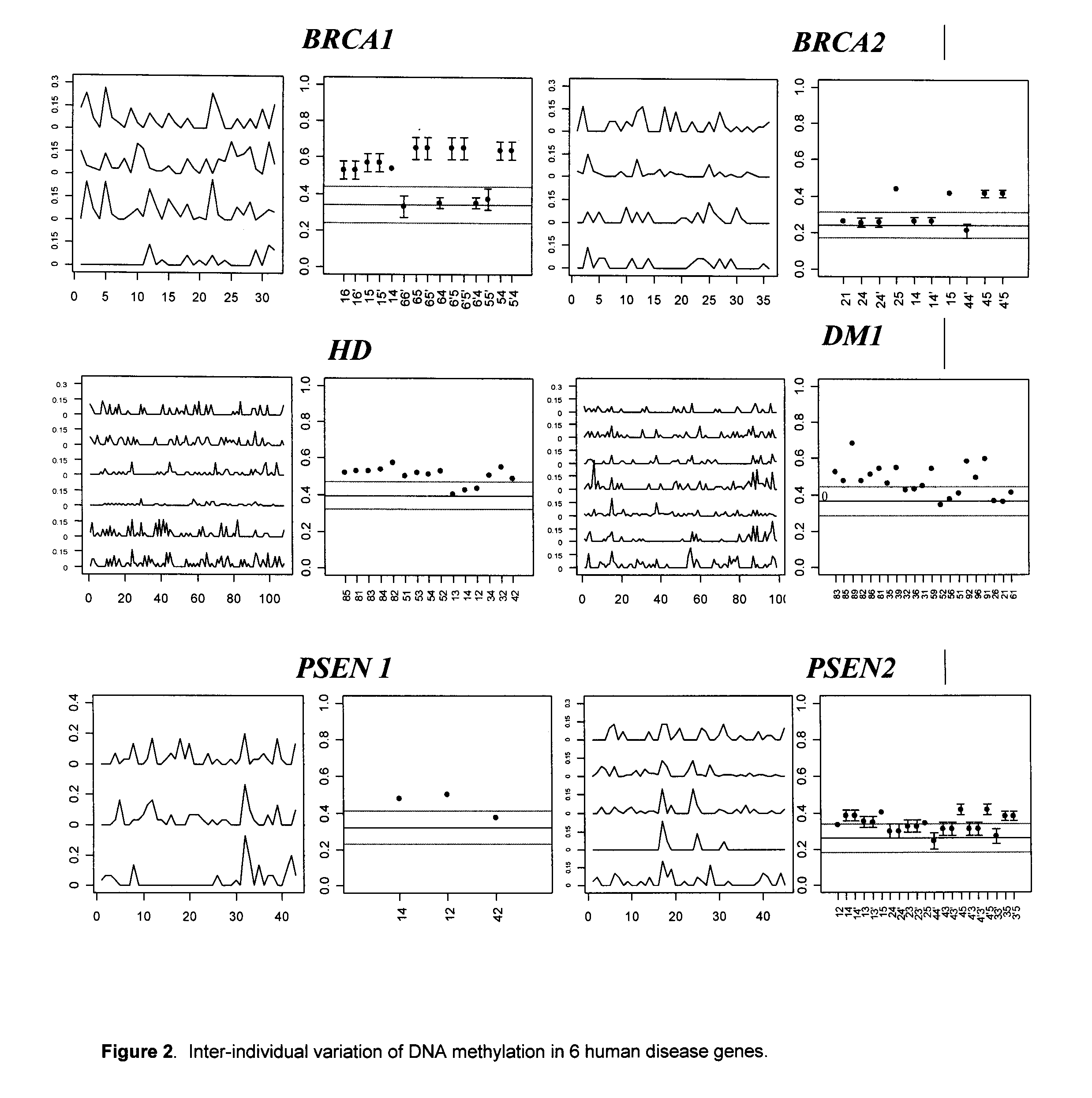
Epigenetic markers are potent diagnostic, prognostic and predictive biomarkers for colorectal cancer. But also, it is a potential pharmacological target due to its reversible nature. Normally, DNA methylation and histone modification act as targeted drug therapies.
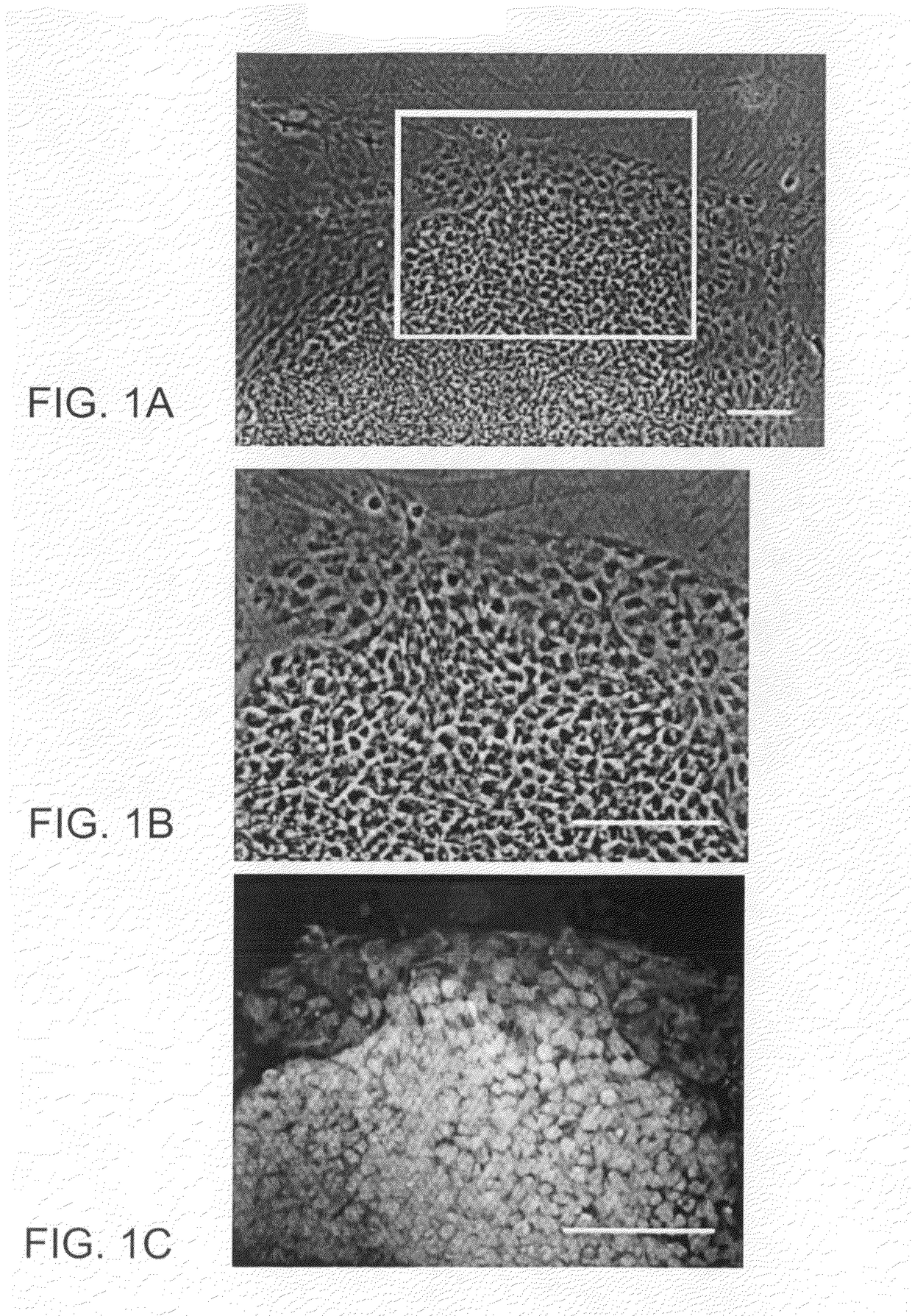
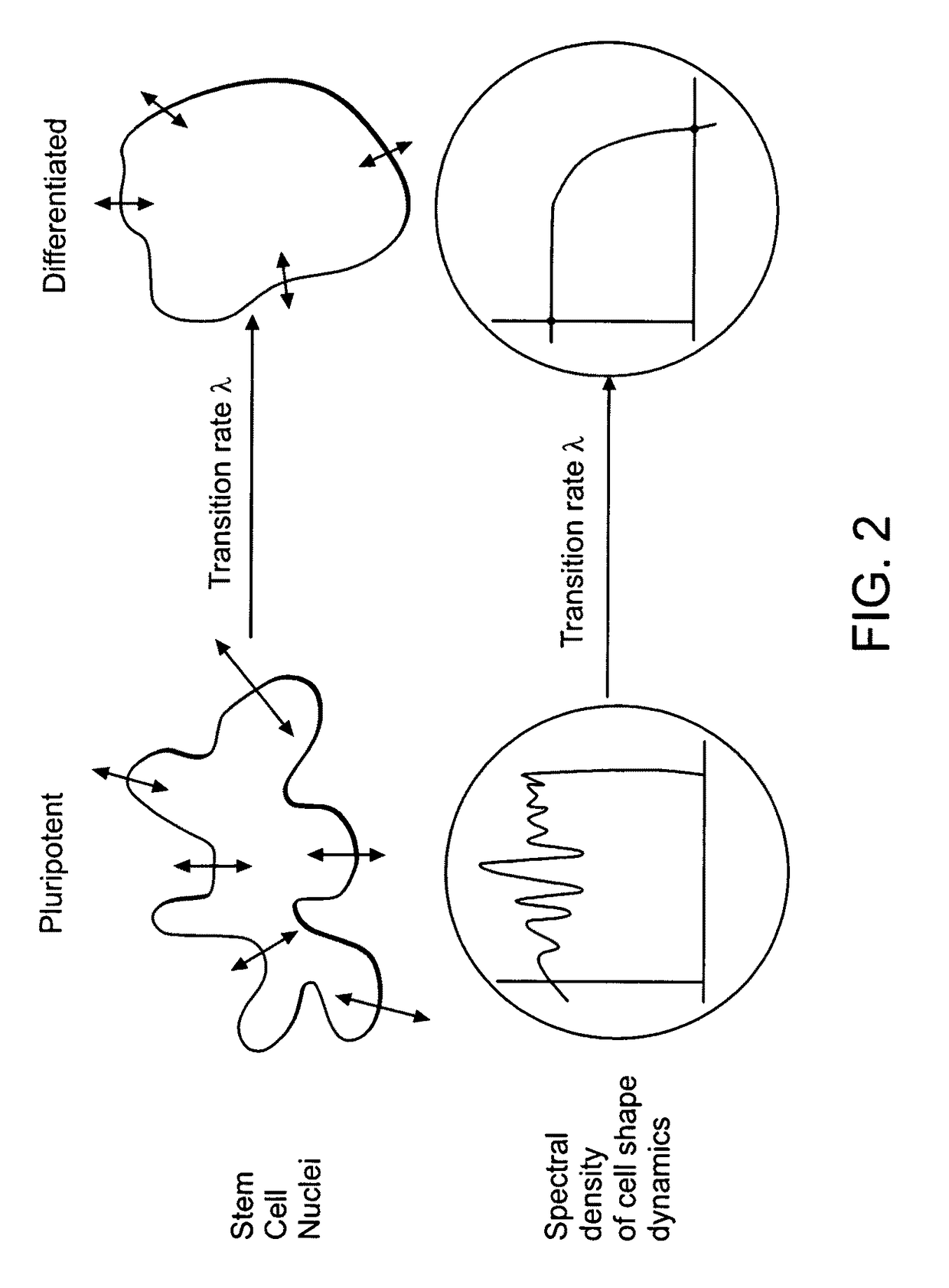
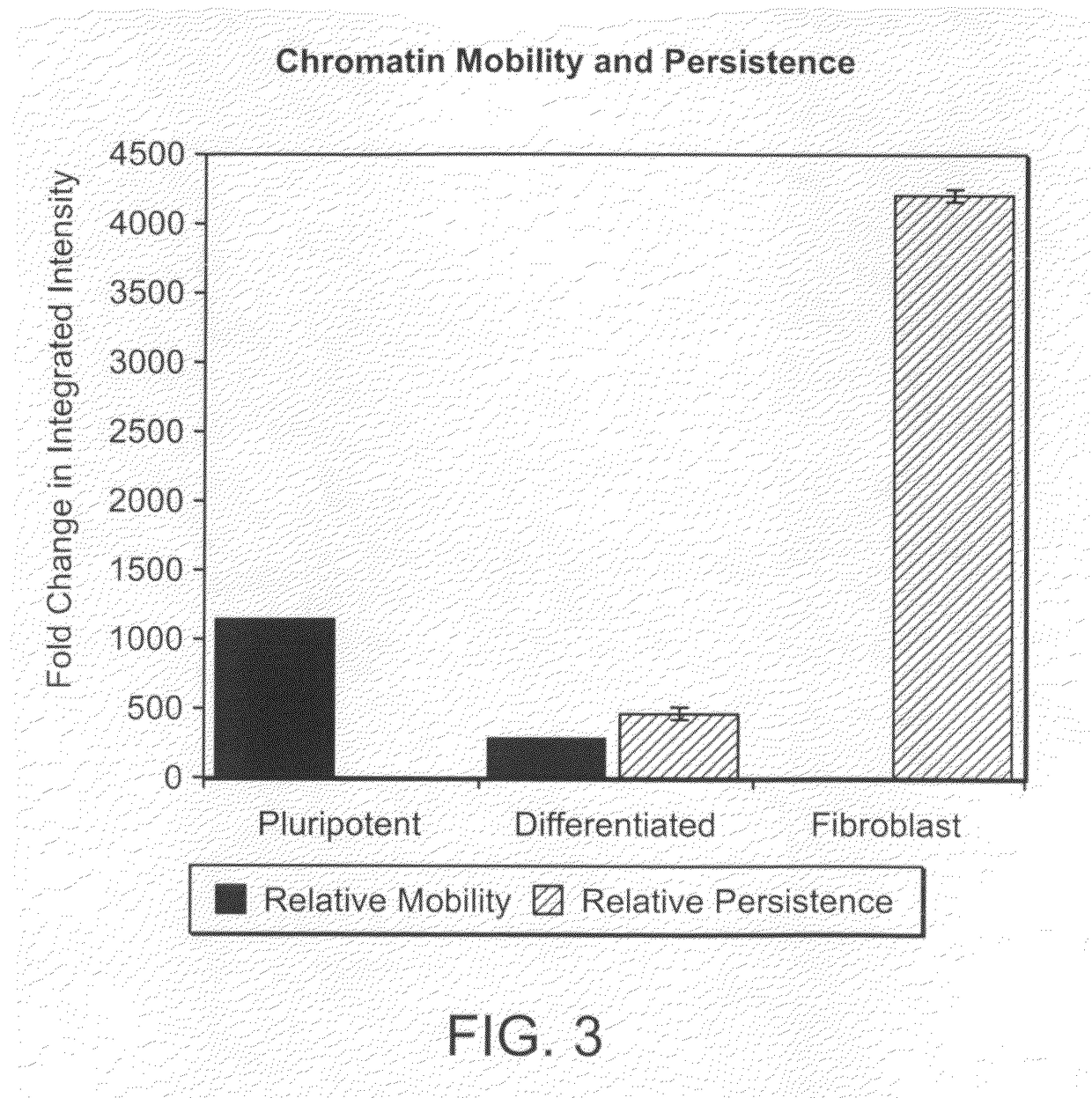
Image-based methods for measuring global nuclear patterns as epigenetic markers of cell differentiation
The invention provides methods for determining the differentiation state of cells. The methods include non-invasive, non-perturbing, automatable, and quantitative methods of analysis of cell colonies, individual cells, and / or cellular structures.
Owner:UNIVERSITY OF PITTSBURGH +1
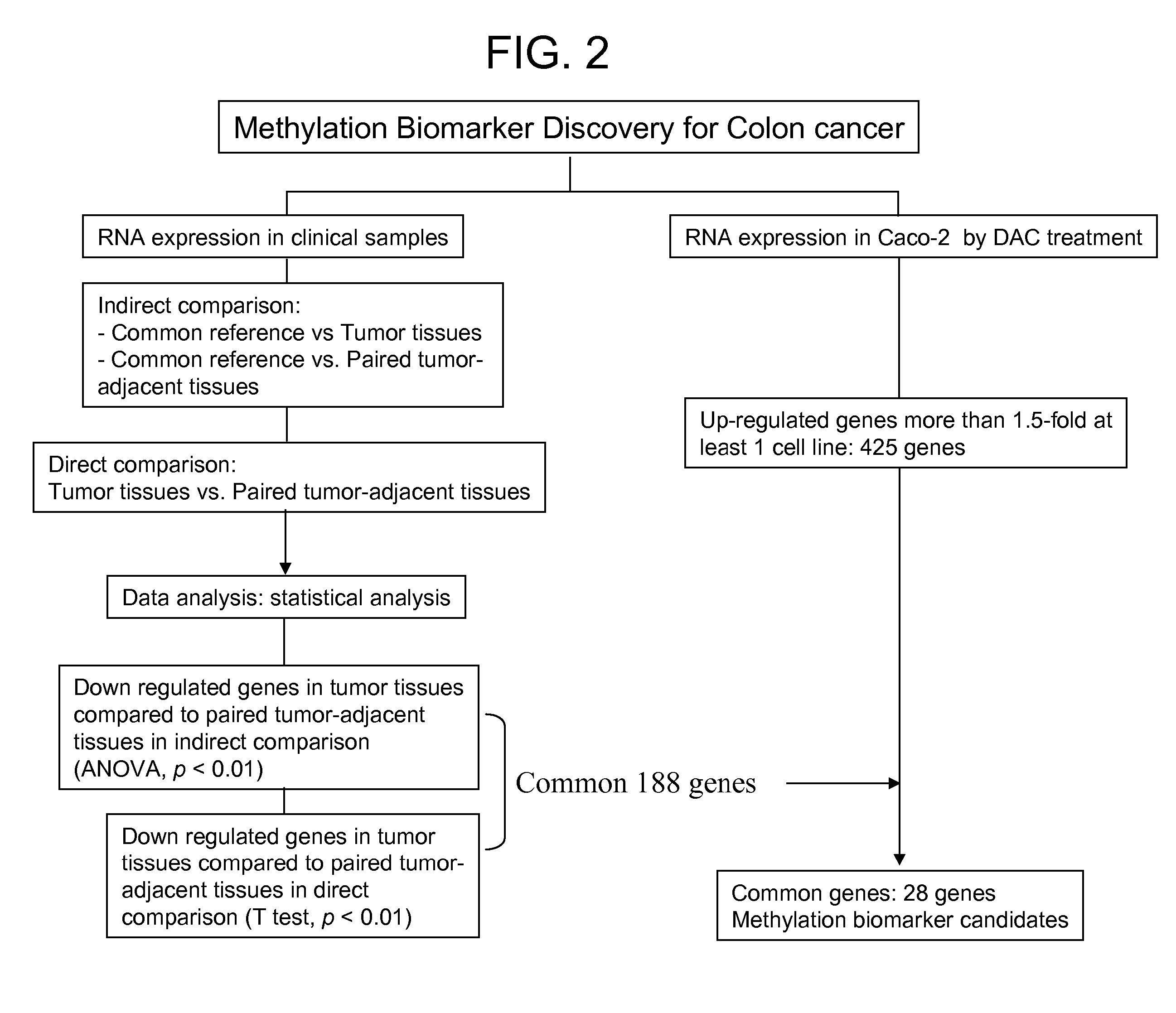
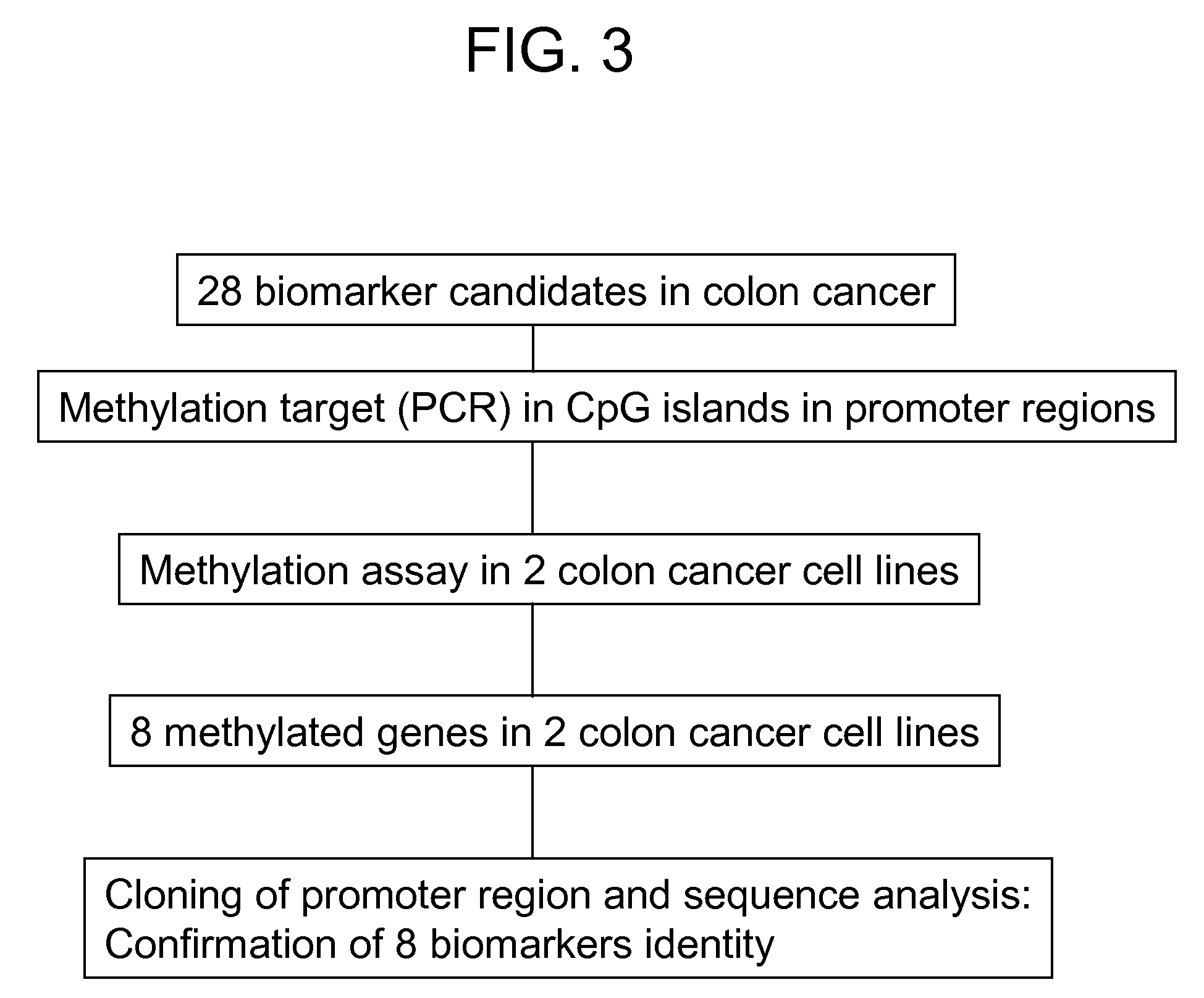
Colon cancer biomarker discovery
InactiveUS20060234254A1Early detectionAccurate diagnostic systemSugar derivativesMicrobiological testing/measurementTumor BiomarkersOncology
Owner:GENOMICTREE
Methods and compositions for differentiating tissues for cell types using epigenetic markers
InactiveUS20060183128A1Accurate and efficientMicrobiological testing/measurementFermentationGenomic DNAEpigenetic Profile
The present invention provides, inter alia, a method for generating a genome-wide epigenomic map, comprising a correlation between methylation variable CpG positions (MVP) and genomic DNA sample types. MVP are those CpG positions that show a variable quantitative level of methylation between sample types. Particular genomic regions of interest (ROI) provide preferred marker sequences that comprise multiple, and preferably proximate MVP, and that have novel utility for distinguishing sample types. The epigenic maps have broad utility, for example, in identifying sample types, or for distinguishing between and among sample types. In a preferred embodiment the epigenomic map is based on methylation variable regions (MVP) within the major histocompatibility complex (MHC), and has utility, for example, in identifying the cell or tissue source of a genomic DNA sample, or for distinguishing one or more particular cell or tissue types among other cell or tissue types. Analysis of epigenetic characteristics of one, or of a set of nucleic acid sequences, in the context of an inventive epigenomic map, allows for the determination of an origin of the nucleic acids.
Owner:EPIGENOMICS AG
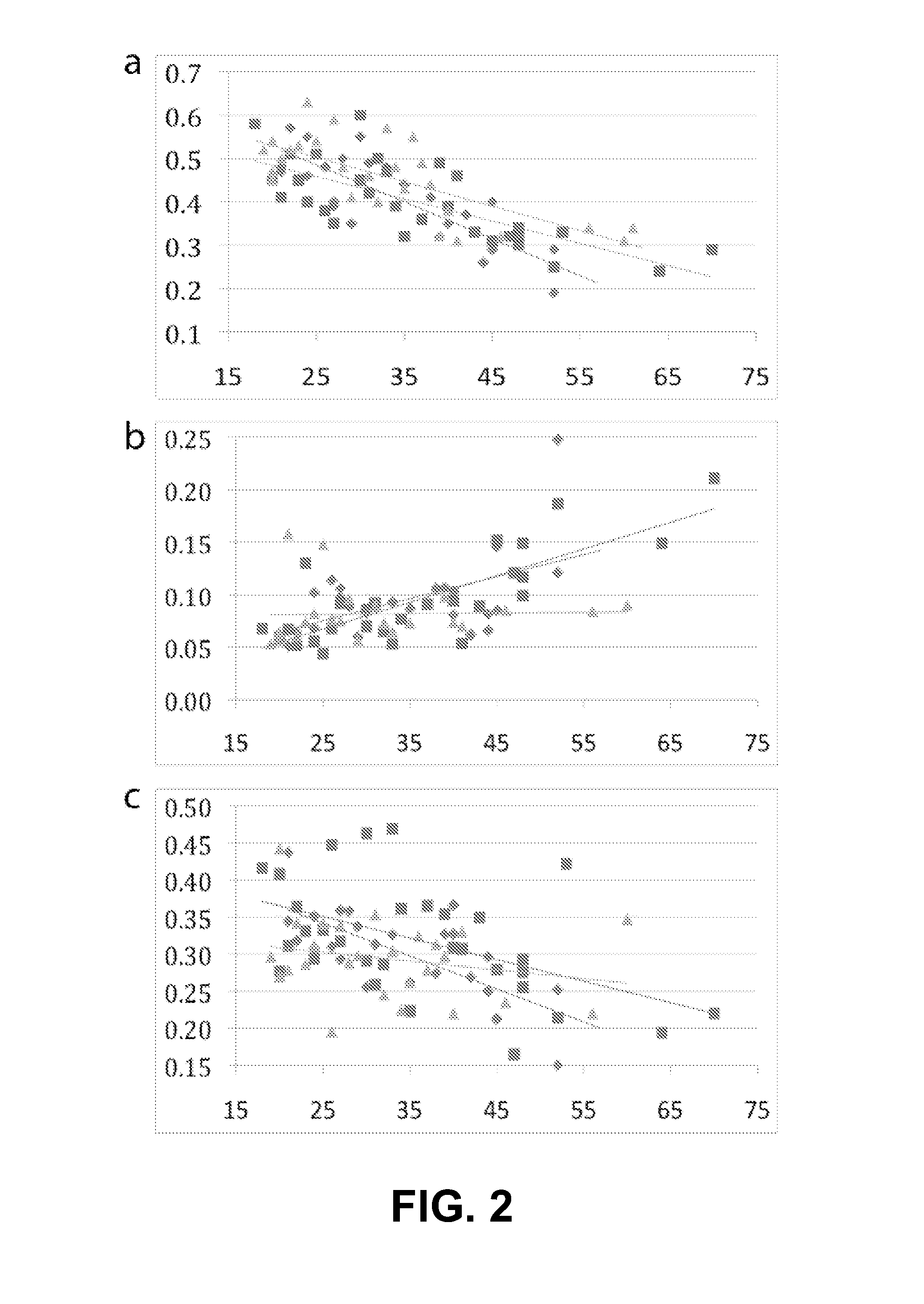
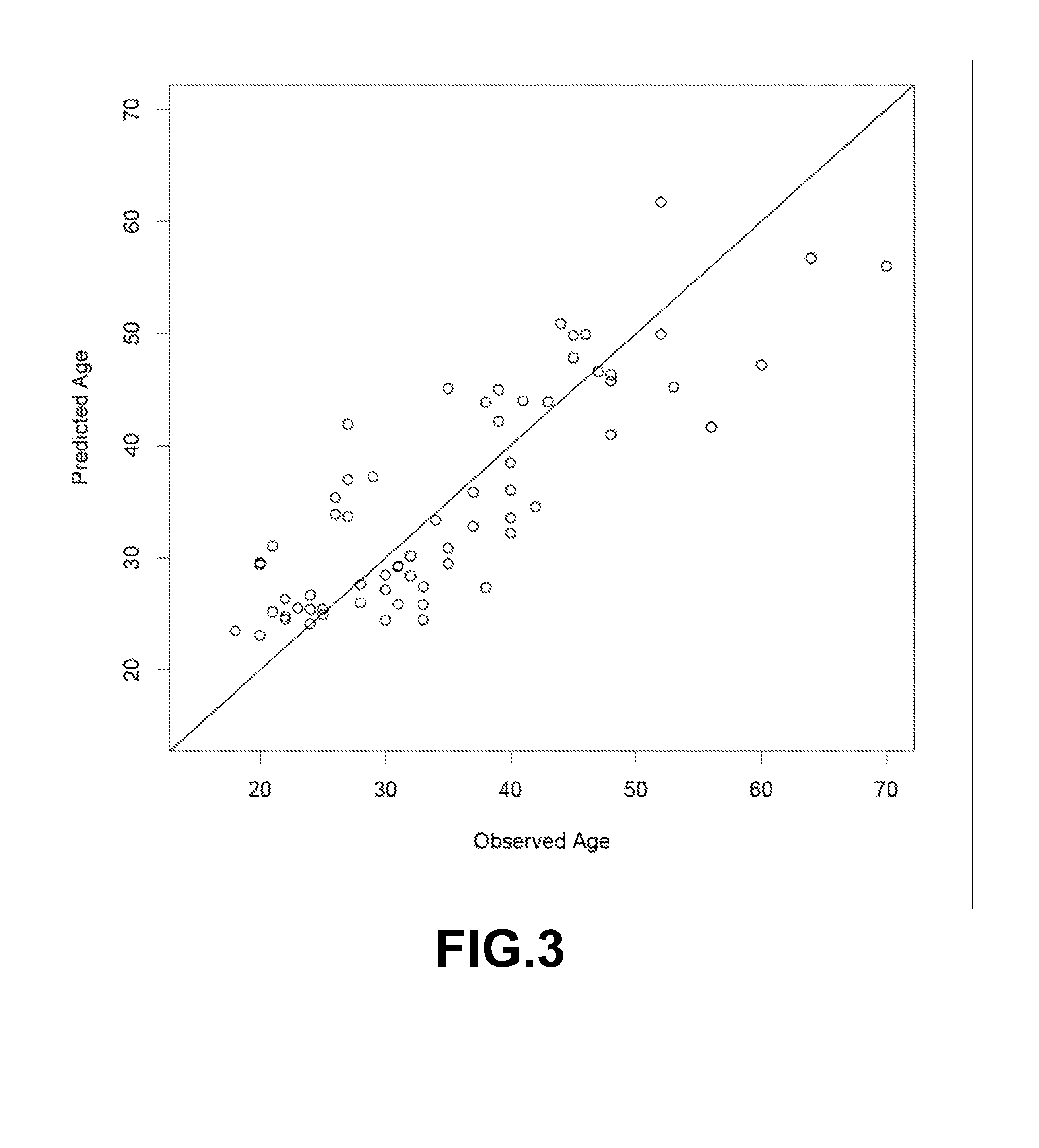
Method to estimate age of individual based on epigenetic markers in biological sample
ActiveUS20140228231A1Nucleotide librariesMicrobiological testing/measurementCytosineWhite blood cell
The invention provides methods and materials that use observation of DNA characteristics to obtain information relating to the age of individuals. The instant disclosure identifies 88 sites in or near 80 genes for which the degree of cytosine methylation in epithelial and / or white blood cells is significantly correlated with age. In illustrative embodiments of the invention, cytosine methylation patterns the promoters of the EDARADD, TOMILI, and NPTX2 genes are used to predict the age of an individual with a high degree of accuracy.
Owner:RGT UNIV OF CALIFORNIA
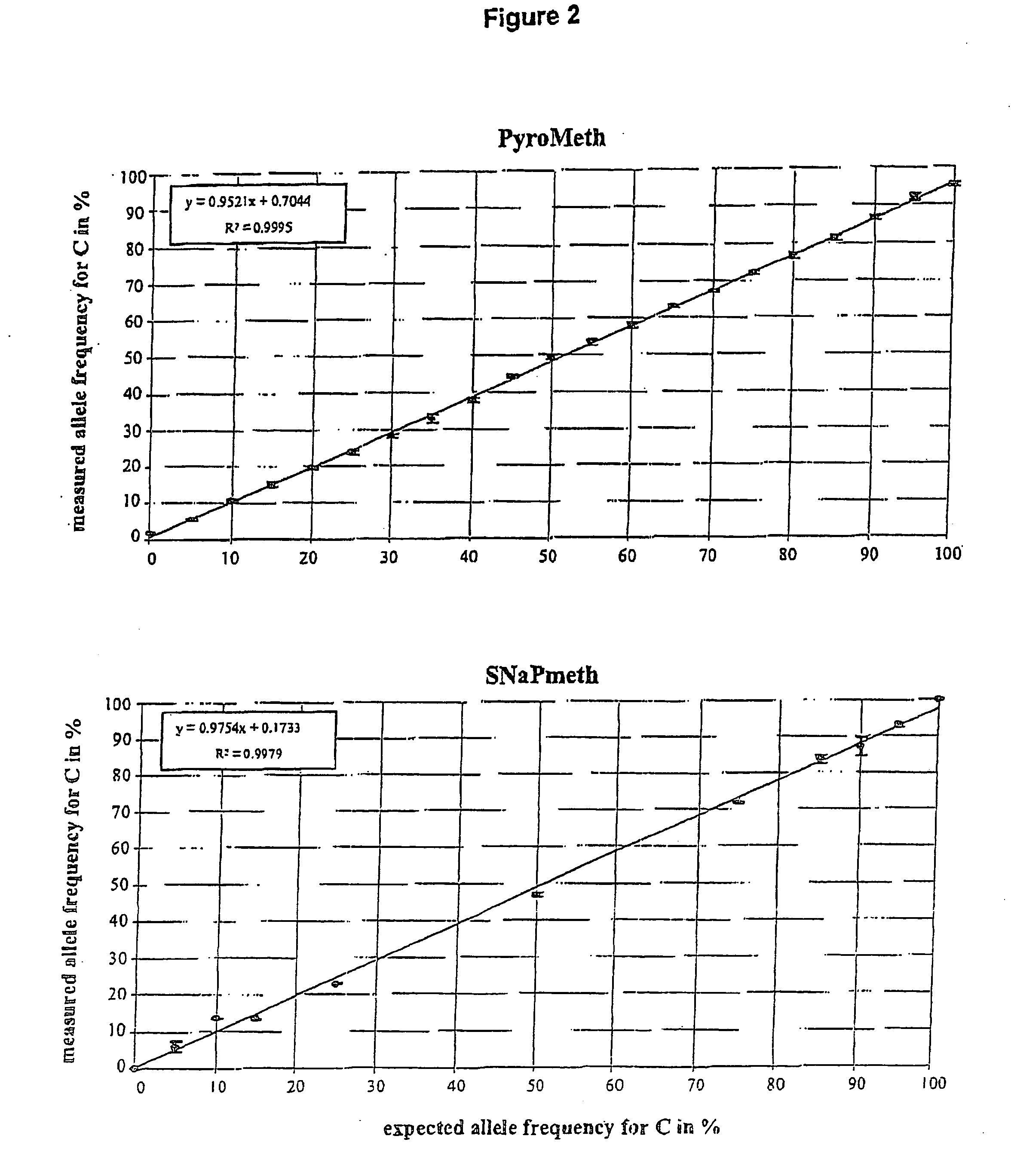
Method of detecting epigenetic biomarkers by quantitative methyISNP analysis
InactiveUS20060024676A1High methylationLow methylationMicrobiological testing/measurementAqueous solutionBiology
Owner:MAX DELBRUECK CENT FUER MOLEKULARE MEDIZIN
Method
A method of screening for genetic or epigenetic markers associated with autism or related disorders comprises the steps of providing a biological sample from a mammal; and testing the sample or genetic material isolated from the sample for genetic polymorphisms / mutations and / or epigenetic alterations. The polymorphism may be located in the Xq / Yq pseudoautosomal gene region and extends into the adjacent Xq28 gene region.
Owner:UNIV COLLEGE CORK NAT UNIV OF IRELAND CORK
Methods for diagnosing cancer based on DNA methylation status in NBL2
InactiveUS20070087358A1Raise the possibilityMinimize the possibilityMicrobiological testing/measurementNucleotideCancers diagnosis
The present invention relates to methods for diagnostic or prognostic assay for cancer based on analysis of altered methylation status at specific CpG dinucleotide sequences within the epigenetic marker, NBL2. The methods of the invention comprise determining the methylation status of a subregion of genomic CpG dinucleotide sequences within the DNA repeat, NBL2, in a sample of a subject and comparing the methylation status of the genomic CpG dinucleotide sequences in the sample to the methylation status of the genomic CpG dinucleotide sequences in a reference, wherein a difference in the methylation status of the genomic CpG dinucleotide sequences in the sample as compared to the reference indicates an association of the subject with cancer or cancer progression. The invention further relates to genomic DNA sequences that exhibit altered CpG methylation status in disease state as compared to normal state. The invention also provides nucleic acids, nucleic acid arrays and kits useful for practicing the methods of the present invention.
Owner:TULANE UNIVERSITY
Lung cancer biomarker discovery
InactiveUS20070264659A1Early detectionAccurate diagnostic systemMicrobiological testing/measurementBiomarker discoveryOncology
Owner:GENOMICTREE
Epigenetic biomarkers for early detection, therapeutic effectiveness, and relapse monitoring of cancer
InactiveUS20100151468A1Strong specificityHigh sensitivityMicrobiological testing/measurementDisease diagnosisEarly Cancer DetectionHistone H4
The present invention provides methods of detection, including early detection, for cancer or other diseases and normal physiologic processes mediated by global epigenetic changes, by using one or more of the following biomarkers: a global DNA methylation index, a global histone H4 acetylation index, and a global histone H4 trimethylation index. These methods are useful for, among other things, assessing the effectiveness of treatment, monitoring relapse, and clinical staging of cancer and other chronic as well as acute diseases. These methods are also useful for among other things monitoring the effectiveness of strategies and therapies used to modify lifestyle and contextual effects to prevent disease, foster wellness and enable health promotion.
Owner:PRESTON RAFAEL GUERRERO
Methods of diagnosing cancer using epigenetic biomarkers
InactiveUS20140213475A1Easy to quantifyEasy to distinguishMicrobiological testing/measurementLibrary screeningCancer cellEpigenetic Profile
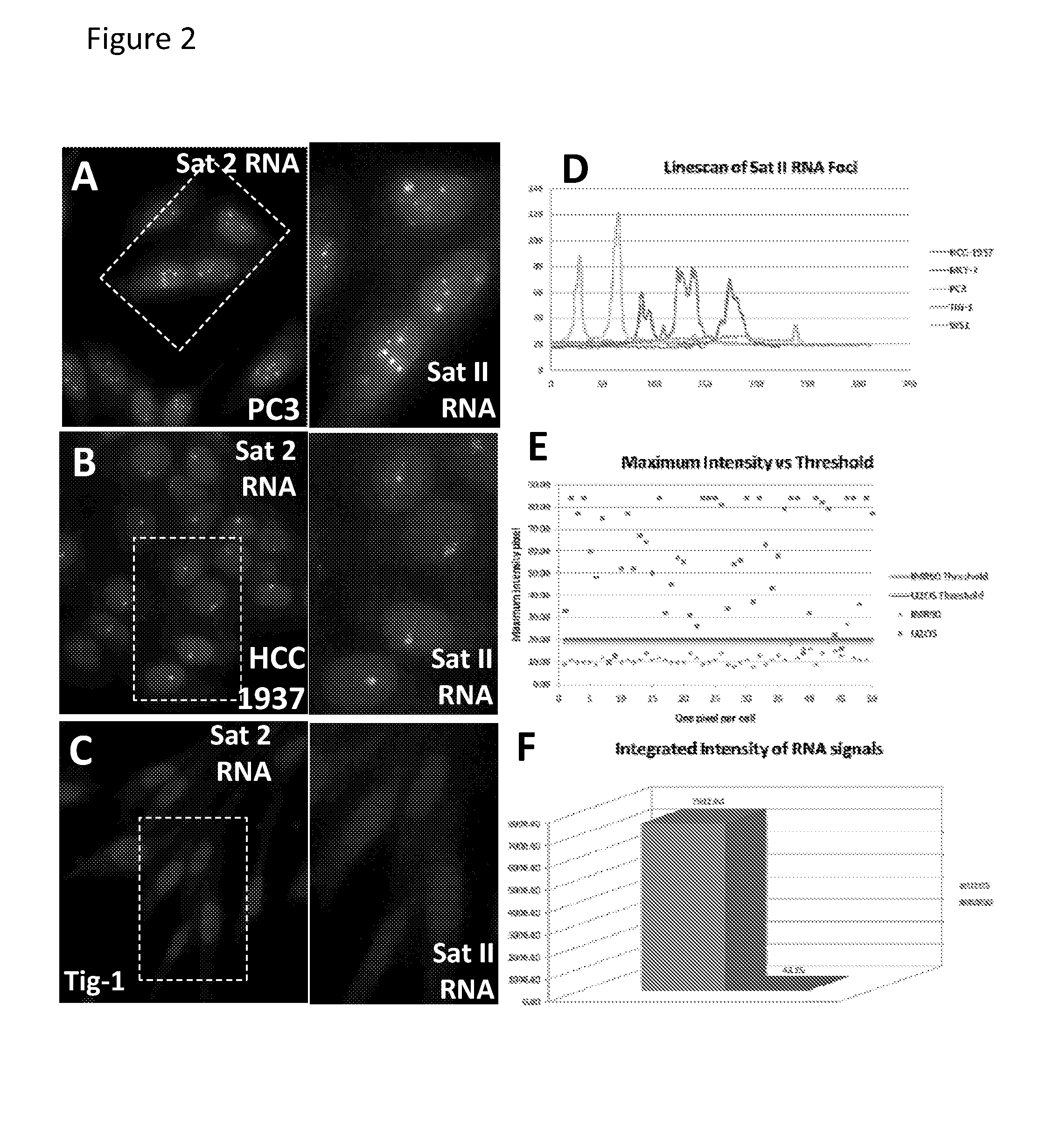
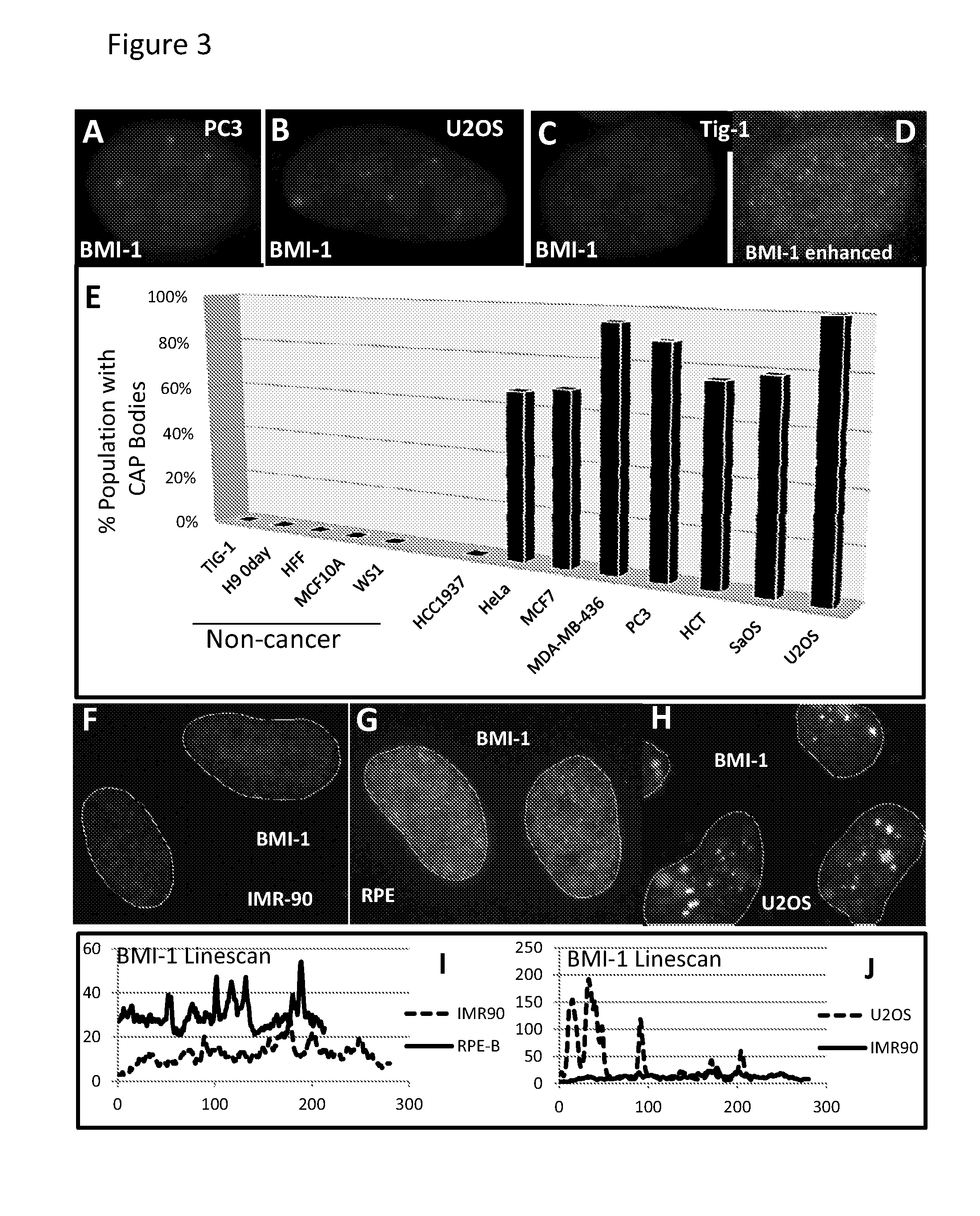
The invention features methods of diagnosing cancer in a mammal (e.g., a human) by detecting a biomarker selected from a satellite II ribonucleic acid (RNA) molecule, a cancer-associated polycomb group (CAP) body, a cancer-associated satellite transcript (CAST) body, and UbH2A. Also featured is a method for identifying an agent for treating cancer in a mammal by contacting a cancer cell having a biomarker selected from a CAP body, a CAST body, and a satellite II RNA molecule with a test agent and determining whether the test agent reduces the level of the biomarker in the cancer cell. Other inventions featured are a method for determining whether a chemotherapeutic agent increases epigenetic imbalance of a cell and a method for detecting epigenetic imbalance by determining a copy number of a satellite II DNA locus at chromosome 1q12 in a cell.
Owner:UNIV OF MASSACHUSETTS
Device and methods for epigenetic analysis
ActiveUS8735065B2Bioreactor/fermenter combinationsBiological substance pretreatmentsEpigenetic AnalysisGenetic Materials
Provided herein are methods and devices for single object detection. The methods and devices can be used to identify a plurality epigenetic markers on a genetic material, or a chromatin, encompassing fragments thereof. The invention provides for the characterization of the genetic material flowing through a channel in a continuous body of fluid based on detection of one or more properties of the genetic material. The methods and systems provided herein allow genome-wide, high-throughput epigenetic analysis and overcome a variety of limitations common to bulk analysis techniques.
Owner:CORNELL UNIVERSITY
Device and Methods for Epigenetic Analysis
ActiveUS20120244532A1Easy to implementFast data collectionBioreactor/fermenter combinationsBiological substance pretreatmentsEpigenetic AnalysisGenetic Materials
Provided herein are methods and devices for single object detection. The methods and devices can be used to identify a plurality epigenetic markers on a genetic material, or a chromatin, encompassing fragments thereof. The invention provides for the characterization of the genetic material flowing through a channel in a continuous body of fluid based on detection of one or more properties of the genetic material. The methods and systems provided herein allow genome-wide, high-throughput epigenetic analysis and overcome a variety of limitations common to bulk analysis techniques.
Owner:CORNELL UNIVERSITY
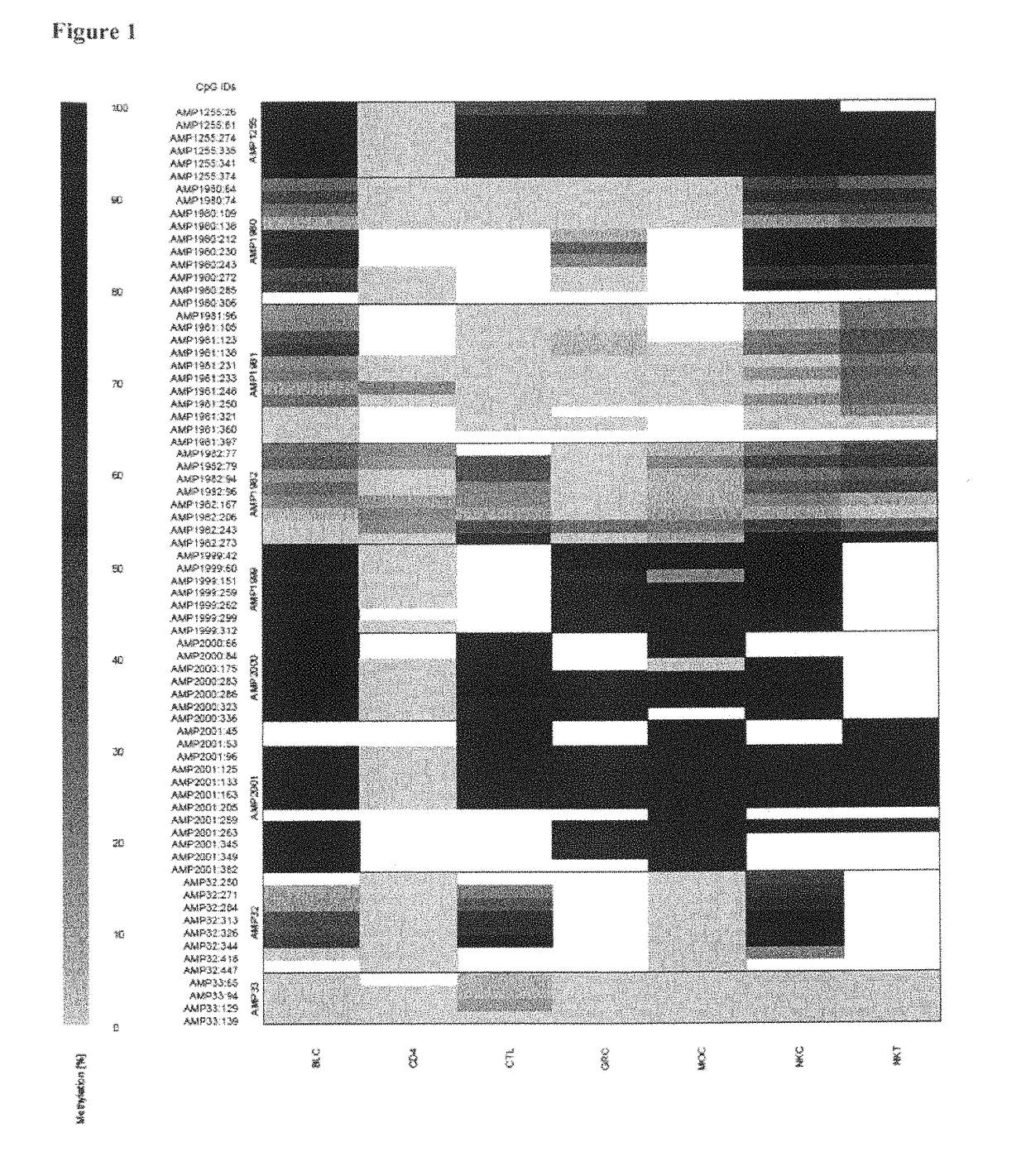
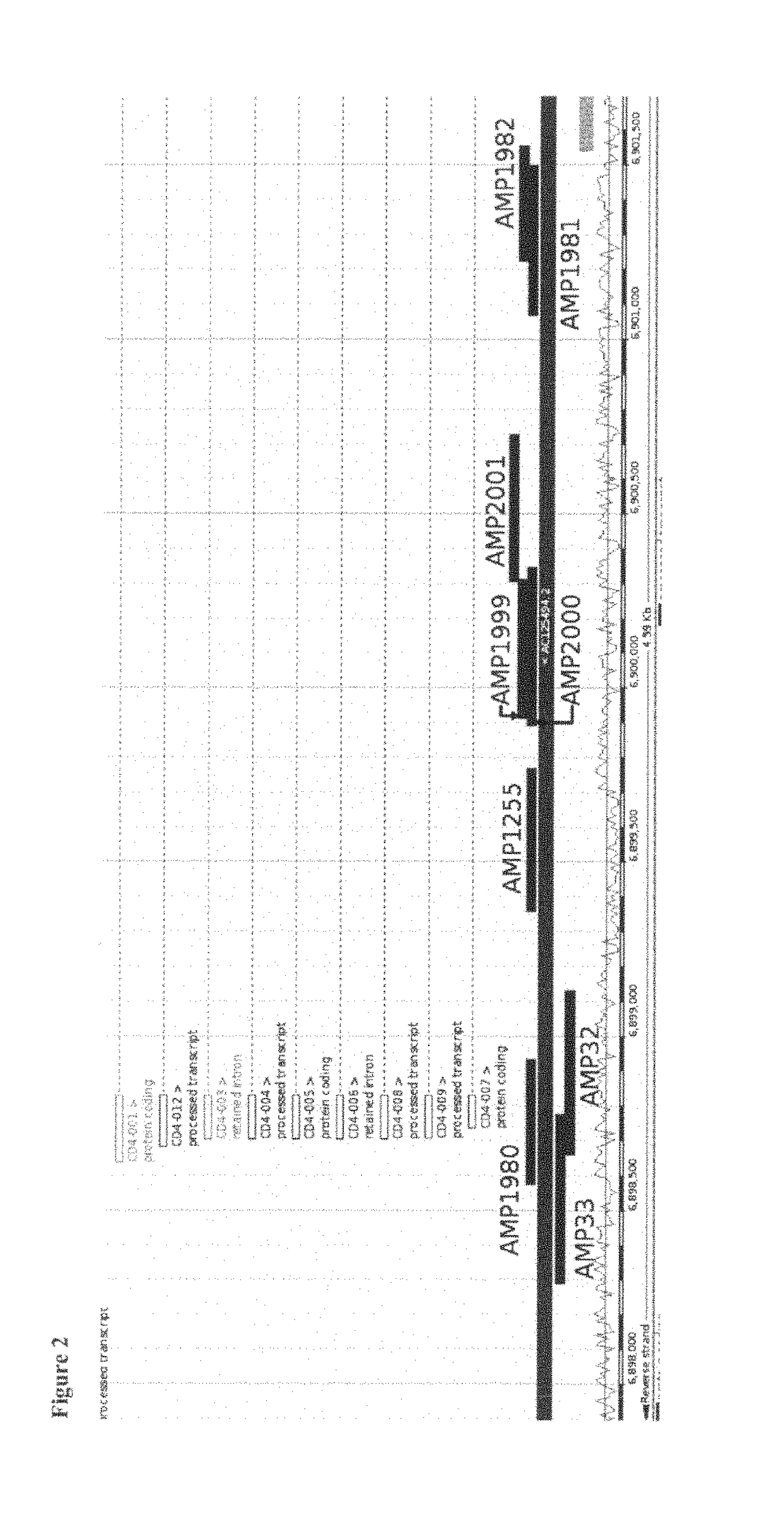
Epigenetic Markers for the Identification of Blood Sub-Cells of Type 1
InactiveUS20120107810A1Accurate analysisMore robustSugar derivativesMicrobiological testing/measurementSerum igeSerum samples
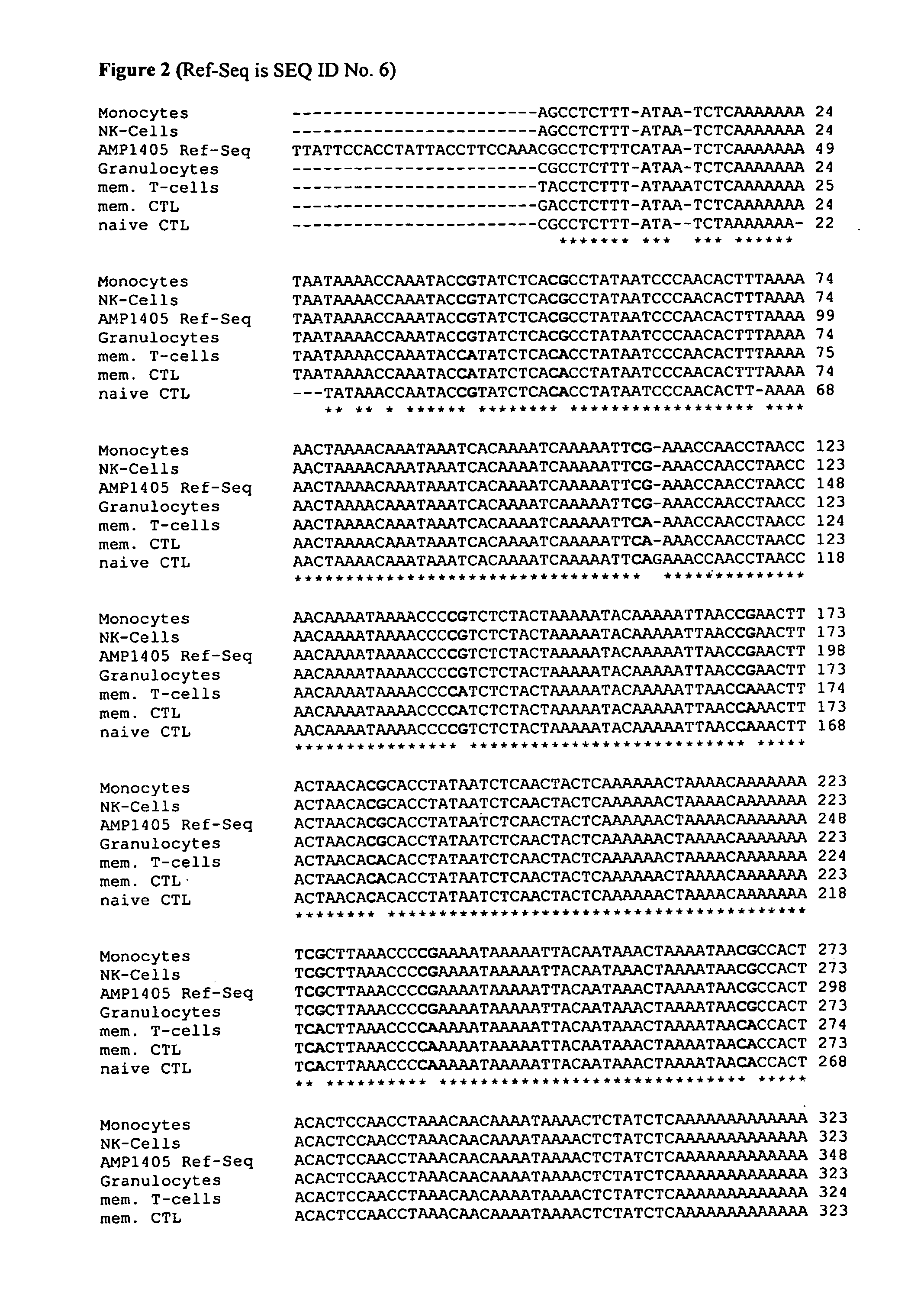
The present invention relates to a method, in particular an in vitro method, for identifying CD3CD4 positive T lymphocytes of a mammal, wherein said method comprises analysing the methylation status of at least one CpG position in the CD3a / b / c / d / g genes, in particular their “upstream” regulatory regions, and in particular the promoter and other conserved regions of the gene cd3, wherein a demethylation of at least one CpG in the analyzed sample to at least 90% is indicative for memory and naive CD4 or / and memory and / or native T lymphocytes. Furthermore, the present invention is directed at the use of DNA-methylation analysis of the genes CD3a / b / c / d for the detection and quality assurance and control of T lymphocytes. Furthermore, the present invention relates to a kit for performing the above methods as well as respective uses thereof. In a preferred embodiment, the present invention furthermore provides an improved method for analysing the methylation status of at least one CpG position in the gene CD3, allowing for a precise analysis even from sub-optimal quality samples, such as non-freshly obtained blood or serum samples.
Owner:EPIONTIS GMBH
DNA Methylation Changes Associated with Major Psychosis
Owner:CENT FOR ADDICTION & MENTAL HEALTH
Hypermethylation biomarkers for detection of cervical cancer
ActiveUS8859468B2Nucleotide librariesMicrobiological testing/measurementPotential biomarkersOncology
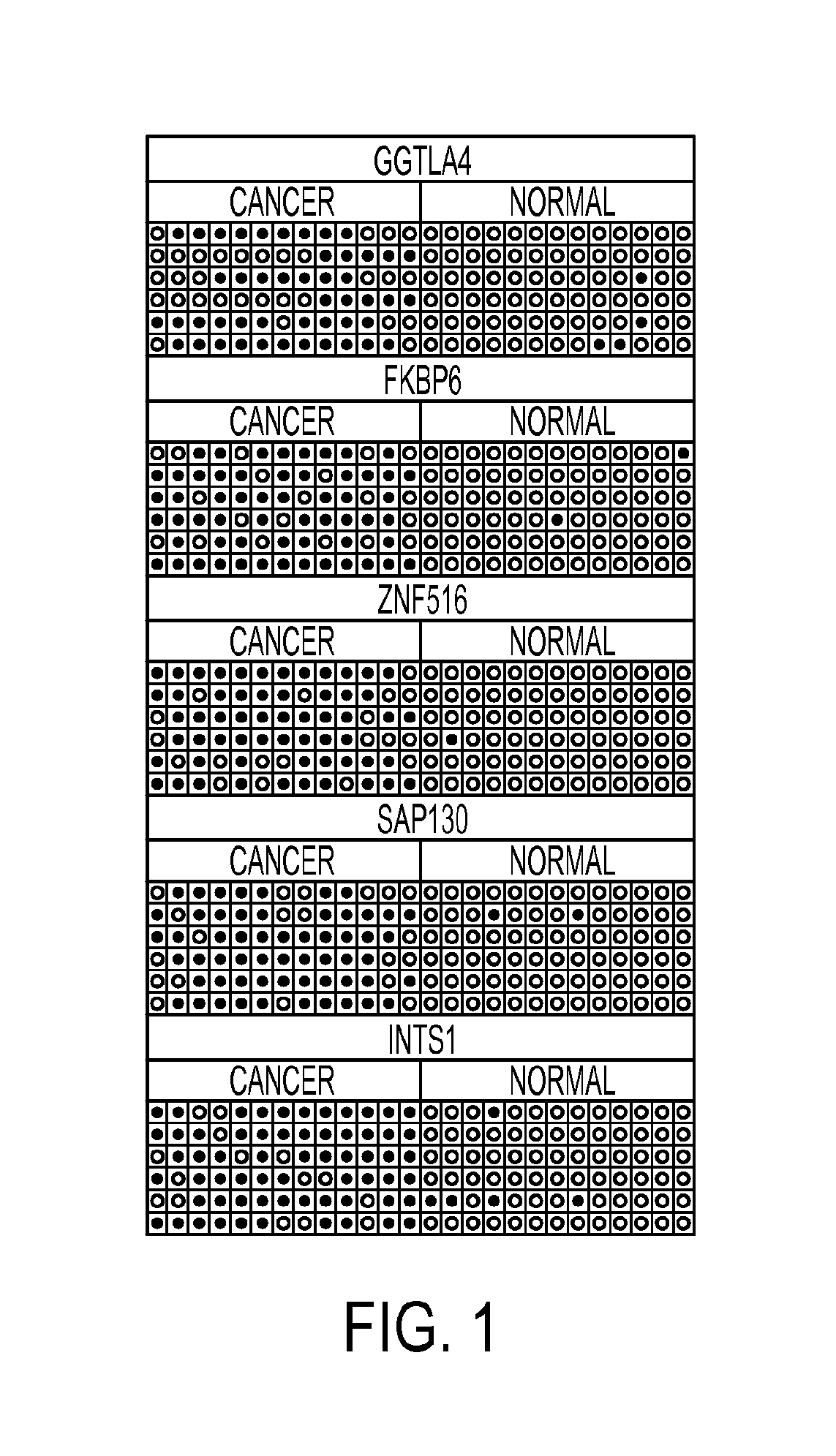
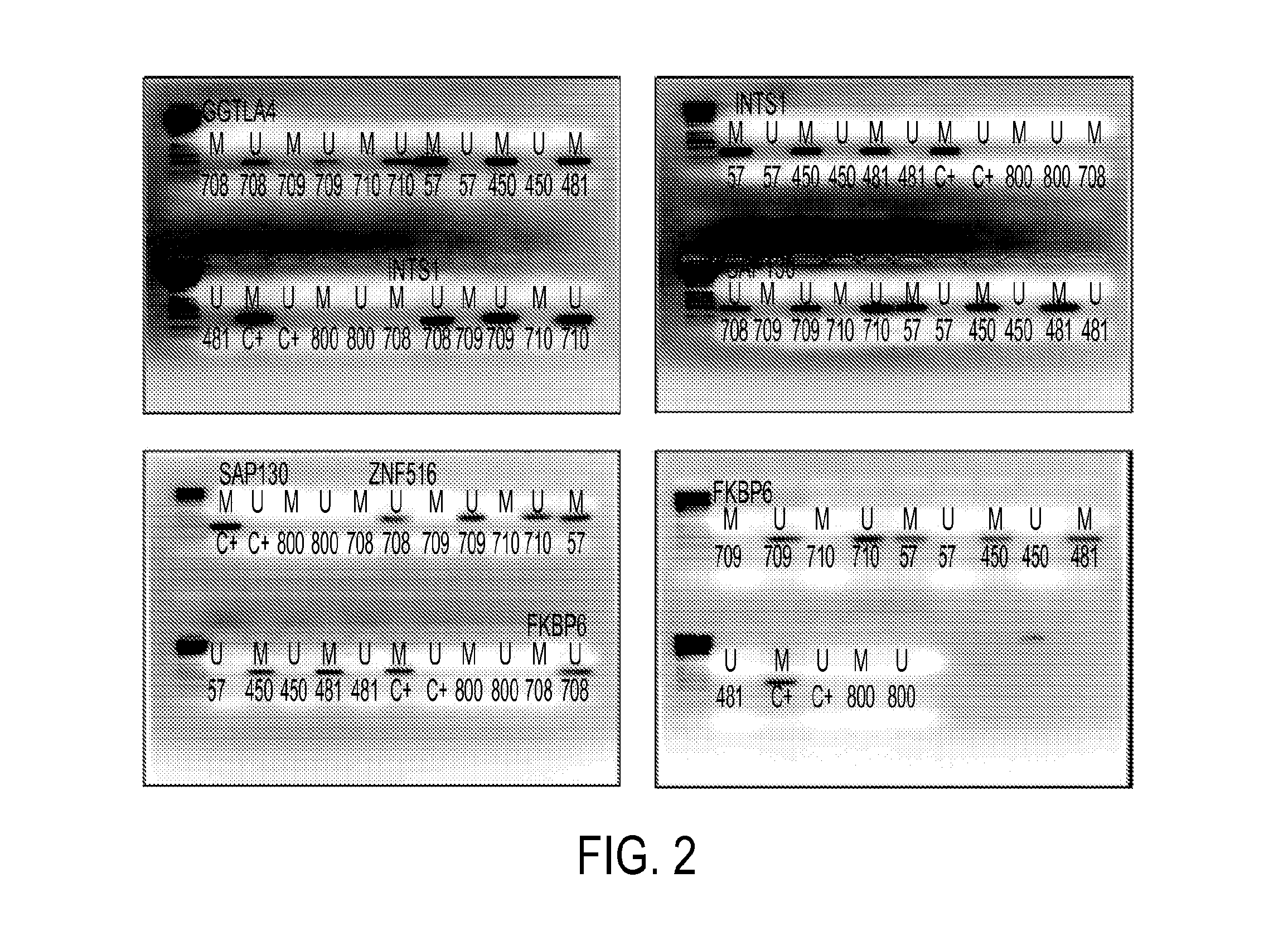
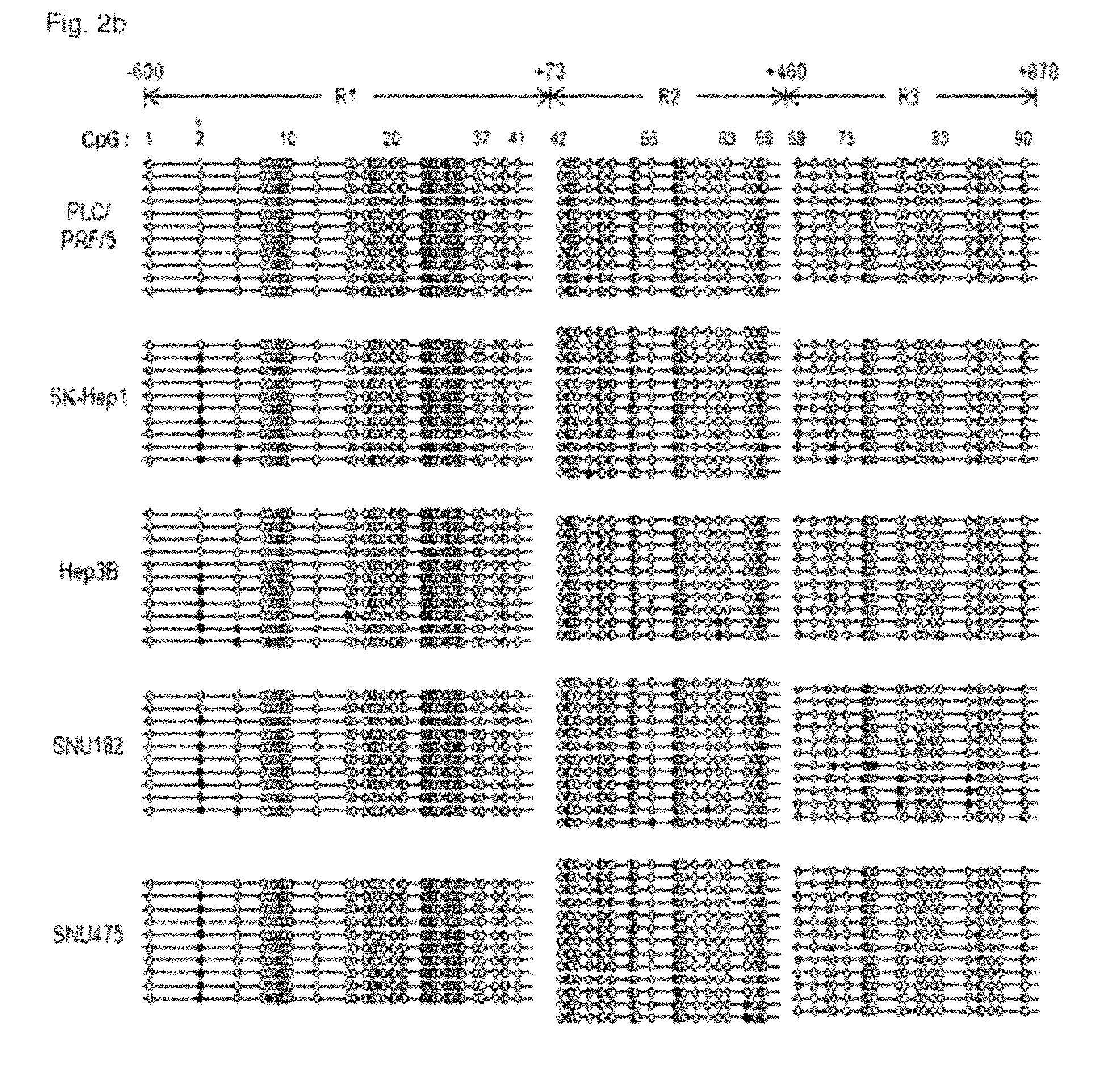
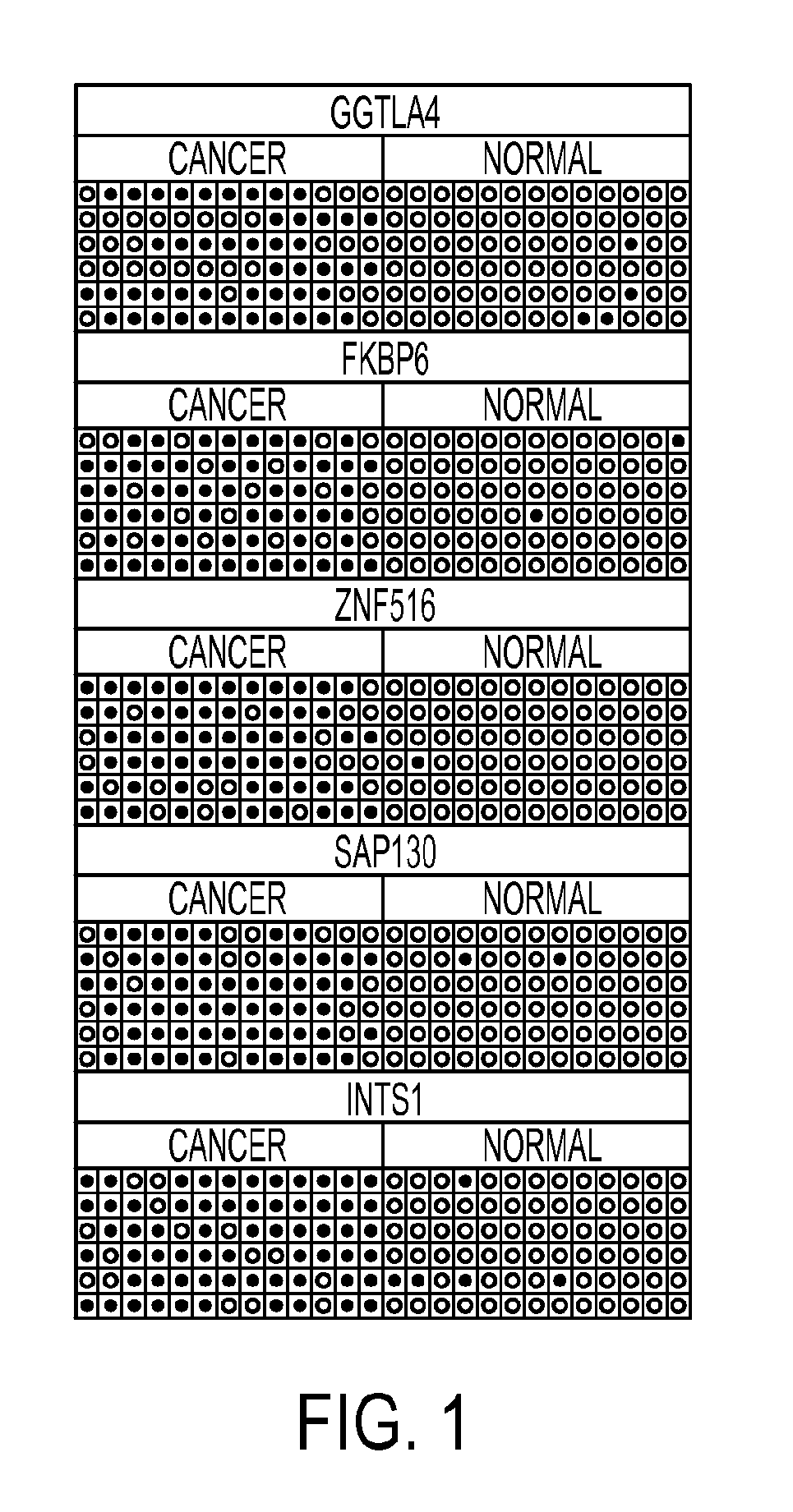
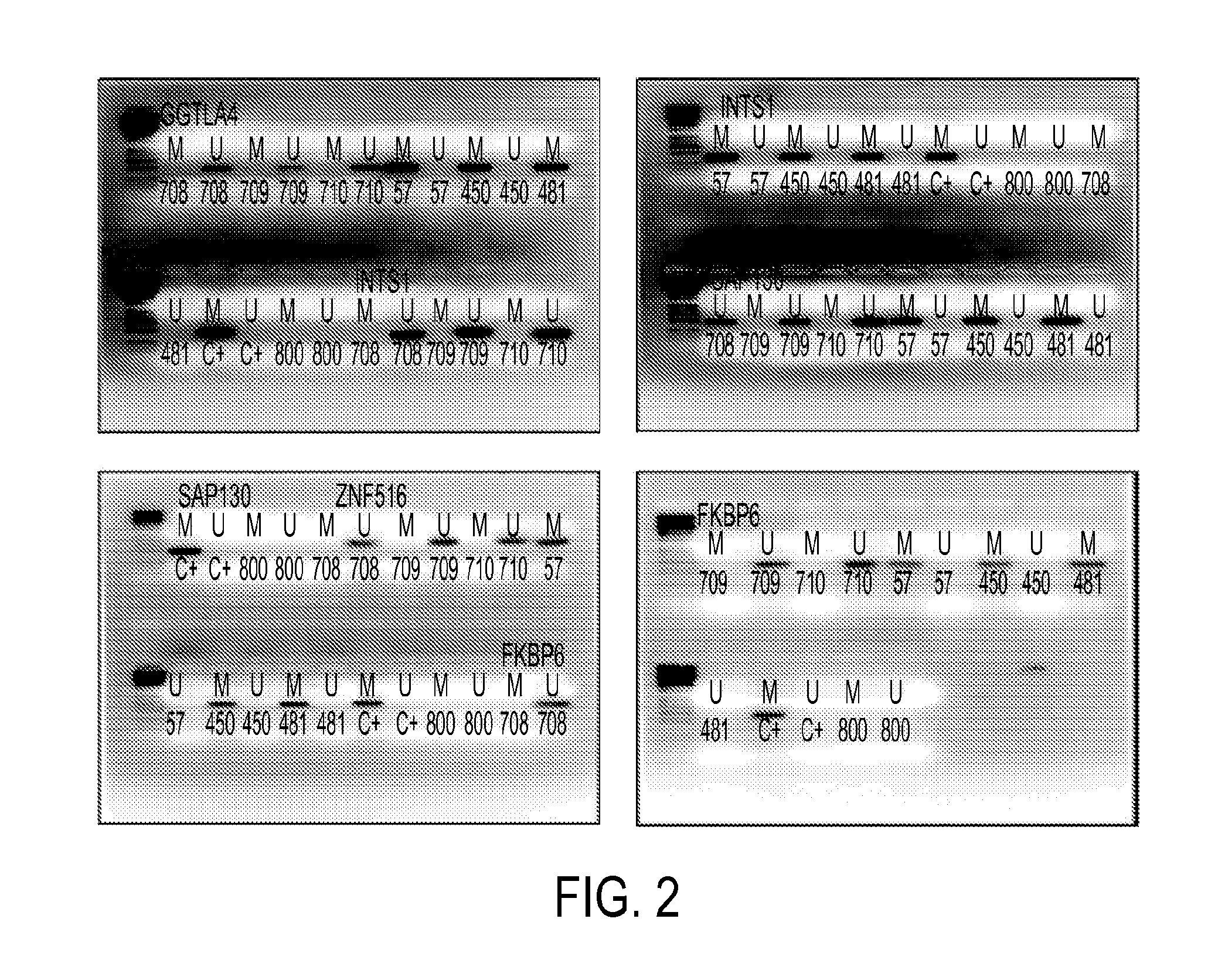
Pap smears and HPV infection tests do not distinguish between lesions that will progress to an invasive carcinoma and those that will not. We aimed to identify epigenetic biomarkers for diagnosis and progression monitoring of premalignant lesions in cervical cancer. Hypermethylated genes were identified as potential biomarkers after validation by MSP, including GGTLA4 and ZNF516. The methylation frequency for these two genes was higher in tumor: GGTLA4 (100%) and ZNF516 (96%); than in normal samples: GGTLA4 (12%) and ZNF516 (16%). The methylation status of GGTLA4 showed a progression in methylation frequency from normal samples to invasive carcinoma. The immunohistochemical expression was lower in tumor for both: GGTLA4 (50.8%) and ZNF516 (66.2%); than in normal samples: GGTLA4 (71.2%) and ZNF516 (88.1%) (p<0.05). In conclusion, we identified methylation biomarkers for the molecular screening and characterization of cervical cancer.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Methods and biomarkers for detection of gastrointestinal cancers
InactiveUS20140031257A1Nucleotide librariesMicrobiological testing/measurementGastrointestinal cancerBlood plasma
The present invention relates to methods and biomarkers (e.g., epigenetic biomarkers) for detection of gastrointestinal cancers (e.g., colorectal cancer, gastric cancer, pancreatic cancer, liver cancer, cancer of the gall bladder and / or bile ducts (e.g., cholangiocarcinoma)) in biological samples (e.g., tissue samples, stool samples, blood samples, plasma samples, cell samples, gall samples, bile samples, serum samples).
Owner:UNIV OSLO HF
DNA Methylation Changes Associated with Major Psychosis
The present invention provides a method of identifying one or more epigenetic markers associated with psychosis-associated diseases such as bipolar disease or schizophrenia, the method comprising a) obtaining a first group of samples comprising genomic DNA from a plurality of bipolar or schizophrenic subjects and a second group of samples comprising genomic DNA from a plurality of control subjects; b) performing DNA methylation analysis to determine methylation differences in one or more DNA regions between the first group and second group of samples, wherein a methylation difference in a DNA region is indicative of an epigenetic marker associated with bipolar disease or schizophrenia. The invention also provides one or more epigenetic markers associated with psychosis-associated diseases such as bipolar disease or schizophrenia.
Owner:CENT FOR ADDICTION & MENTAL HEALTH
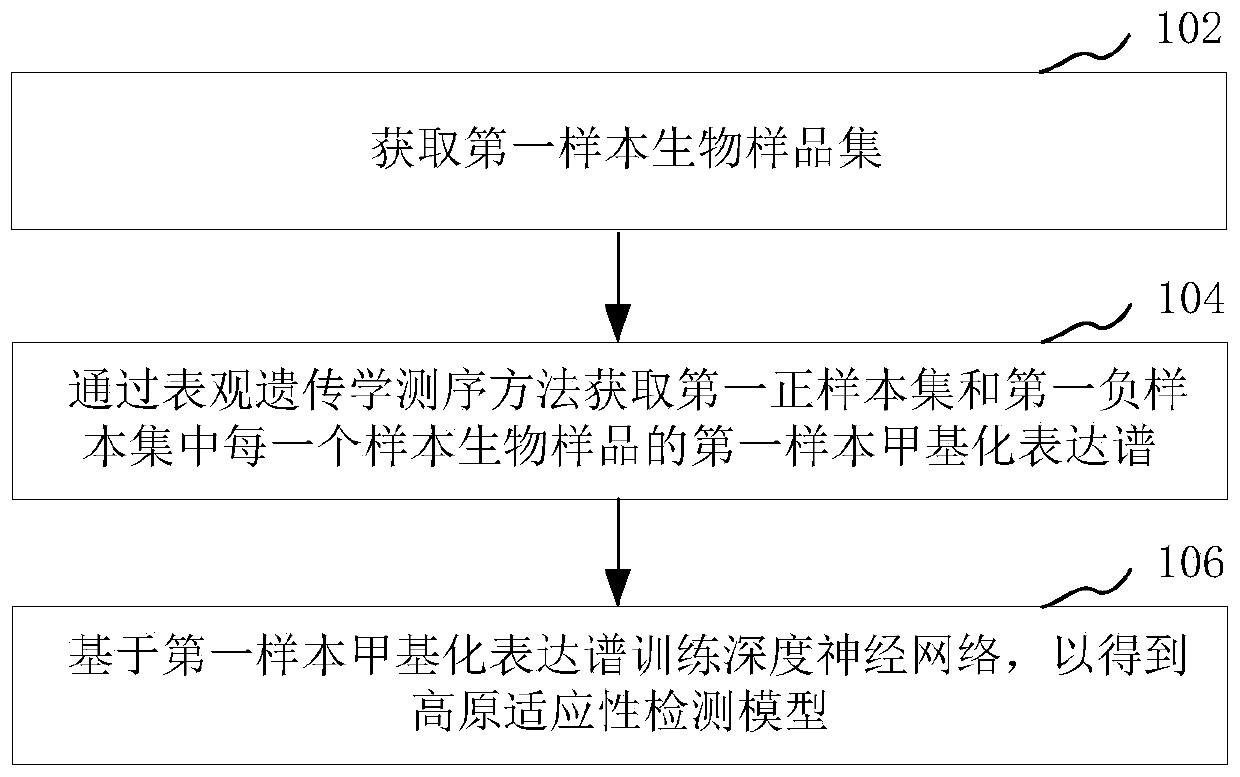
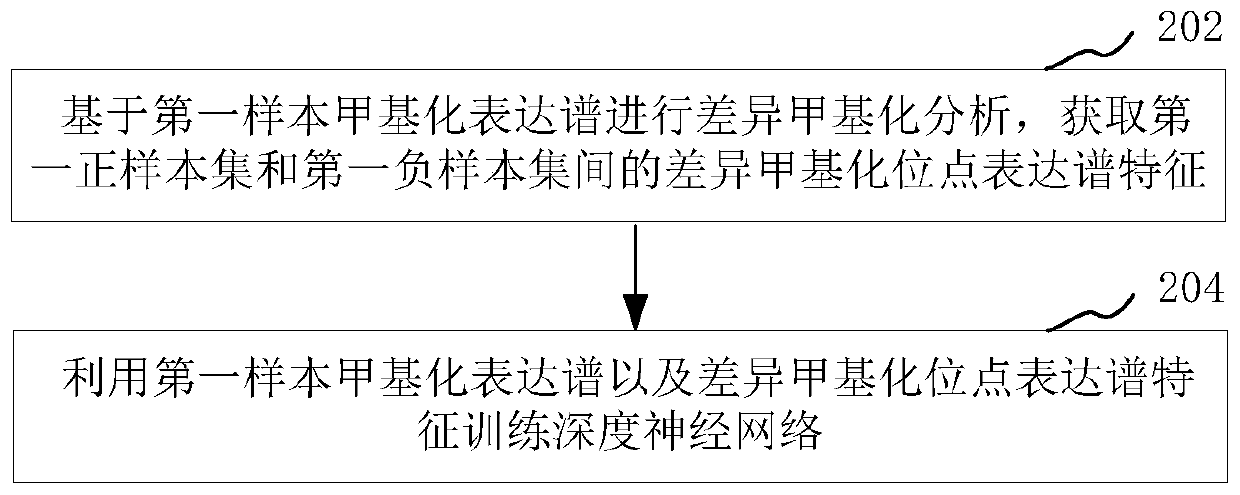
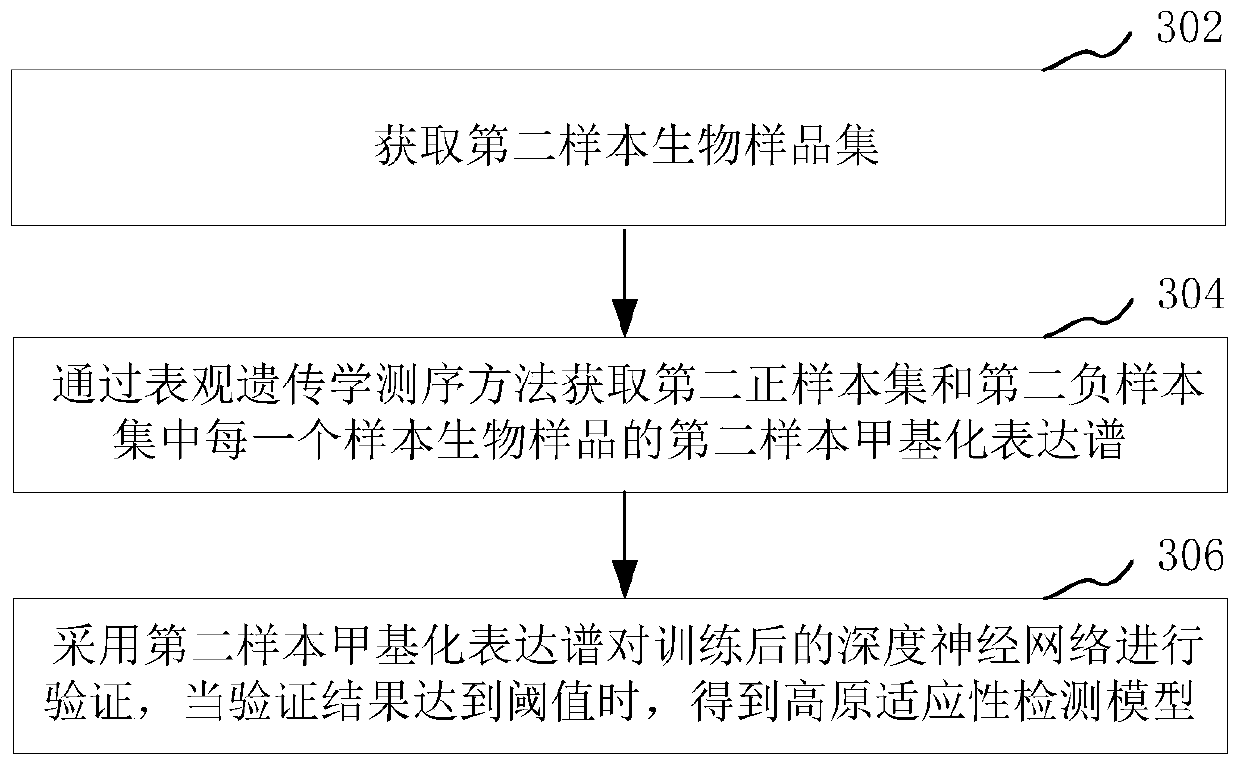
Plateau adaptability detection model training method and adaptability identification method and device
PendingCN111009290APrediction and prevention of high altitude hypoxic diseaseBiostatisticsProteomicsDiseaseData mining
The invention relates to a plateau adaptability detection model training method and an adaptability recognition method and device, and the model training method comprises the steps: obtaining a firstsample biological sample set, and obtaining a first sample methylation expression profile of each sample biological sample in the first sample biological sample set through an epigenetics sequencing method; and training a deep neural network based on the first sample methylation expression profile to obtain a plateau adaptability detection model. The method comprises the following steps: a deep neural network is trained through epigenetic markers of a first sample biological sample set to obtain a plateau adaptability detection model; the methylation expression profile of the biological sampleto be identified is identified by using the plateau adaptability detection model so as to obtain the plateau adaptability identification result of the biological sample to be identified, such that the important significance is provided for predicting and preventing plateau hypoxic diseases.
Owner:GENERAL HOSPITAL OF PLA
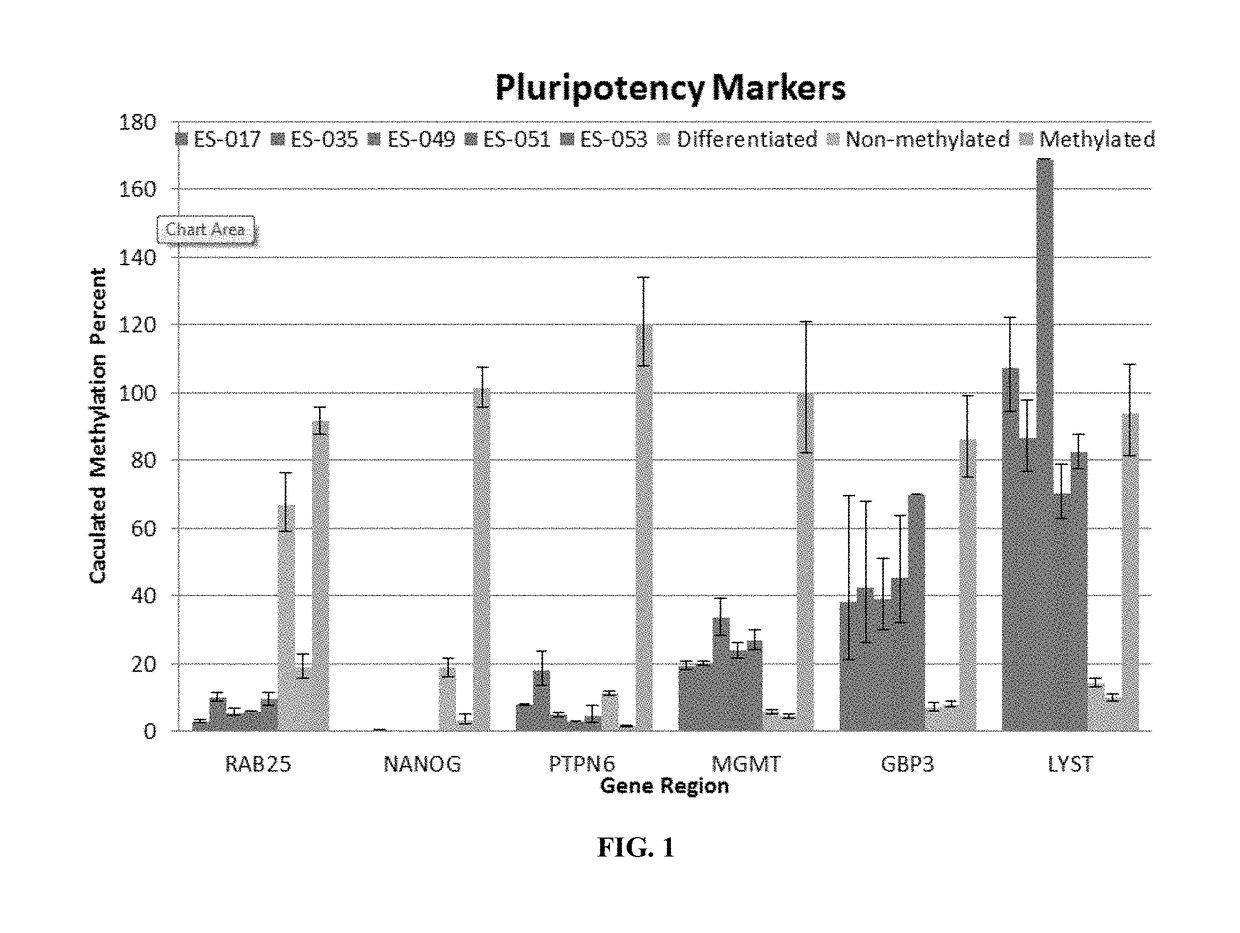
Epigenetic markers of pluripotency
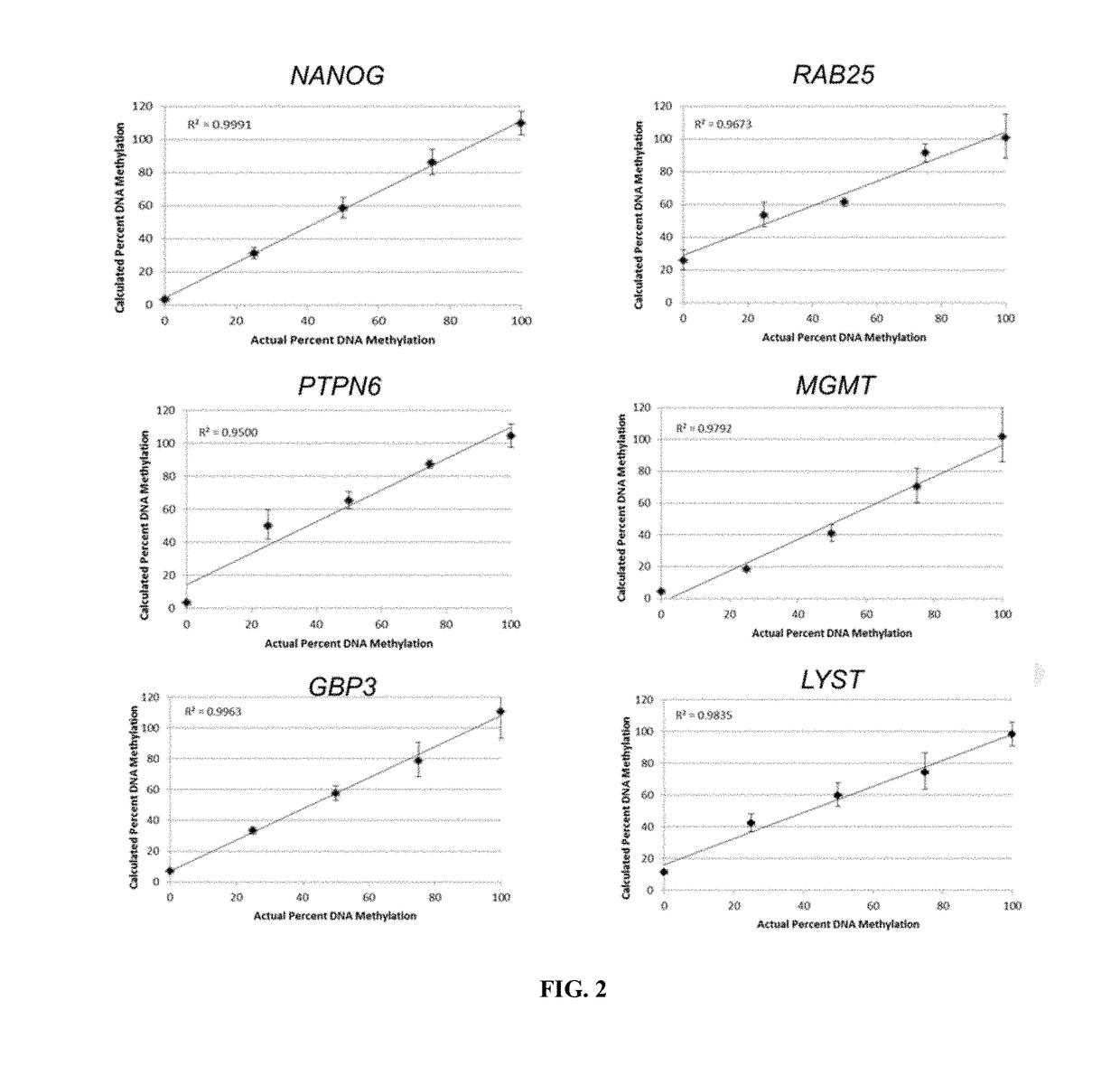
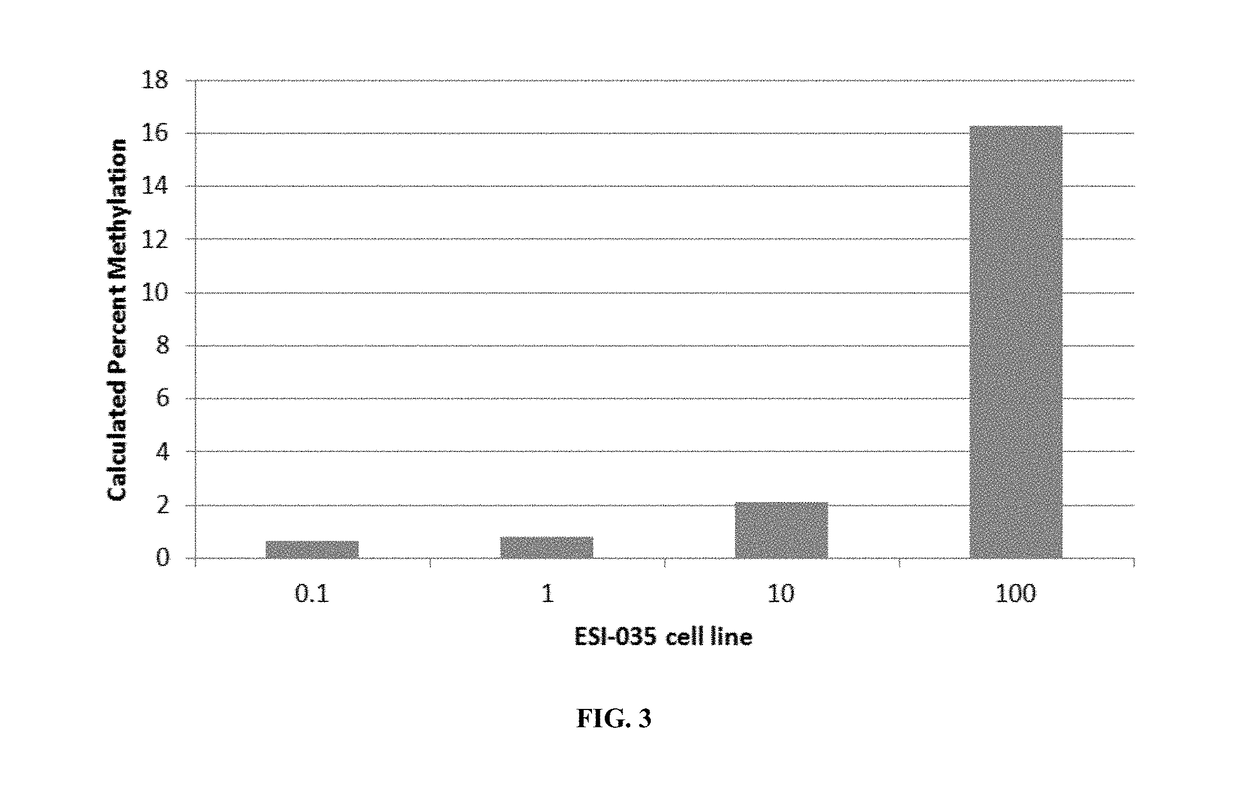
InactiveUS20170321274A1Accurate assessment of pluripotencyImprove the level ofMicrobiological testing/measurementEpigenetic ProfilePTPN6
Epigenetic methods for assessing pluripotentcy of a cell population, such as a stem cell culture are provided. For example, pluripotency can be assessed by determining DNA methylation status at the RAB25, NANOG, PTPN6, MGMT, GBP3 and / or LYST gene regions. Kits and reagents for testing cells are likewise provided.
Owner:ZYMO RES CORP
Compositions and Methods for the Identification and Use of Epigenetic Markers Useful in the Study of Normal and Abnormal Mammalian Gametogenesis
ActiveUS20110152122A1Low efficiencyPeptide/protein ingredientsLibrary screeningGametogenesisInfertility
The invention includes compositions comprising a S. cerevisiae yeast library, and methods of identifying an epigenetic marker for the diagnosis of infertility or a disorder associated with gametogenesis in an individual.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Epigenetic marker for the identification of CD3CD4 positive T lymphocytes
Owner:EPIONTIS GMBH
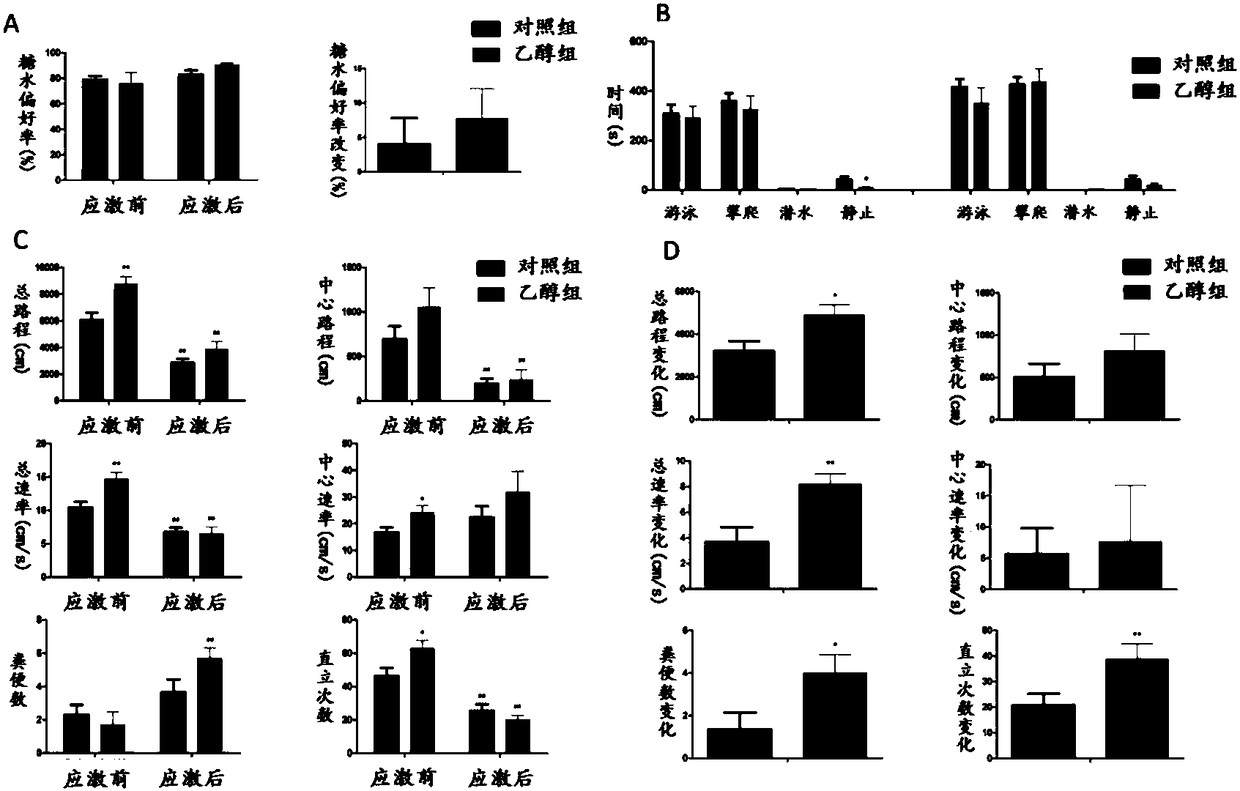
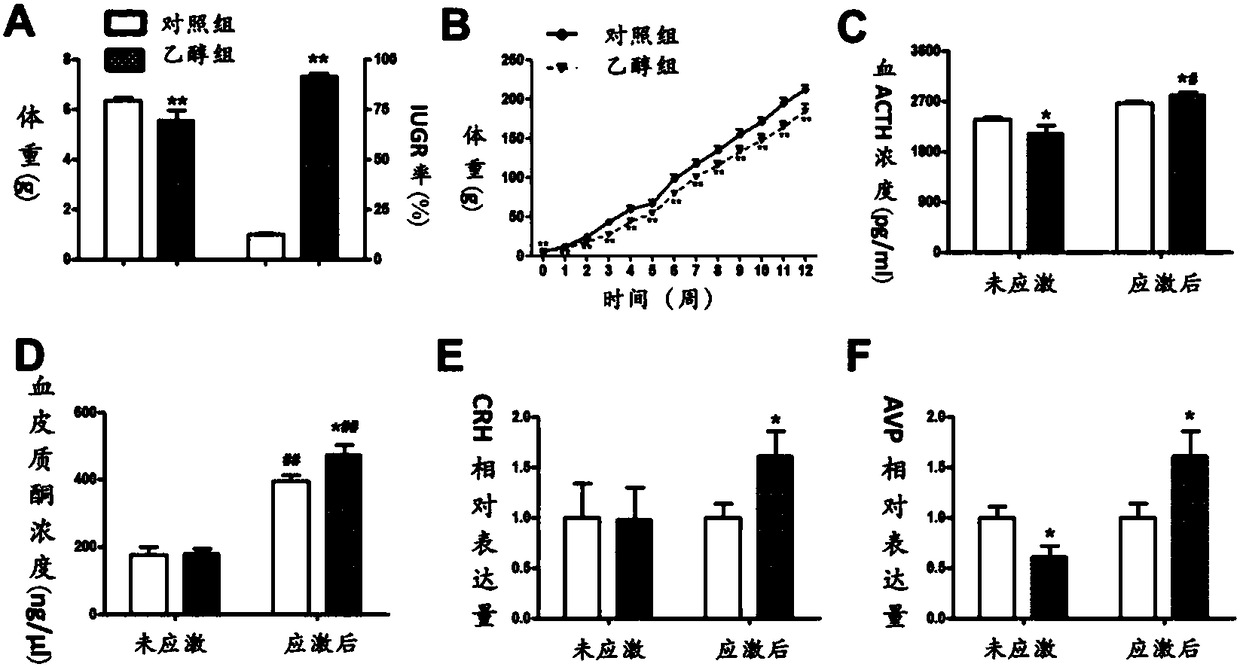
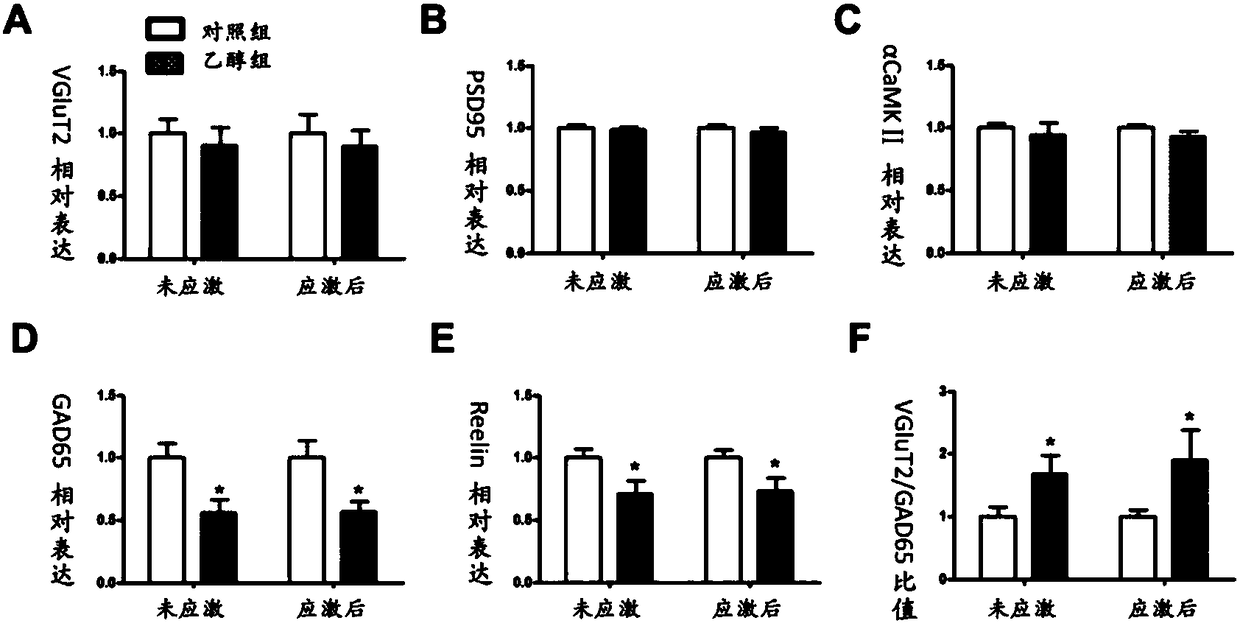
Application of GAD67 and epigenetic mark thereof which serve as early marker of fetal-original bipolar affective disorder susceptibility
ActiveCN108192967AQuick collectionQuick storageMicrobiological testing/measurementWhite blood cellEpigenetic Profile
The invention discloses application of GAD67 and an epigenetic mark thereof which serve as early markers of fetal-original bipolar affective disorder susceptibility, and belongs to the field of gene functions and application. Research results show that in hippocampi and white blood cells of fetal rats in a pregnancy ethanol exposure group, compared with a normal control group, the methylation level of a GAD67 promoter region is significantly reduced, and the gene expression of the GAD67 promoter region is improved; after growing up, the rats in the xenobiotic exposure group show high hypothalamic-pituitary-adrenal (HPA) axis stress sensibility and bipolar affective disorder susceptibility under the condition of chronic unpredictable stress. Therefore, risks that offspring have bipolar affective disorder after growing up can be judged by detecting the epigenetic modification and expression levels of the GAD67 genes in the white blood cells of fetal blood / umbilical cord blood. Research bases are provided for an early warning technology of the fetal-original bipolar affective disorder, and meanwhile, possibilities are provided for effectively treating the fetal-original bipolar affective disorder in early stages.
Owner:WUHAN UNIV
Epigenetic marker for the identification of cd3cd4 positive t lymphocytes
ActiveUS20150004602A1Sugar derivativesMicrobiological testing/measurementLymphocytic cellT lymphocyte
The present invention relates to a method, in particular an in vitro method, for identifying CD3CD4 positive T lymphocytes of a mammal, wherein said method comprises analyzing the bisulfite convertibility of at least one CpG position in the CD3+CD4+ T helper cell specific non-methylated bisulfite convertible region according to SEQ ID No. 1, wherein a bisulfite convertibility of at least one CpG position to at least 90%, preferably to at least 91% and more preferably to at least 92% and most preferred to at least 95% in said sample is indicative for a CD4+ T-lymphocyte cell, in particular a CD3+ CD4+ T-lymphocyte cell. The present invention further relates to analyzing the bisulfite convertibility of at least one CpG position in the genes FLJ00060, FLJ38379, PPP6C, CD226, ZBTB7B and TNFAIP8 that are capable of positively identifying CD4 expressing cells in whole blood and segregate between CD4 and CD8 positive CD3 positive cells. Furthermore, the present invention relates to a kit for performing the above methods as well as respective uses thereof.
Owner:EPIONTIS GMBH
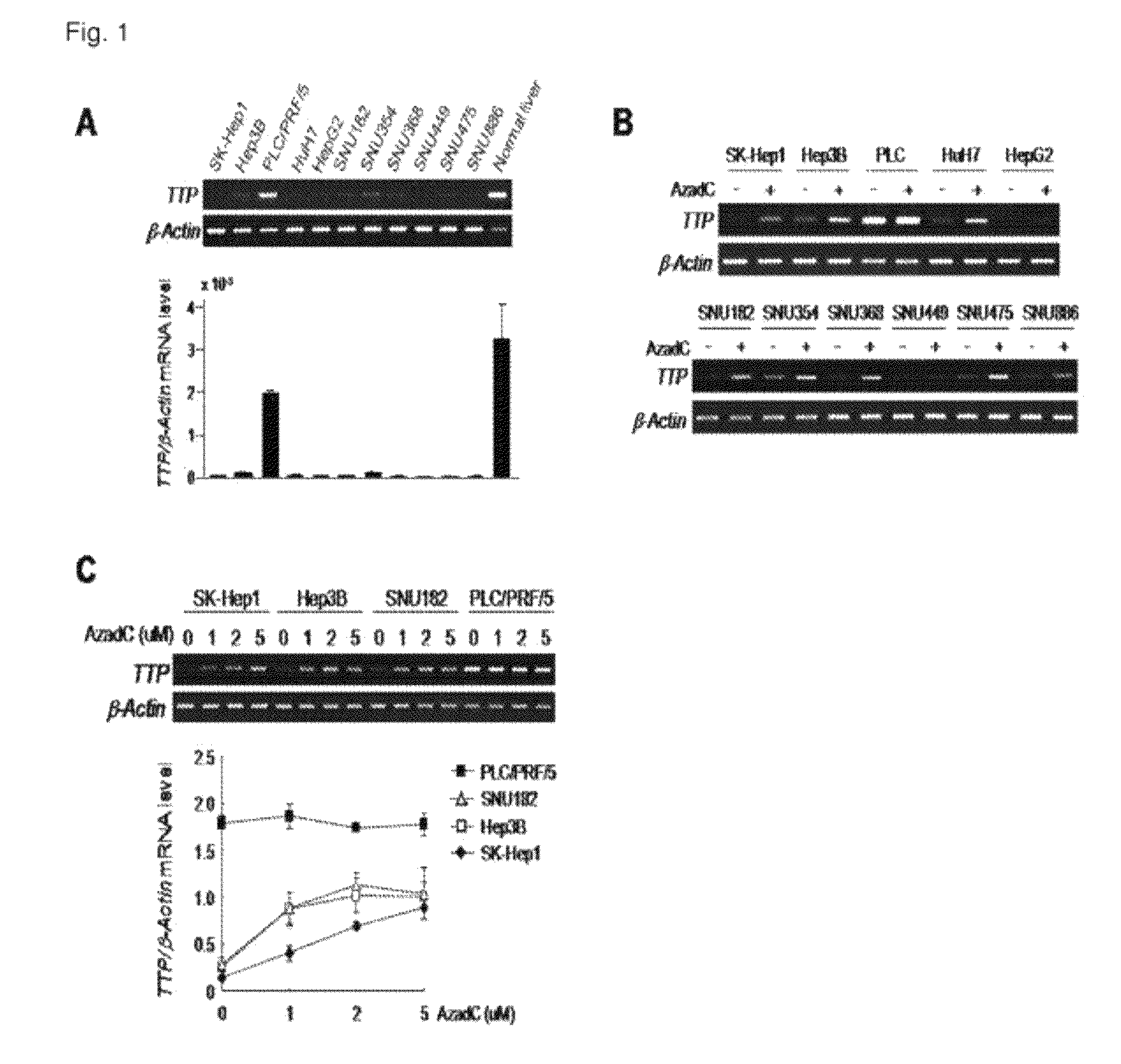
Method for diagnosis/prognosis of cancers using an epigenetic marker consisting of a specific single CpG site in TTP promoter and treatment of cancers by regulating its epigenetic status
The present invention relates to a method for the diagnosis and prognosis of cancers using an epigenetic marker consisting of a specific single CpG site in TTP (Tristetraprolin) promoter and treatment of cancers by regulating its epigenetic status. Particularly, the present invention relates to a method for the diagnosis and prognosis of liver cancer by measuring specific methylation of C, the 32nd residue of the nucleic acid sequence represented by SEQ. ID. NO. 41, and a method for treatment of cancer by regulating the same. The present invention can be effectively used for the diagnosis and / or treatment of liver cancer characterized by TTP down-regulation and methylation of C, the 32nd residue of the nucleic acid sequence represented by SEQ. ID. NO. 41. The present invention can be further applied for the diagnosis and treatment of other cancers or inflammatory diseases that are characterized by TTP down-regulation and methylation of C, the 32nd residue of the nucleic acid sequence represented by SEQ. ID. NO. 41.
Owner:KOREA RES INST OF BIOSCI & BIOTECH
Hypermethylation Biomarkers for Detection of Cervical Cancer
ActiveUS20130109584A1Nucleotide librariesMicrobiological testing/measurementParanasal Sinus CarcinomaEpigenetic Profile
Pap smears and HPV infection tests do not distinguish between lesions that will progress to an invasive carcinoma and those that will not. We aimed to identify epigenetic biomarkers for diagnosis and progression monitoring of premalignant lesions in cervical cancer. Hypermethylated genes were identified as potential biomarkers after validation by MSP, including GGTLA4 and ZNF516. The methylation frequency for these two genes was higher in tumor: GGTLA4 (100%) and ZNF516 (96%); than in normal samples: GGTLA4 (12%) and ZNF516 (16%). The methylation status of GGTLA4 showed a progression in methylation frequency from normal samples to invasive carcinoma. The immunohistochemical expression was lower in tumor for both: GGTLA4 (50.8%) and ZNF516 (66.2%); than in normal samples: GGTLA4 (71.2%) and ZNF516 (88.1%) (p<0.05). In conclusion, we identified methylation biomarkers for the molecular screening and characterization of cervical cancer.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Epigenetic markers for detection of autism spectrum disorders
InactiveUS20140349977A1Increased and decreased level of methylationBiocideTetracycline active ingredientsMedicineAutism spectrum disorder
Methods for determining if a patient has, or is at risk of having, and autism spectrum disorder by detecting epigenetic changes in the genome of the patient. For example, a method can comprise determining the methylation status of one or more genes in a blood sample.
Owner:ZYMO RES CORP
Methods and biomarkers for detection of bladder cancer
The invention relates to methods and biomarkers (e.g., epigenetic biomarkers) for detection of bladder cancer in biological samples (e.g., tissue samples, urine samples, urine sediments). In some embodiments, methods and biomarkers of the present invention find use in discriminating between bladder cancer, prostate cancer and renal epithelial tumors.
Owner:UNIV OSLO HF
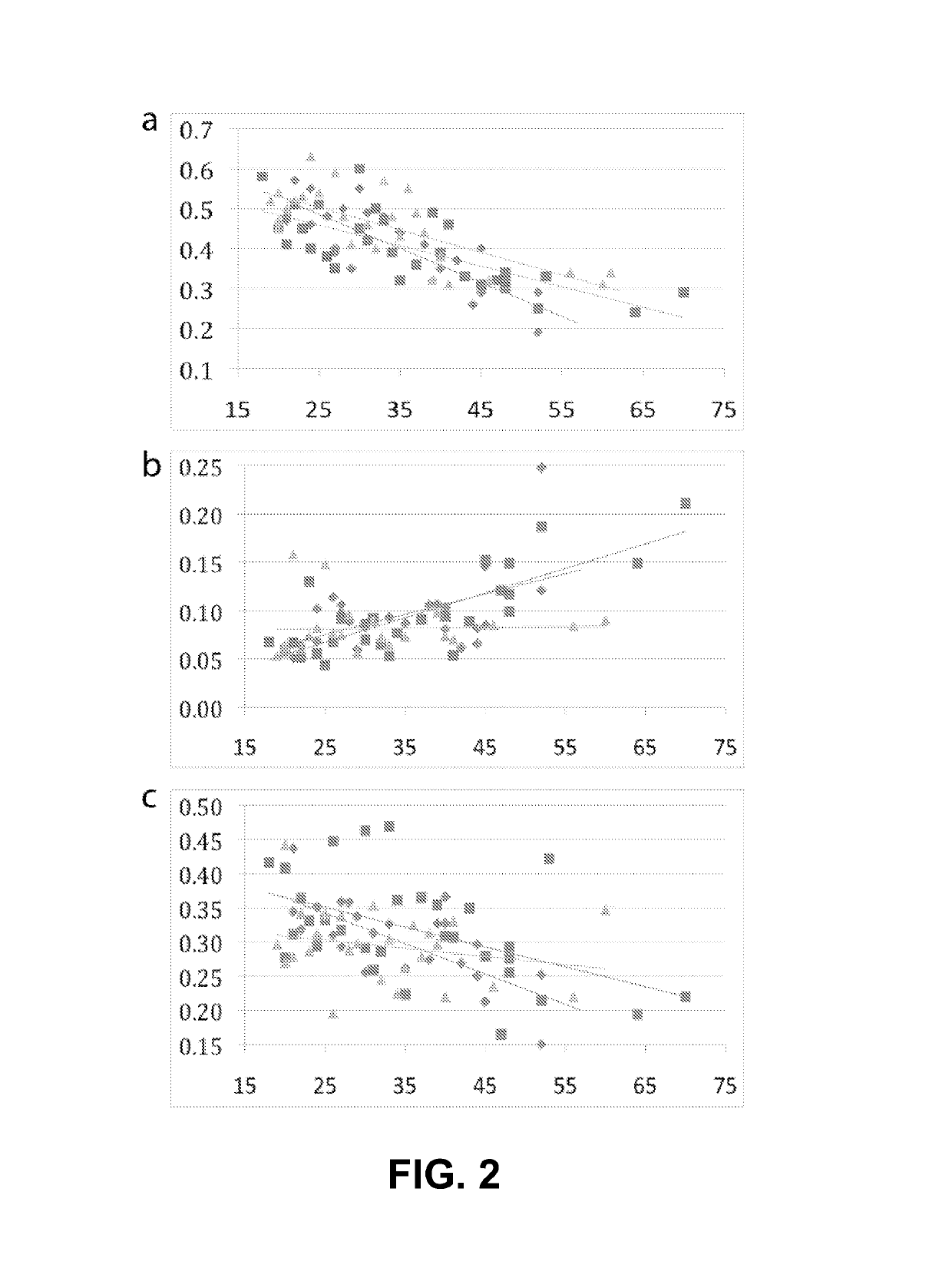
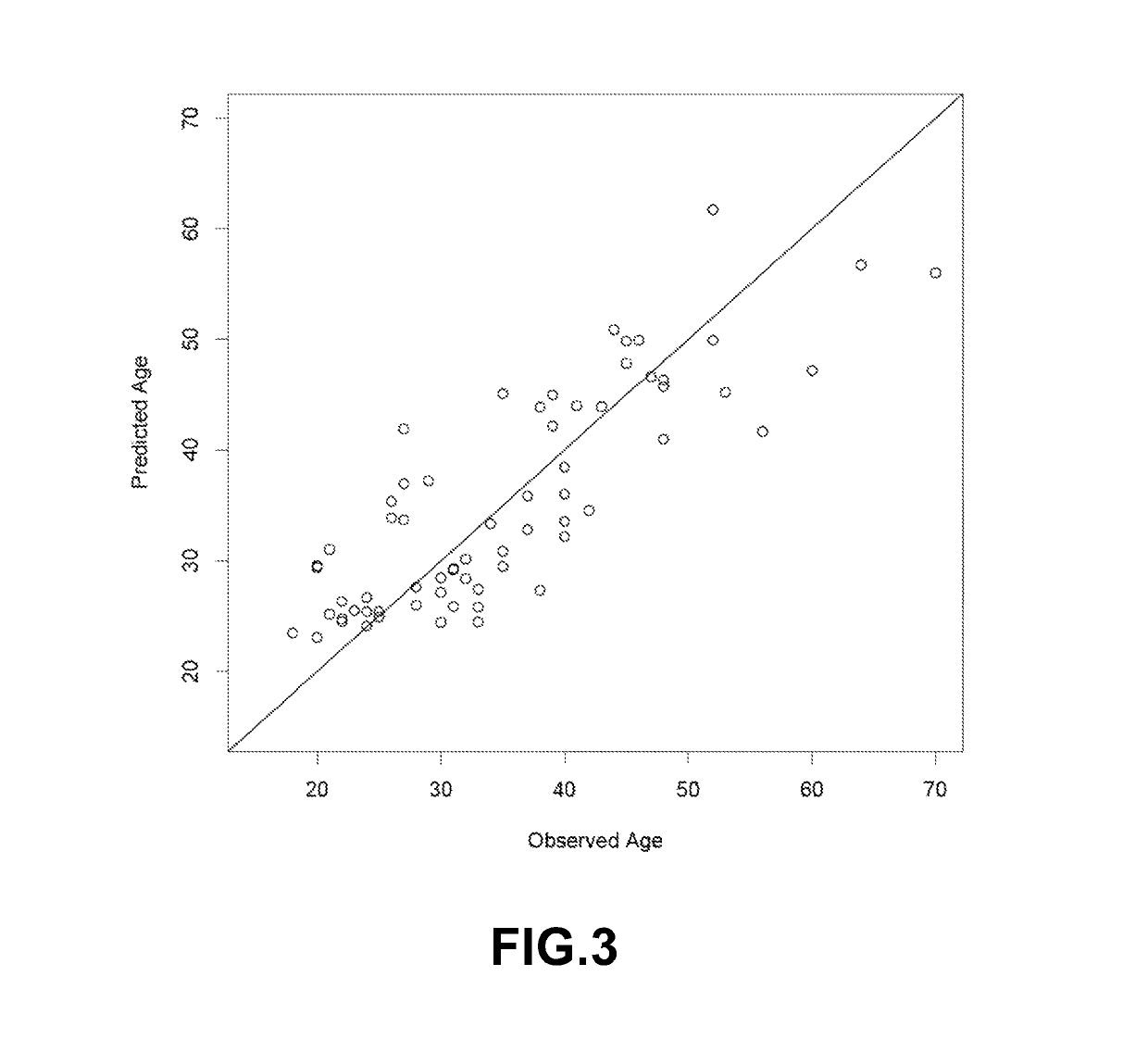
Method to estimate age of individual based on epigenetic markers in biological sample
The invention provides methods and materials that use observation of DNA characteristics to obtain information relating to the age of individuals. The instant disclosure identifies 88 sites in or near 80 genes for which the degree of cytosine methylation in epithelial and / or white blood cells is significantly correlated with age. In illustrative embodiments of the invention, cytosine methylation patterns the promoters of the EDARADD, TOMILI, and NPTX2 genes are used to predict the age of an individual with a high degree of accuracy.
Owner:RGT UNIV OF CALIFORNIA
Classification of epigenetic biomarkers and/or DNA conformational superstructures via use of atomically thin nanopores
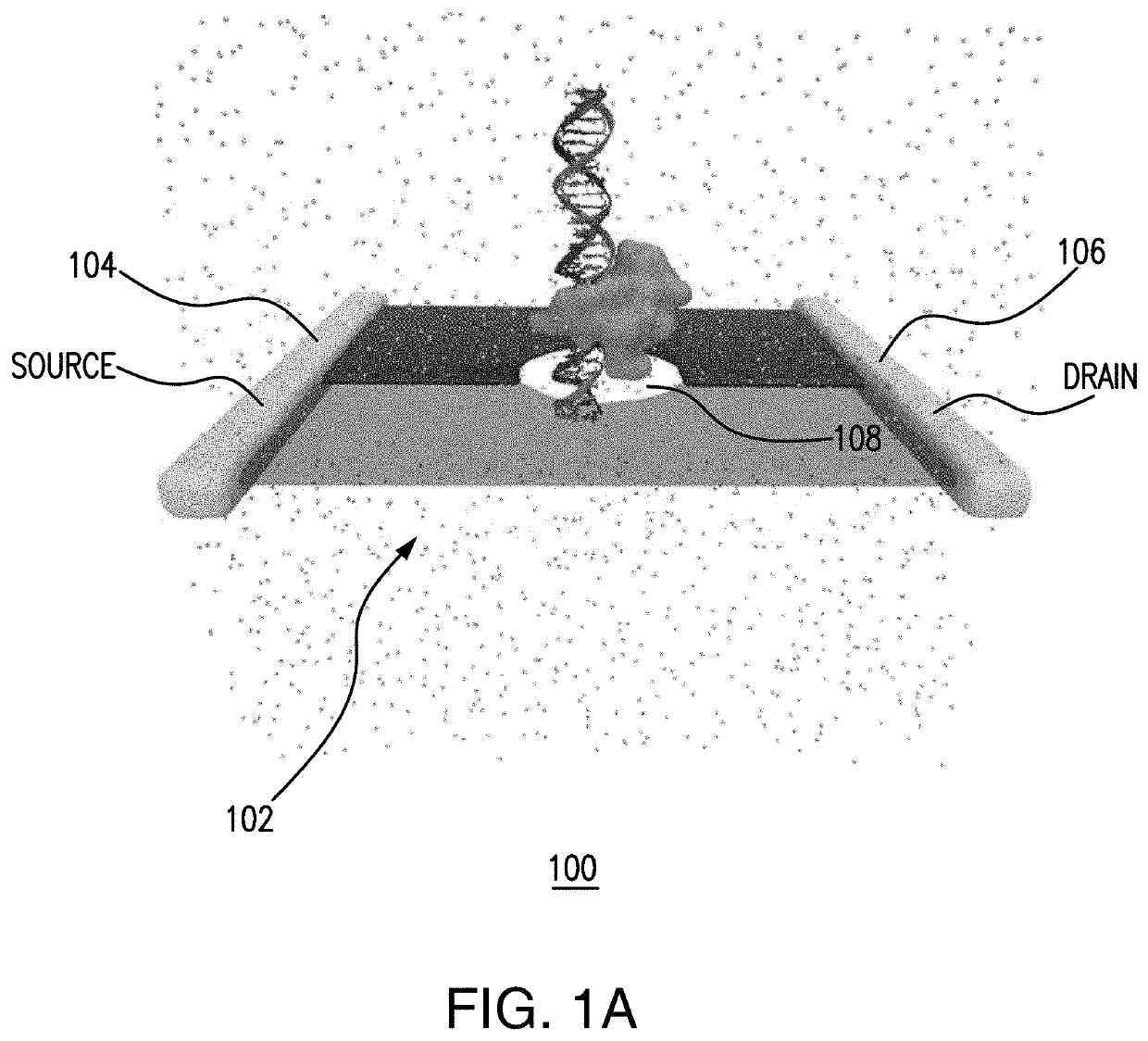
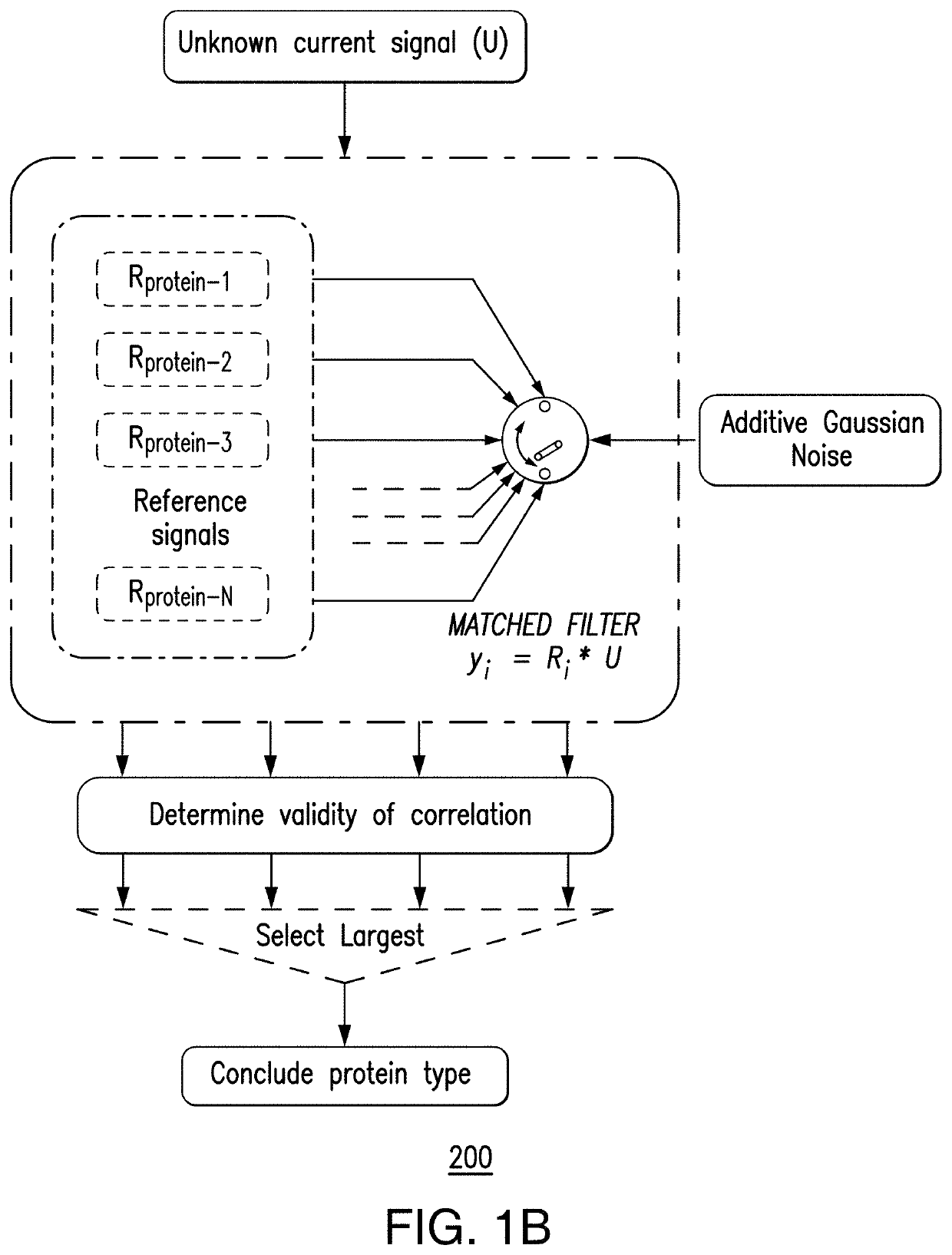
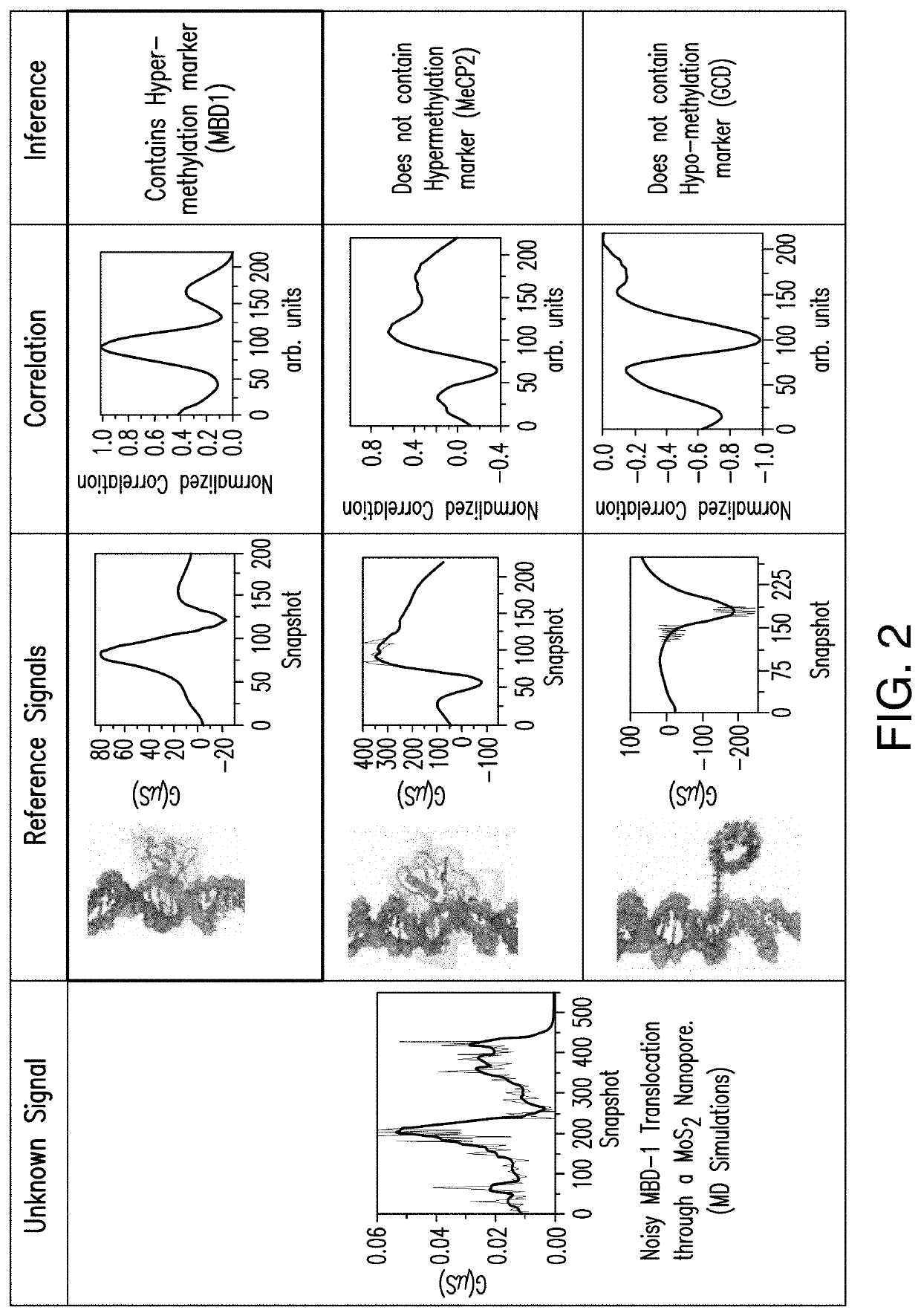
PendingUS20200333290A1Microbiological testing/measurementMaterial analysis by electric/magnetic meansEpigenetic ProfileHandling system
Aspects of the subject disclosure may include, for example, an apparatus comprising: a membrane with a first side, a second side, and a pore that extends through the membrane; a processing system including a processor; and a memory that stores executable instructions that, when executed by the processing system, facilitate performance of operations, the operations comprising: electronic sensing of an electrical characteristic associated with a translocation of a test DNA through the pore, resulting in a sensed electrical characteristic; comparing the sensed electrical characteristic with a plurality of reference electrical characteristics, wherein each of the plurality of reference electrical characteristics is associated with a respective one of a plurality of reference features, and wherein the comparing results in a comparison result; and determining, based upon the comparison result, with which of the plurality of reference features a feature of the test DNA corresponds. Additional embodiments are disclosed.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
Epigenetic Markers for the Identification of Blood Sub-cells of Type 1
InactiveUS20170233807A1Accurate analysisSimple methodMicrobiological testing/measurementMammalDna methylation profiling
The present invention relates to a method, in particular an in vitro method, for identifying CD3CD4 positive T lymphocytes of a mammal, wherein said method comprises analysing the methylation status of at least one CpG position in the CD3a / b / c / d / g genes, in particular their “upstream” regulatory regions, and in particular the promoter and other conserved regions of the gene cd3, wherein a demethylation of at least one CpG in the analyzed sample to at least 90% is indicative for memory and naive CD4 or / and memory and / or naïve T lymphocytes. Furthermore, the present invention is directed at the use of DNA-methylation analysis of the genes CD3a / b / c / d for the detection and quality assurance and control of T lymphocytes. Furthermore, the present invention relates to a kit for performing the above methods as well as respective uses thereof. In a preferred embodiment, the present invention furthermore provides an improved method for analysing the methylation status of at least one CpG position in the gene CD3, allowing for a precise analysis even from sub-optimal quality samples, such as non-freshly obtained blood or serum samples.
Owner:EPIONTIS GMBH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com