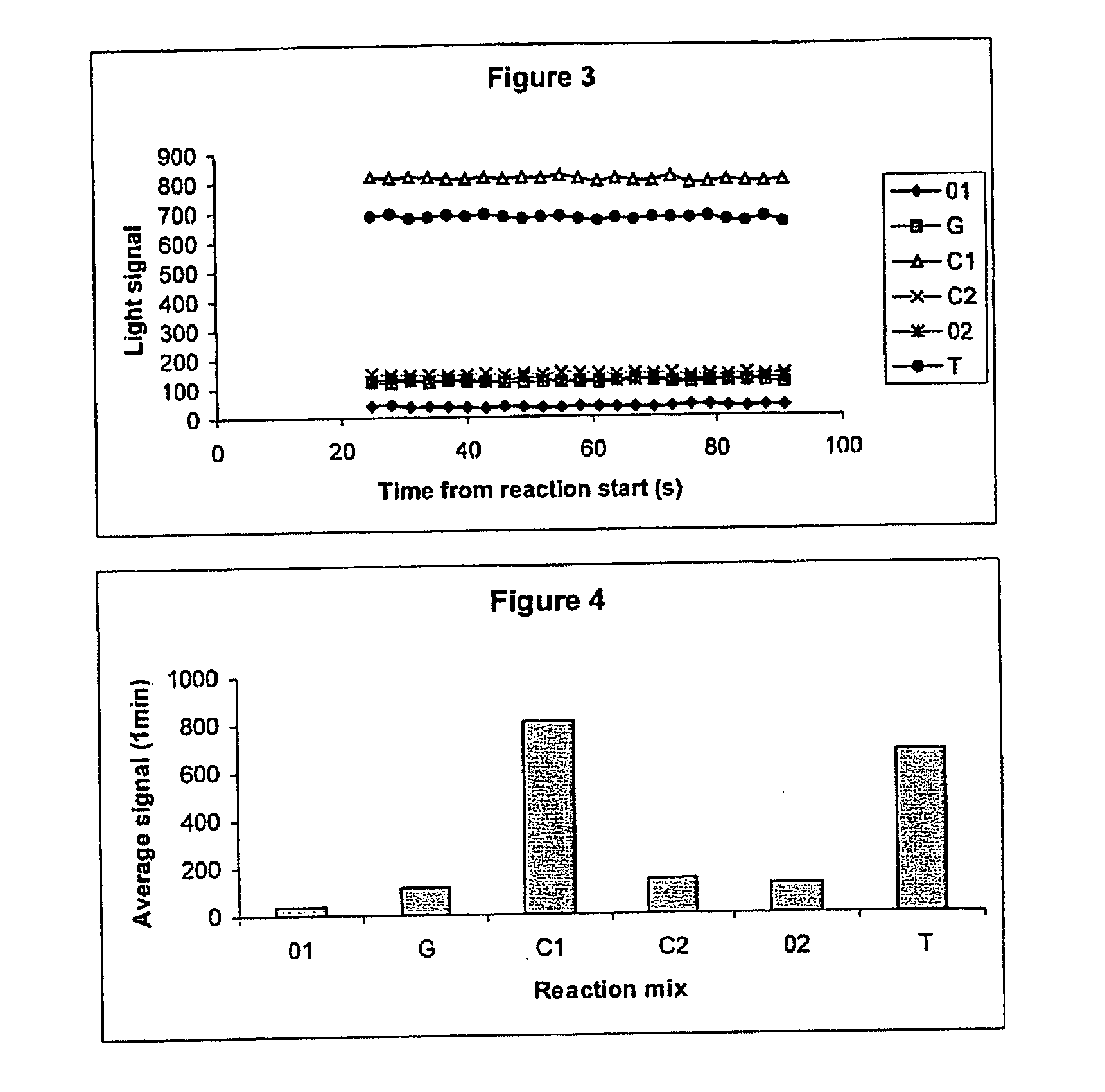
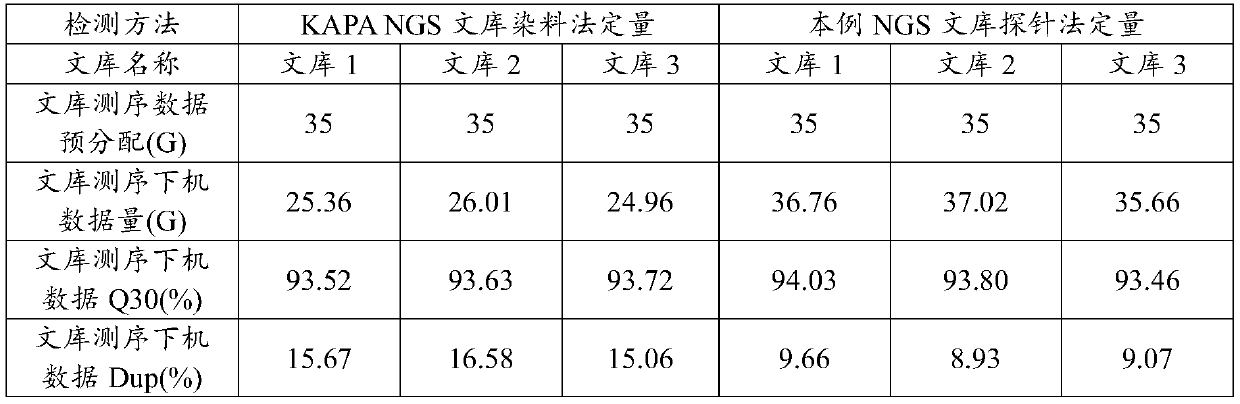
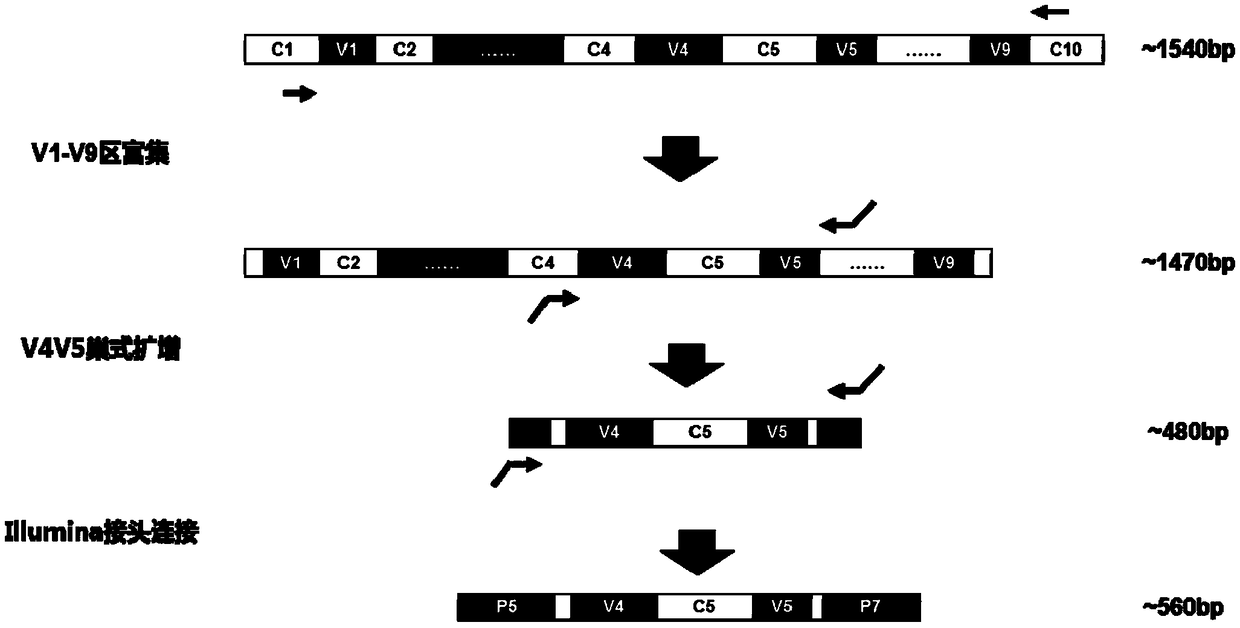
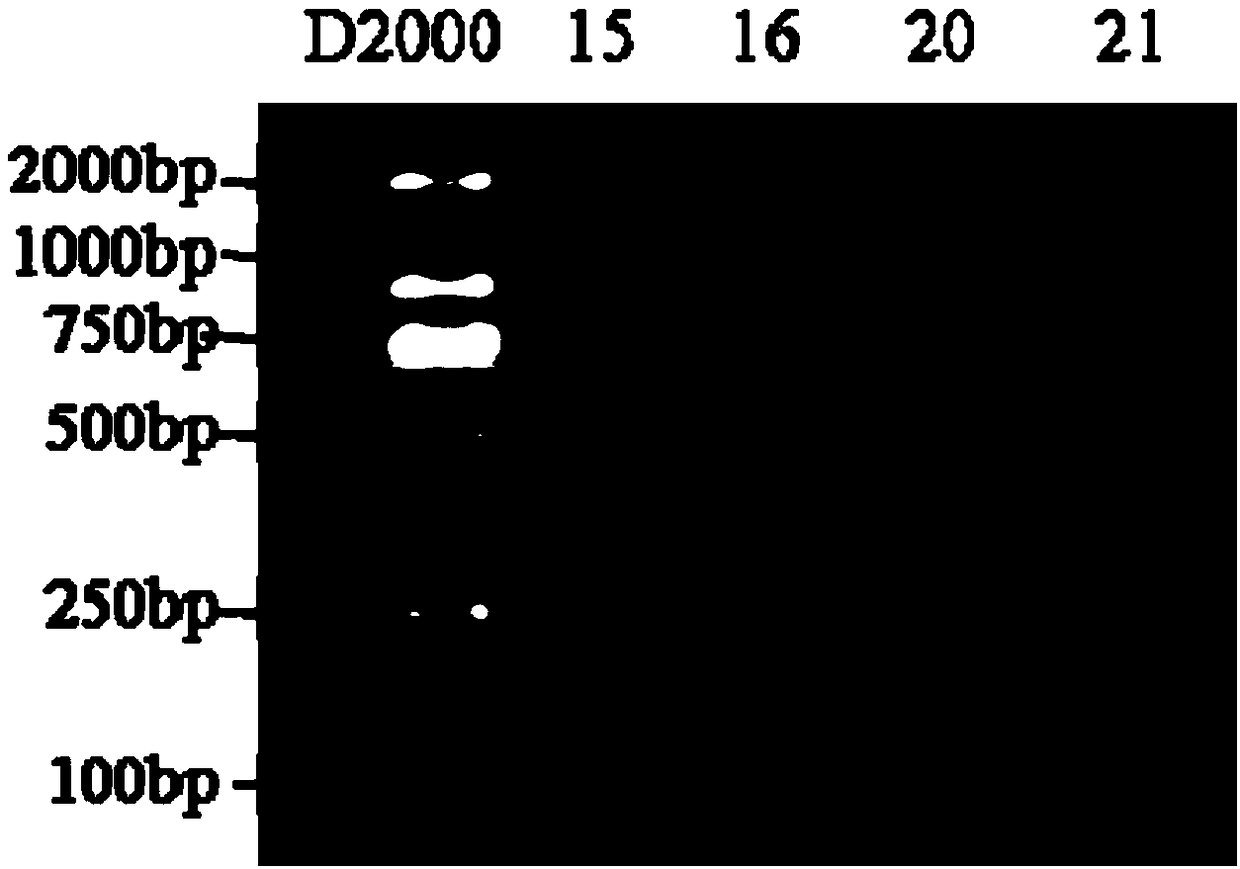
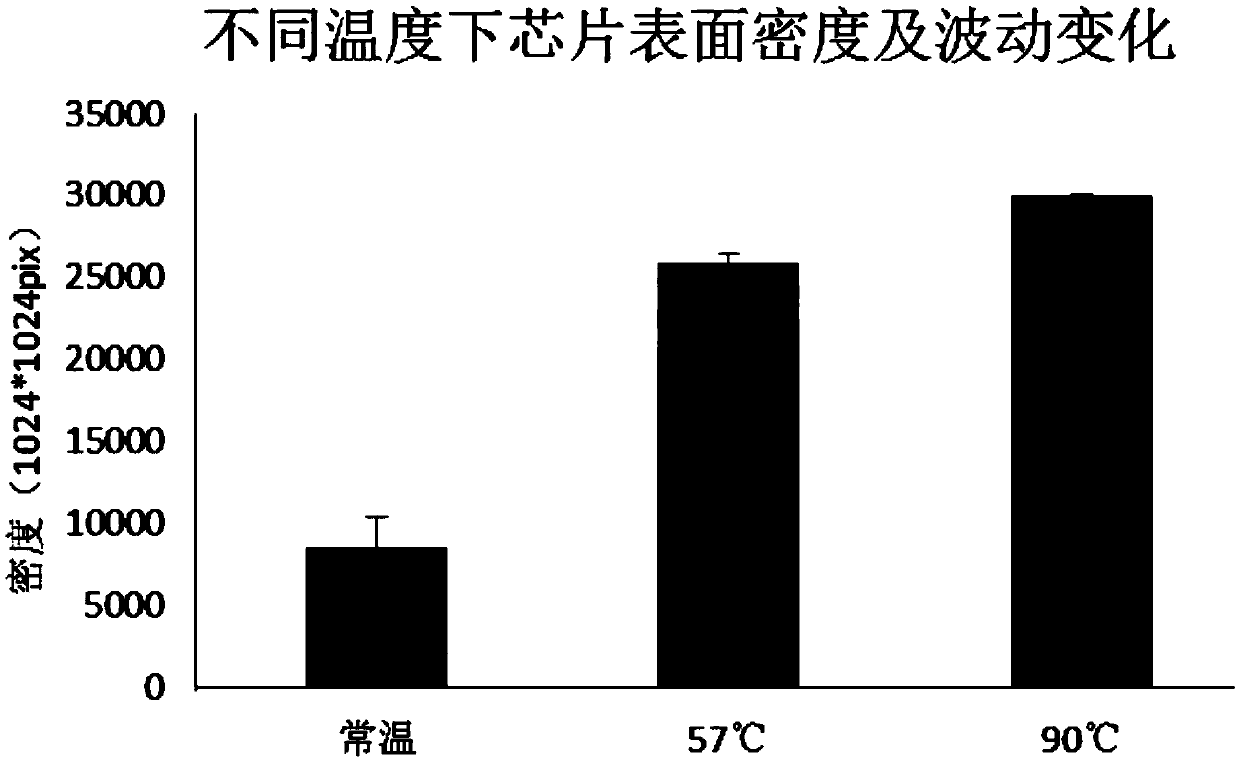
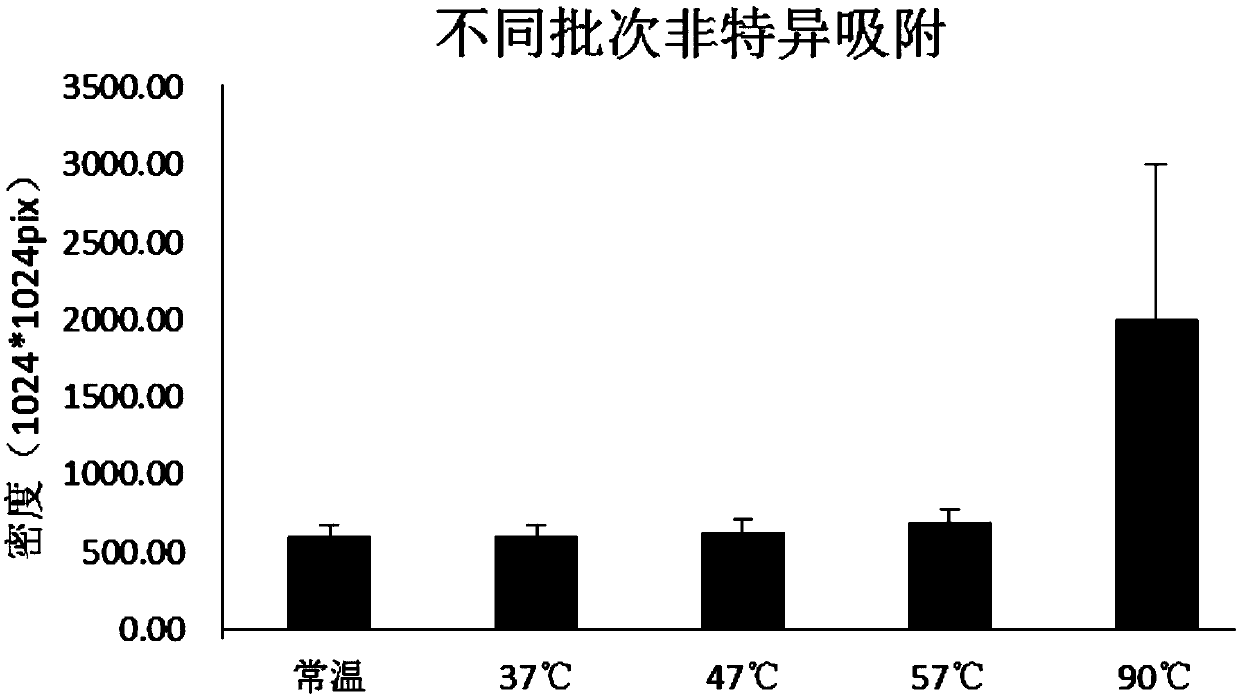
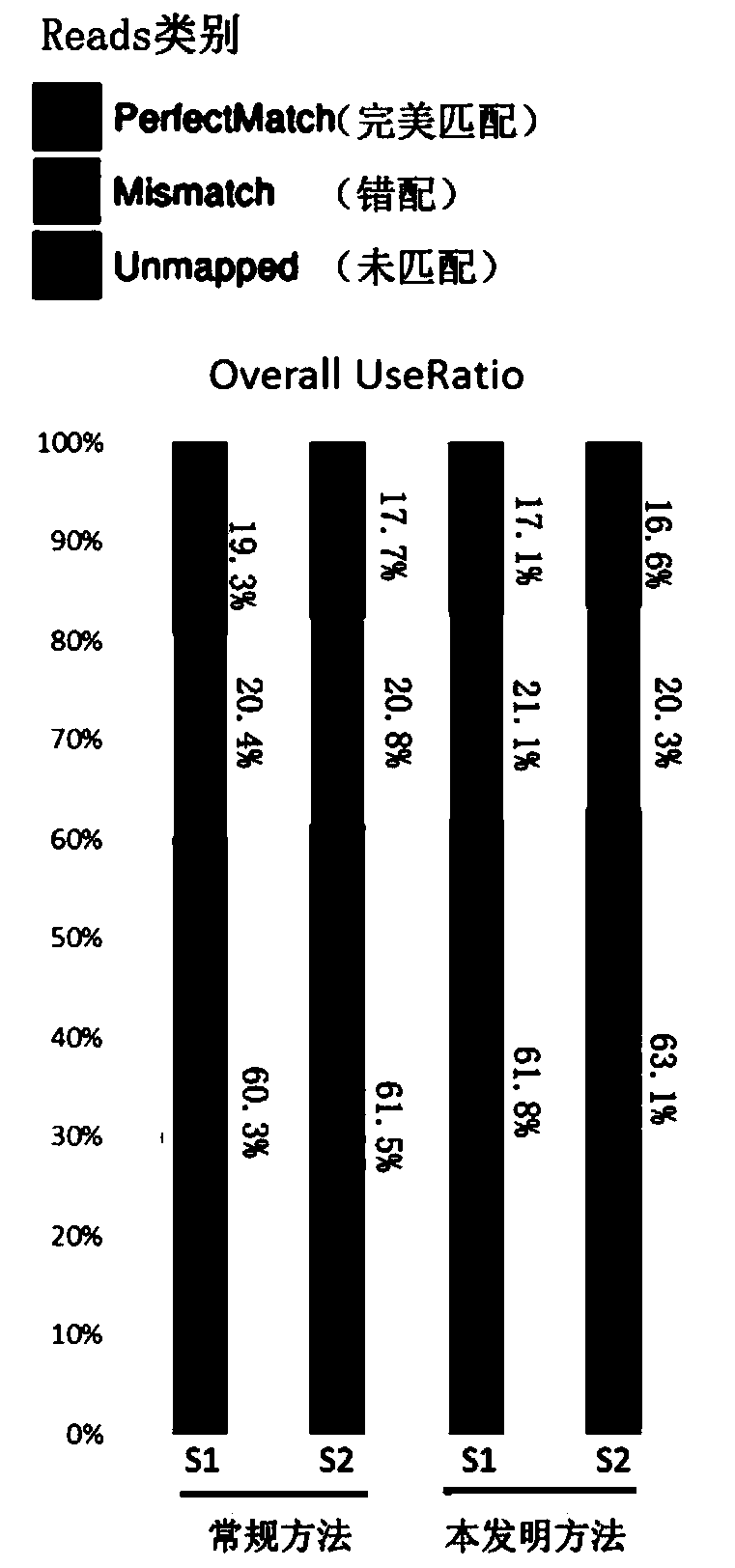
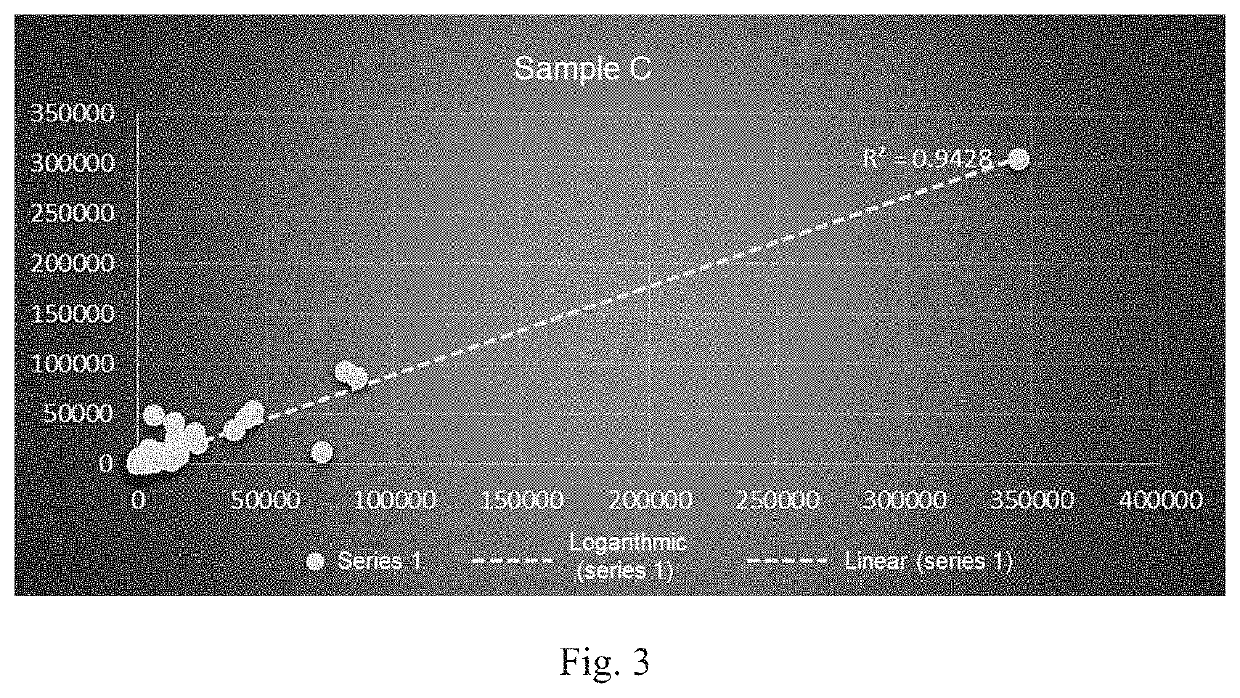
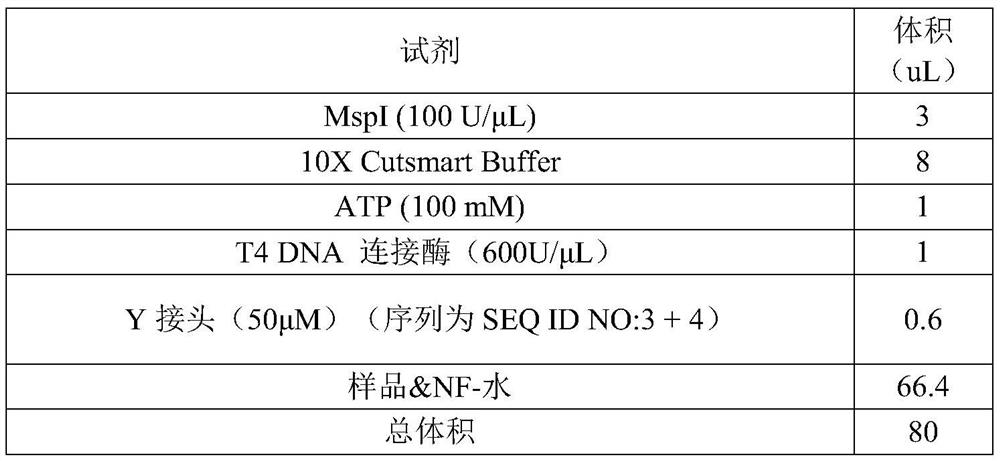
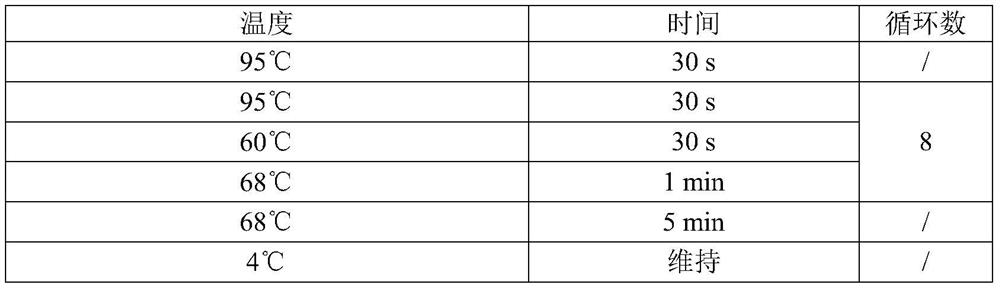
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
32results about How to "Improve sequencing quality" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for constructing high-flux simplified genome sequencing library
ActiveCN104694635AReduce the impact of quantitative accuracy before mixingImprove uniformityMicrobiological testing/measurementLibrary creationGenomic sequencingPhosphorylation
The invention provides a method for constructing a high-flux simplified genome sequencing library. The method comprises the following steps: digesting genome DNA of different samples by using restriction enzymes, thereby obtaining blunt-end DNA fragments with phosphorylation modification at the 5' end; adding the base A at the 3' end, connecting the DNA fragments of different samples with 5' phosphorylation and 3' cohesive ends A with joints containing different bar code sequences, connecting the products PCR, purifying and mixing the amplification products, performing low-cycle PCR amplification, thereby obtaining a target fragment amplification product of which the ratio of the joints to joint dimers is greatly reduced; and separating, purifying, and forming the high-flux simplified genome sequencing library from the purified products. The method disclosed by the invention has the advantages that the flux is high, the cost is low, and the generated library is high in sequencing quality and has very wide application prospects in researches such as high-flux SNP detection and marker development, genetic map drawing and functional gene location.
Owner:BIOMARKER TECH
Method for simultaneously sequencing a plurality of nucleic acid samples
InactiveCN103602726AThe sequencing results are accurateGood repeatabilityMicrobiological testing/measurementPhosphorylationNucleic acid sequencing
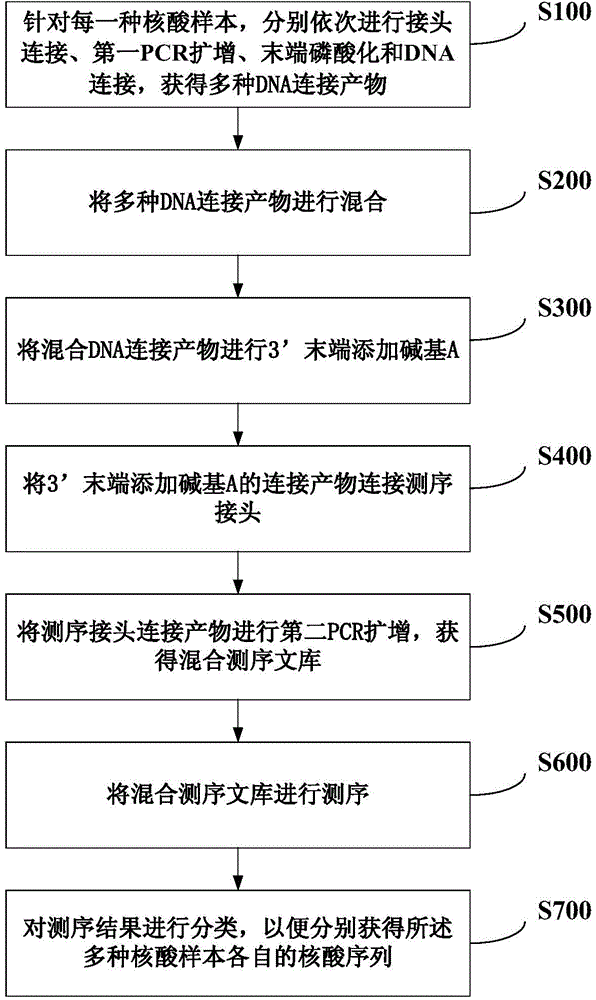
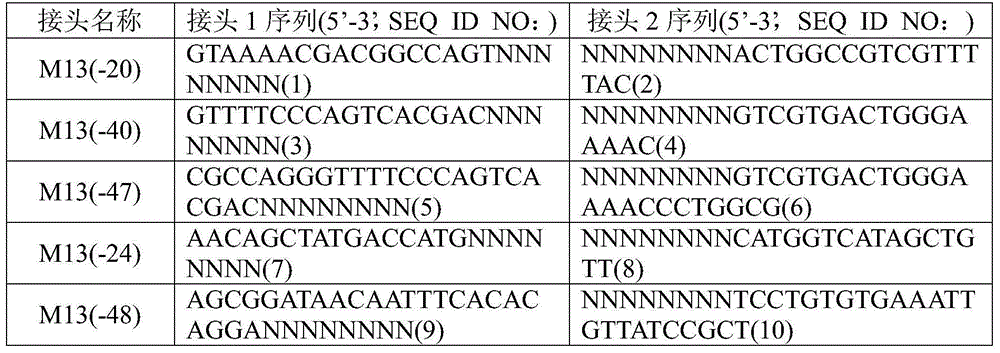
The invention discloses a method for simultaneously sequencing a plurality of nucleic acid samples, which comprises: respectively performing adaptor ligation, first PCR amplification, terminal phosphorylation, and DNA ligation in order of the plurality of nucleic acid samples; mixing the plurality of DNA ligation products; adding a base A at the 3'-terminal of the mixed DNA ligation products; connecting the ligation products with the 3'-terminal added with the base A to a sequencing interface; performing second PCR amplification of the sequencing interface-connected products to obtain a mixed sequencing library of the plurality of nucleic acid samples; sequencing the mixed sequencing library; and classifying the sequencing results to respectively obtain respective nucleic acid sequences of the plurality of nucleic acid samples. The method of the invention can simultaneously perform sequencing of a plurality of nucleic acid samples, is accurate in sequencing results, good in repeatability, low in cost, and high in efficiency, makes full use of sequencing flux, and is especially suitable for blood free DNA samples.
Owner:田埂
Method for analyzing APA (Alternative Polyadenylation) based on 3T-seq (TT-mRNA Terminal Sequencing) within whole-genome range
InactiveCN103911449AAvoid damageHigh yieldMicrobiological testing/measurementBiotin-streptavidin complexPoly-A RNA
The invention relates to a method for detecting a PAS (Poly A Site) of an RNA (Ribose Nucleic Acid), and provides a method for analyzing APA (Alternative Polyadenylation) based on 3T-seq (TT-mRNA Terminal Sequencing) within a whole-genome range. The method for analyzing APA comprises the following steps: preparing magnetic beads containing oligo dT, i.e., mixing an oligo dT primer modified by 5' end biotin and the magnetic beads with streptavidin; mixing the magnetic beads with total RNA and screening RNA containing Poly A; performing reverse transcription, synthesizing the second strand of a double-stranded cDNA and breaking the double-stranded cDNA; removing the Poly A structure; after terminal modification, connecting a sequencing linker; and recovering libraries. According to the method for analyzing APA, RNA horizontal operation is reduced, a Poly A sequence is horizontally removed from the DNA, and the influence of the Poly structure on sequencing is eliminated; double-stranded DNA breaking enzyme is selected for treatment, and on the premise that the quality of the libraries is not affected, the DNA is broken horizontally; the experiment process is simplified, the experiment difficulty and cost are lowered and the method for analyzing APA is wide in application range.
Owner:SHANGHAI JIAO TONG UNIV
Method for sequencing-by-synthesis
InactiveUS20050227261A1Easy to disassembleImprove sequencing qualityBioreactor/fermenter combinationsBiological substance pretreatmentsPipetteSequencing by synthesis
The invention refers to a tubular member, such as a pipette tip, for binding nucleic acid molecules for a subsequent biomolecular reaction, to methods for at least partly performing a sequencing-by-synthesis reaction in a tubular member, to an automated system using the tubular member for performing the methods of the invention, and kits and computer programs for use in relation to the methods of the invention. By performing a sequencing-by-synthesis reaction in a tubular member, such as a pipette tip, a new format for enzymatic reactions of this type, i.e. sequencing by synthesis, is provided.
Owner:454 CORP
Test kit and detection method for quantitative detection of high-throughput sequencing library
PendingCN111088328AAchieving absolute quantificationGet the actual concentrationMicrobiological testing/measurementDNA/RNA fragmentationFluorophoreA-DNA
The invention discloses a test kit and a detection method for quantitative detection of a high-throughput sequencing library. The test kit comprises a detection primer and a detection probe which aredesigned for the joint sequence of a to-be-detected high-throughput sequencing library, wherein upstream and downstream primers of the detection primer have sequences shown as Seq ID No.1 and Seq ID No.2; the detection probe has a sequence shown as Seq ID No.3; the 5'end of the detection probe is modified by a fluorophore; and the 3'end of the detection probe is modified by a fluorescence quenching group. According to the test kit, an NGS library is quantitatively detected through a TaqMan probe method, and absolute quantification of the NGS library can be achieved; and compared with a conventional dye method for QPCR quantitative detection, the test kit can obtain the actual concentration of the NGS library more accurately; the size of a DNA fragment based on the NGS library does not needto be corrected, which is of great significance to high-throughput sequencing; and the sequencing quality can be improved, so an optimal sequencing data result is obtained.
Owner:深圳海普洛斯医学检验实验室 +1
Library construction method and reagent for pooling of large-sample-size PCR products based on high-throughput sequencing
ActiveCN108504651AReduce the cost of building a libraryIncrease profitMicrobiological testing/measurementLibrary creationSingle sampleProduct base
The invention discloses a library construction method and reagent for pooling of large-sample-size PCR products based on high-throughput sequencing. The library construction method comprises the following steps: separately subjecting each of a plurality of PCR products derived from DNAs of different samples to annealing with a random primer having a tag sequence and carrying out an isothermal amplification reaction under the action of an isothermal amplification enzyme; mixing isothermal amplification products derived from different samples and carrying out piece selection on the mixed products; subjecting the products of the piece selection to repairing of 5' phosphorylated blunt ends and A addition reaction of 3' ends; connecting the products with linker sequences so as to obtain ligation products capable of distinguishing library information; and subjecting the ligation products to PCR amplification to obtain a library applicable to high-throughput sequencing. The method provided bythe invention realizes pooling without dependence on an ultrasonic breaking instrument, is lowered in library construction cost and complexity, reduces data waste, and improves the base randomness and coverage depth uniformity of sequencing data, thereby reducing the sequencing cost of a single sample.
Owner:CHEERLAND BIOTECH CO LTD
Method and detection kit used for nasal cavity flora detection
ActiveCN108866220AIncrease coverageIncreased amplification sensitivityMicrobiological testing/measurementLibrary creationNasal cavityMicrobiology
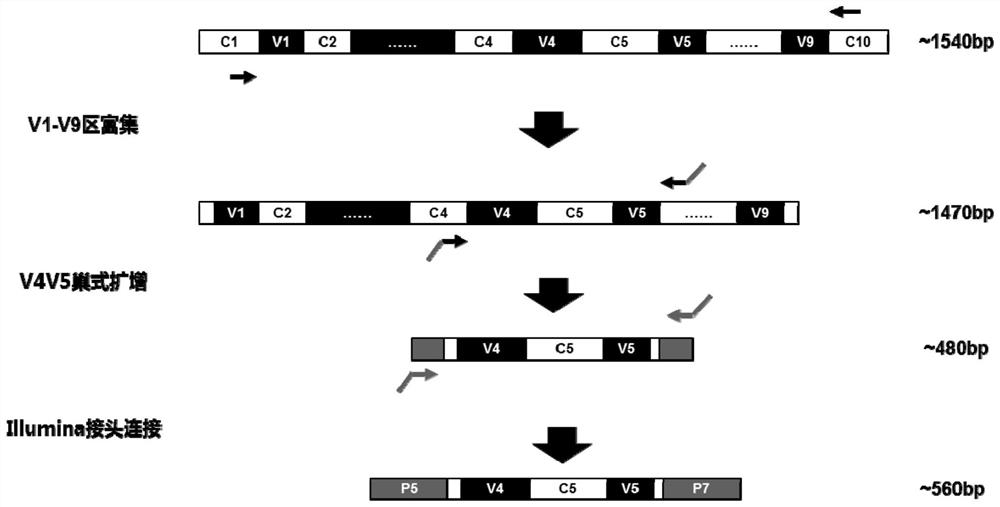
The invention discloses a method and a detection kit used for nasal cavity flora detection. According to the method and detection kit used for nasal cavity flora detection, a whole set of primers usedfor sample flora detection are provided, and the primers comprise the primers for amplifying V4-V5 areas of bacterium 16S rDNA and the primers for amplifying V1-V9 areas of bacterium 16S rDNA. According to the method and detection kit used for nasal cavity flora detection, firstly, the V1-V9 primers which are high in amplification efficiency, high in specificity and low in humanized homology areadopted for enriching 16S rDNA fragments, and then nested PCR is used for amplifying target sections V4V5 double fragments, so that the problem that when the V4V5 sections are used for amplifying primers directly, the homology with a humanized genomic sequence is high, so that when a nasal cavity swab sample with high humanized gDNA content is amplified, a serious non-specific amplification phenomenon appears is solved.
Owner:CAPITALBIO CORP
Genome methylation library, preparation method and application thereof
PendingCN113668068ASolve the problem of comparative analysisAccurate analysisNucleotide librariesMicrobiological testing/measurementGenes mutationBase J
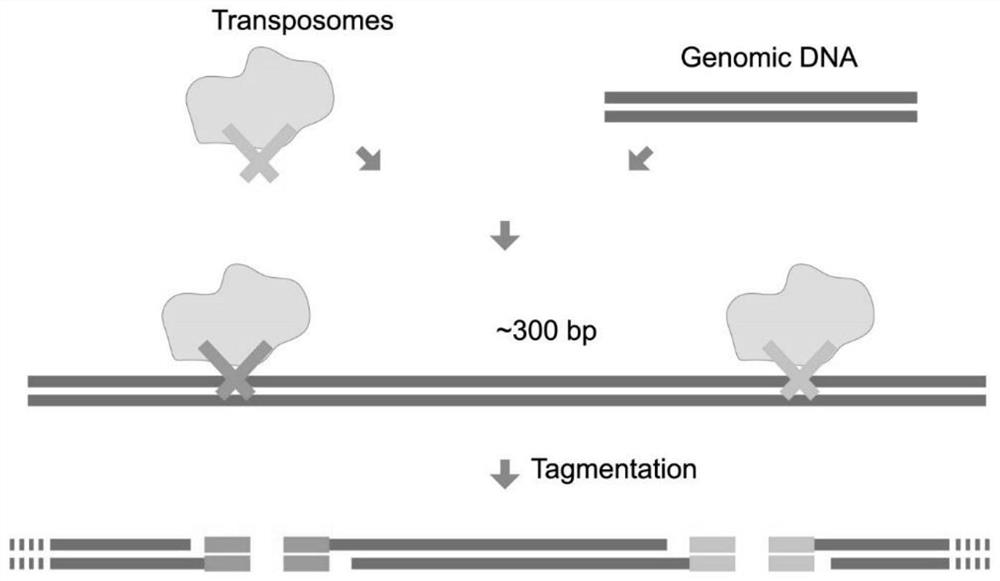
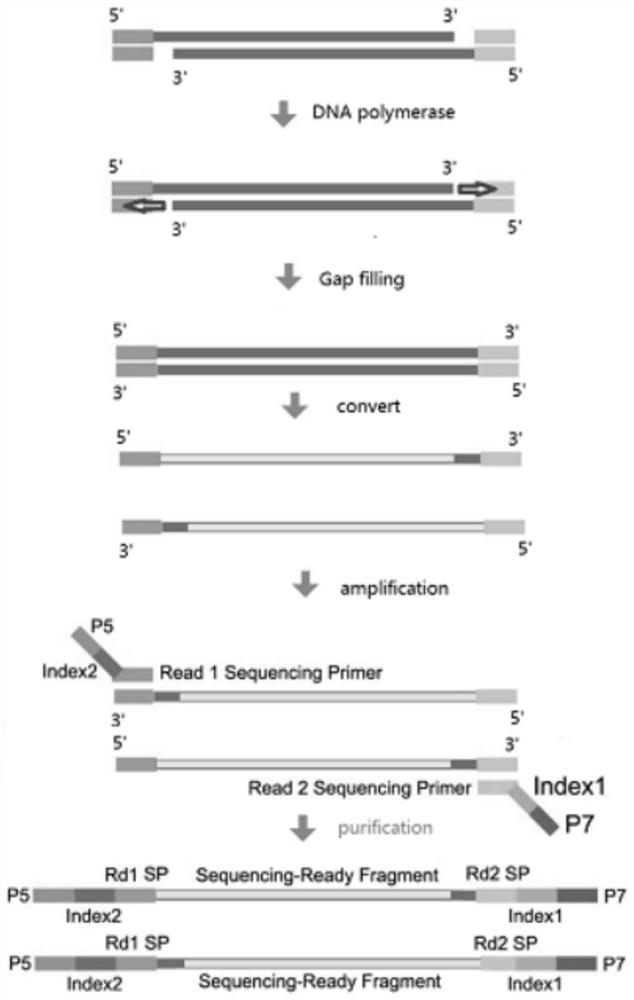
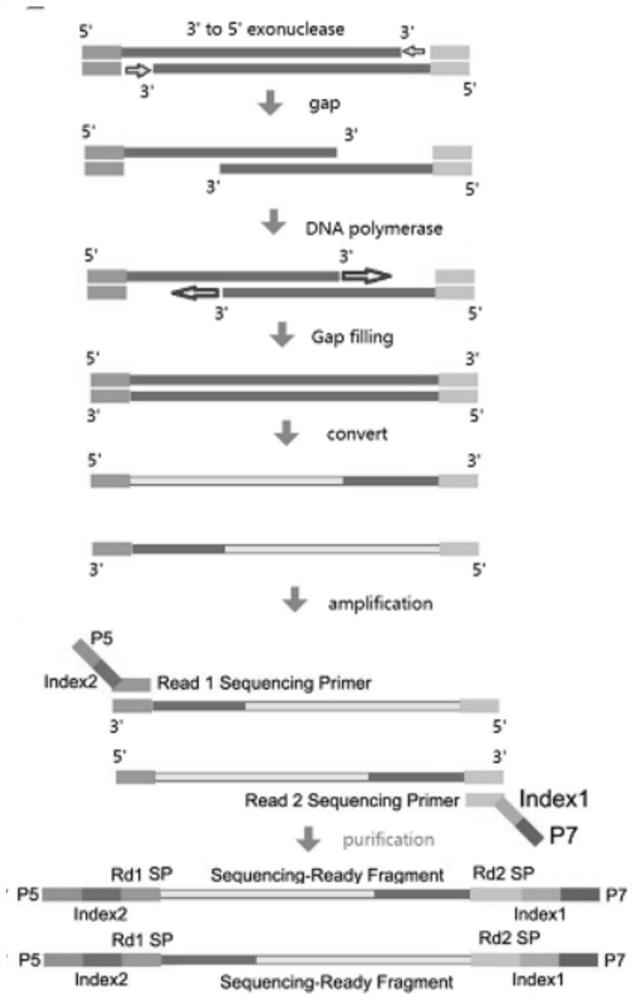
The invention relates to a genome methylation library, a preparation method and application thereof. The preparation method comprises the following steps: providing a transposase complex, wherein the transposase complex is composed of a transposase linker and a transposase, and the transposase linker contains a transposon sequence; carrying out fragmentation on genome DNA by using the transposase complex to obtain a fragmented product added with the transposase linker; carrying out notch completion on the fragmented product by using dNTPs in which dCTP is replaced by mdCTP; carrying out transformation treatment on the fragmented product after the notch completion, so that cytosine which is not subjected to methylation modification is transformed into uracil; and carrying out PCR amplification on the transformed fragmented product to obtain the genome methylation library. According to the invention, the problem of methylation library comparison analysis can be solved, more accurate methylation site analysis is realized, gene mutation site analysis can be carried out, library base balance can be greatly improved, and sequencing quality is greatly improved.
Owner:GUANGZHOU DRIPNANO BIOTECH CO LTD
Solid-phase substrate as well as treatment method and application thereof
ActiveCN111100786AImprove performanceIncrease distribution densityBioreactor/fermenter combinationsBiological substance pretreatmentsPhysical chemistrySilanization
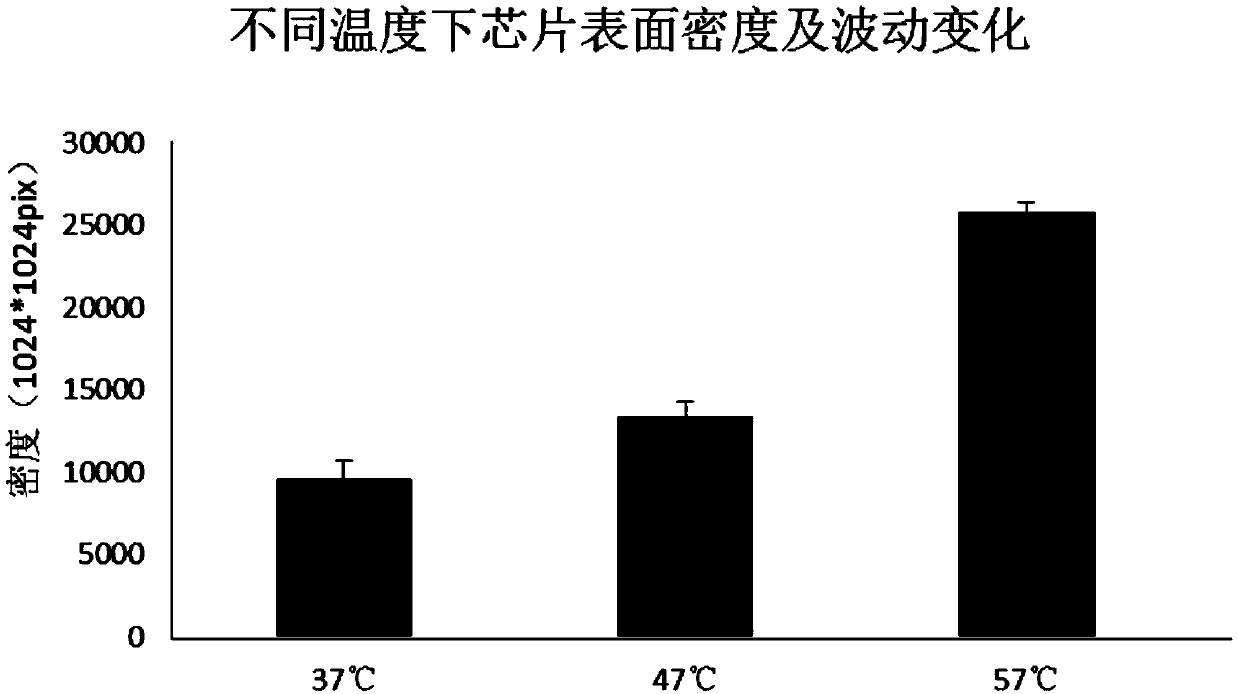
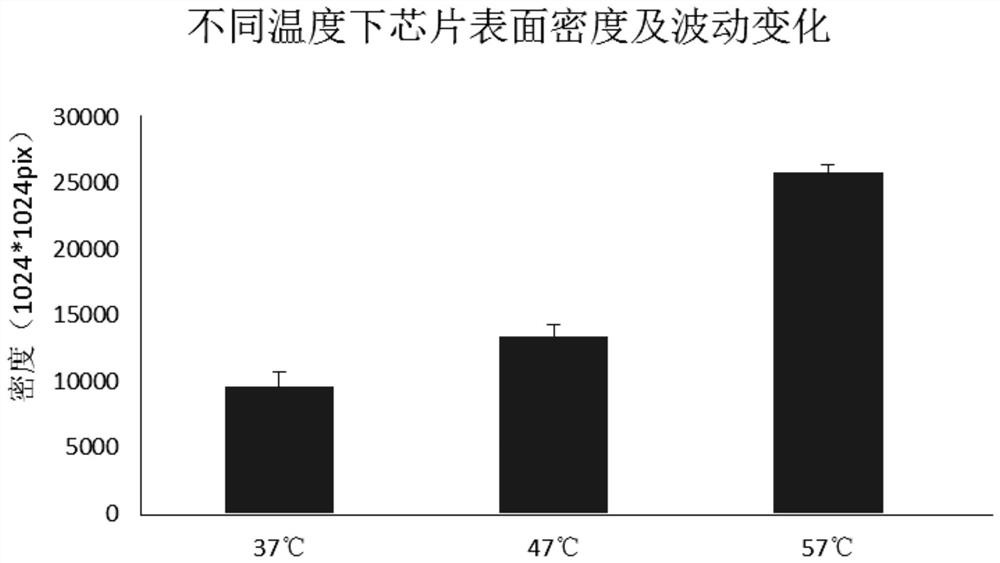
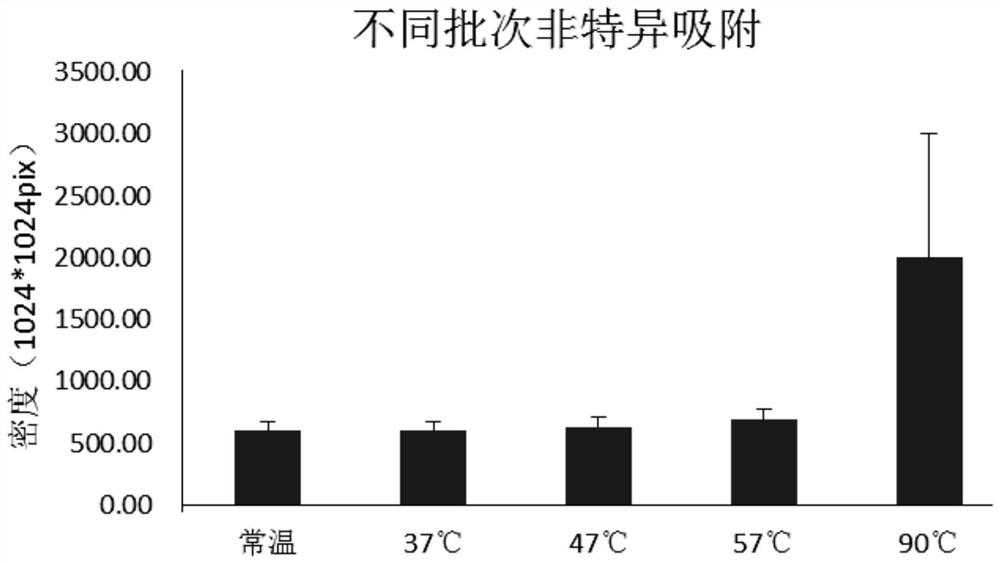
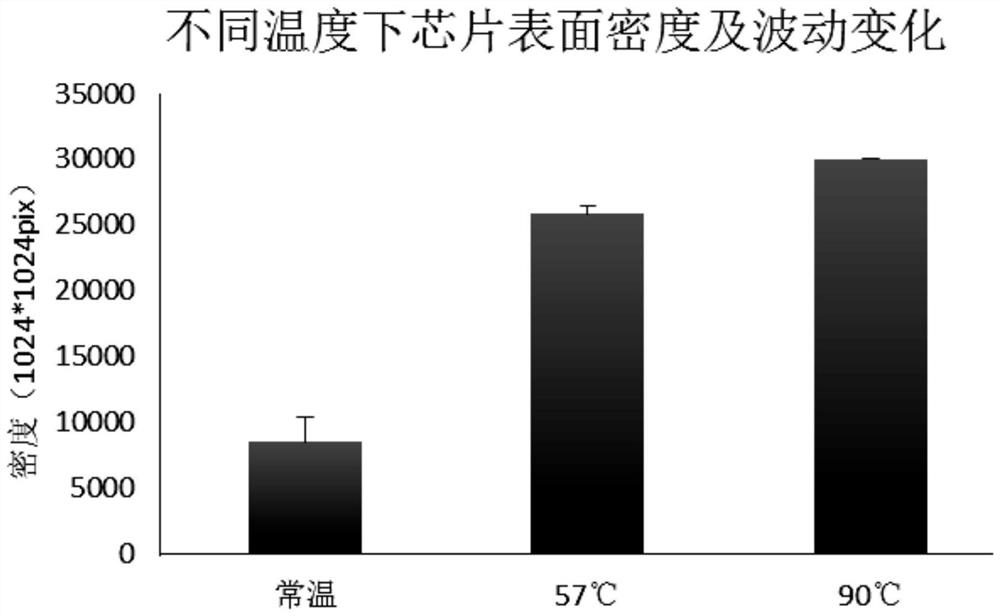
The invention discloses a solid-phase substrate as well as a treatment method and application thereof. The solid-phase substrate is provided with at least one silanized surface, wherein the surface isa surface exposed in a specific environment for a certain period of time, the specific environment is an inert gas environment, the temperature of the specific environment is selected from 37-60 DEGC, and the certain period of time is no less than 2 hours. By adopting the solid-phase substrate, the distribution density and stability of nucleic acid molecules immobilized on the surface of the solid-phase substrate are improved, the quality of a chip can be improved, and sequencing quality can be improved when the solid-phase substrate is applied to sequencing.
Owner:GENEMIND BIOSCIENCES CO LTD
High-fidelity Pfu DNA polymerase mutant, coding DNA of high-fidelity Pfu DNA polymerase mutant and application of high-fidelity Pfu DNA polymerase mutant in NGS
ActiveCN113388596AImprove fidelityThe results obtained by the analysis and identification are true and reliableMicrobiological testing/measurementTransferasesBase JWild type
The invention provides a high-fidelity Pfu DNA polymerase mutant. The high-fidelity Pfu DNA polymerase mutant is characterized in that on the basis of a wild type Pfu DNA polymerase, mutation of one, three or six amino acid sites is carried out, and the mutation sites are selected from the following sites: Y403A, P411H, V438K, A741S, R757Q and S768K. The invention also discloses coding DNA of the high-fidelity Pfu DNA polymerase mutant and application of the high-fidelity Pfu DNA polymerase mutant in NGS. The high-fidelity Pfu DNA polymerase mutant has ultrahigh fidelity and high amplification efficiency, can be stably and efficiently applied to the library enrichment process in next-generation sequencing, effectively improves the library sequencing quality, reduces the base mismatch rate, and can be used for precise analysis and interpretation of genetic information.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
A high-throughput simplified genome sequencing library construction method
ActiveCN104694635BReduce the impact of quantitative accuracy before mixingImprove uniformityMicrobiological testing/measurementLibrary creationGenomic sequencingHigh flux
The invention provides a method for constructing a high-flux simplified genome sequencing library. The method comprises the following steps: digesting genome DNA of different samples by using restriction enzymes, thereby obtaining blunt-end DNA fragments with phosphorylation modification at the 5' end; adding the base A at the 3' end, connecting the DNA fragments of different samples with 5' phosphorylation and 3' cohesive ends A with joints containing different bar code sequences, connecting the products PCR, purifying and mixing the amplification products, performing low-cycle PCR amplification, thereby obtaining a target fragment amplification product of which the ratio of the joints to joint dimers is greatly reduced; and separating, purifying, and forming the high-flux simplified genome sequencing library from the purified products. The method disclosed by the invention has the advantages that the flux is high, the cost is low, and the generated library is high in sequencing quality and has very wide application prospects in researches such as high-flux SNP detection and marker development, genetic map drawing and functional gene location.
Owner:BIOMARKER TECH
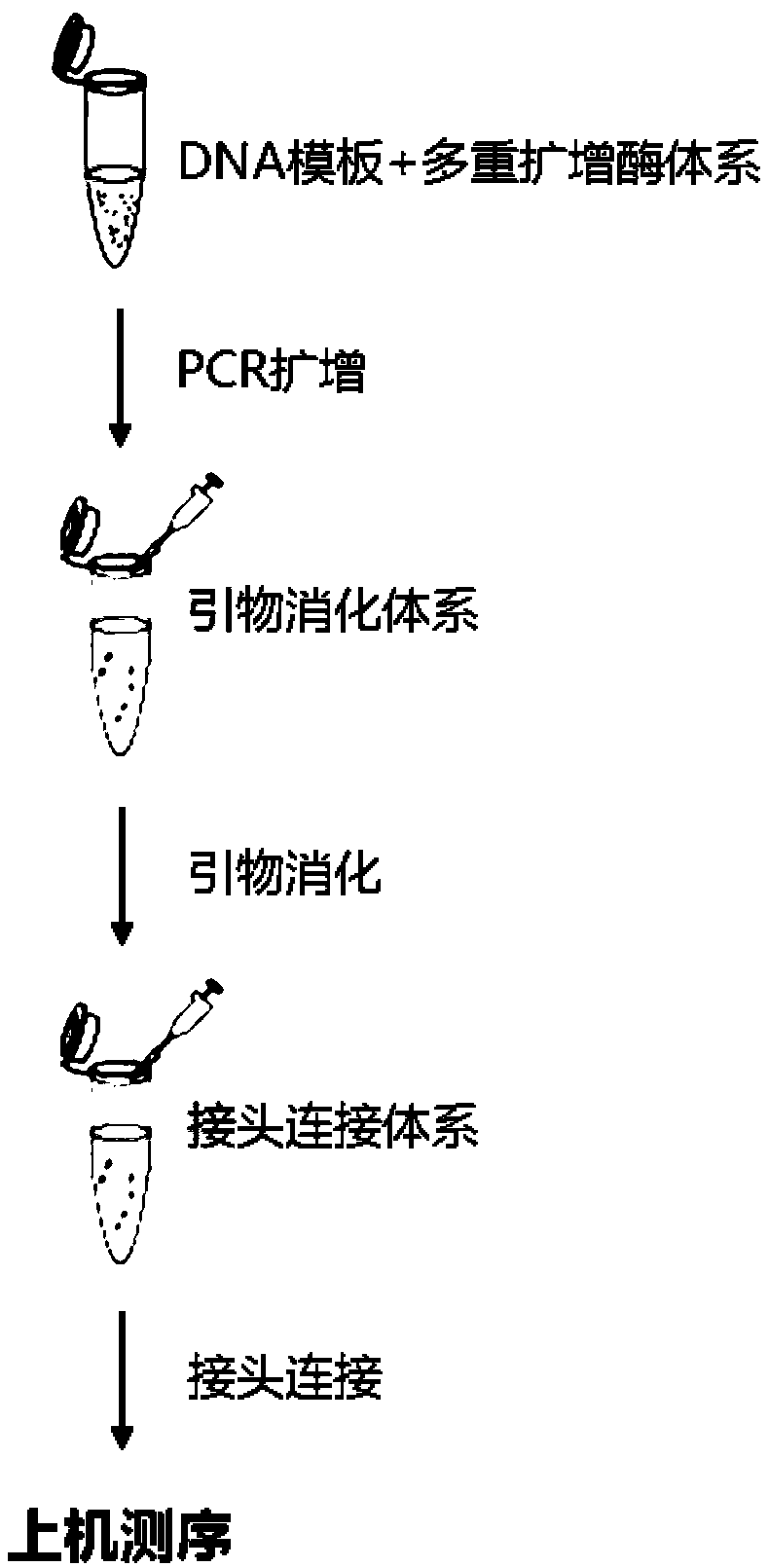
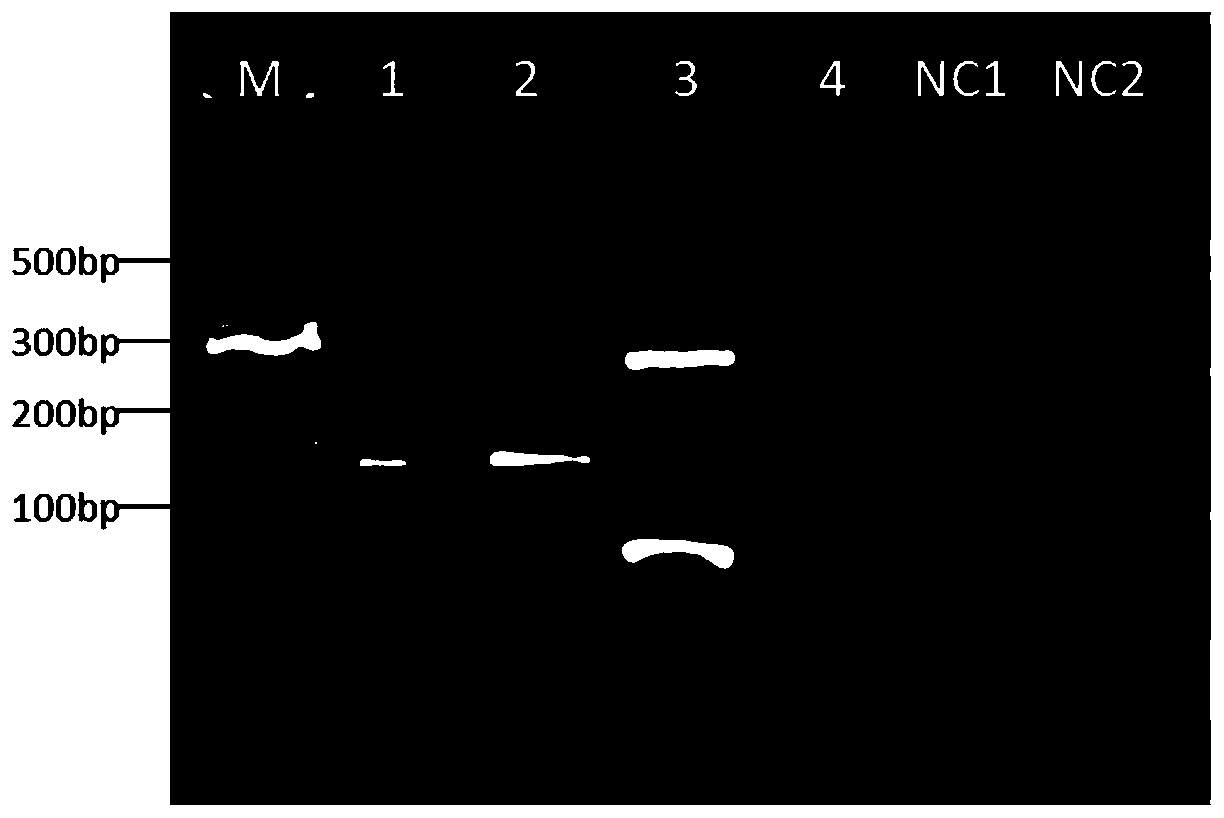
Multi-amplification database building method for trace DNA and special kit
ActiveCN108929901AGood uniformity of amplificationEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationMutation detectionCommercial kit
The invention discloses a multi-amplification database building method for trace DNA and a special kit. The multi-amplification database building method can realize multiple amplification sequencing with trace DNA or even single-cell-level DNA as a template, and the whole reaction process can be realized in a single tube. Experiments prove that the database building method achieves 207-fold one-tube amplifier amplification at the single cell level, has the fidelity higher than that of conventional commercial kits, not only has the advantages of convenient operation, low cost, high sequencing quality and high sensitivity, but also can meet the sequencing requirements of two sequencing platforms, and is suitable for high-throughput and low-frequency mutation detection fields such as the ctDNA mutation detection field and the low-initial-quantity DNA embryonic mutation detection field such as single cell mutation detection and screening.
Owner:CAPITALBIO CORP
Method for preparing circular ssDNA and kit of method
PendingCN112063688AAvoid formingImprove sequencing qualityMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionO-Phosphoric Acid
The invention discloses a method for preparing circular ssDNA and a kit of the method. The method comprises the following steps: 1) breaking sample gDNA; 2) constructing a DNA library, wherein one ofthe amplification primers is modified with 5 '-phosphoric acid; 3) performing enzyme digestion on one chain modified with 5 '-phosphoric acid in double chains of the library; 4) performing 5 'phosphorylation on the other chain; and 5) performing cyclizing by ligase and splint oligo. The kit comprises exonuclease, 5'-terminal phosphorylase, ligase and splint oligo suitable for double-chain DNA of which the substrate is 5' phosphorylation. The method has the advantages that formation of circular dsDNA can be avoided, the sequencing quality of samples is improved, and the sequencing uniformity ofsamples with high GC content is improved.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
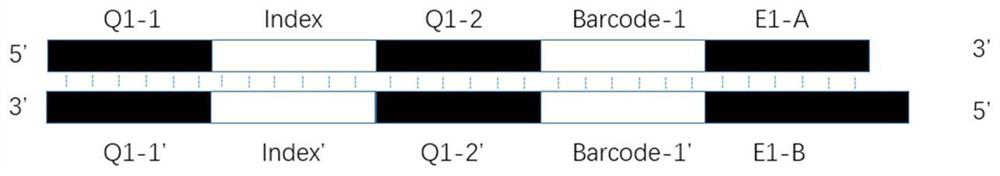
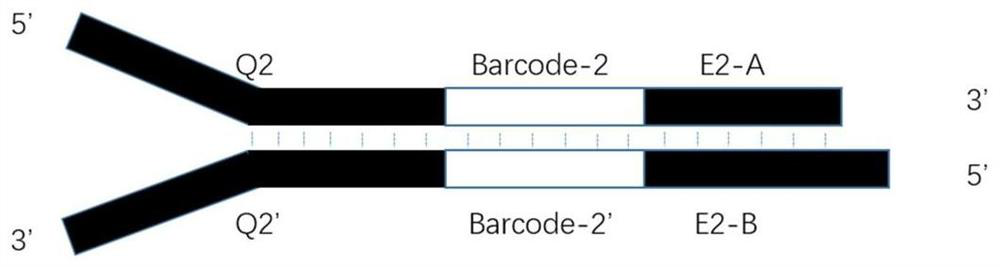
Library building connector and method suitable for simplified genome sequencing of DNBSEQ technology
PendingCN114854825ASimplified sequencingImprove sequencing qualityMicrobiological testing/measurementLibrary linkersGenomic sequencingGenetics
The invention relates to the technical field of biological information, in particular to a library building connector and method suitable for simplified genome sequencing of a DNBSEQ technology. The linker comprises an upstream linker sequence and a downstream linker sequence; the upstream linker sequence comprises an upstream Barcode fragment, a Q1-1 fragment, an Index fragment and a Q1-2 fragment which are connected in sequence; the downstream linker sequence comprises a downstream Barcode fragment and a Q2 fragment; wherein the upstream linker sequence is reversely complementary, and the Q2 fragment part in the downstream linker sequence is reversely complementary; the upstream Barcode fragment and the downstream Barcode fragment are not equal in length. The invention provides a library building joint and designs a simplified genome sequencing method suitable for a DNBSEQ technology based on the library building joint, and genome sequencing can be accurately, quickly and effectively carried out.
Owner:CHINA AGRI UNIV
Primer suitable for sequencing DNA-coding compound sequencing library
PendingCN110964794AHigh data availabilityImprove sequencing qualityNucleotide librariesMicrobiological testing/measurementLibraryBioinformatics
The invention discloses a sequencing primer suitable for sequencing a DNA coding compound library. The sequencing primer is a sequence formed by connecting a DNA coding compound amplification primer sequence, a connector sequence and a sequencing primer sequence. The primer can improve the sequencing quality of the DNA coding compound library, one-step sequencing can further be realized, and the application prospects are excellent.
Owner:HITGEN INC
A kind of method and detection kit for nasal flora detection
ActiveCN108866220BReduce distractionsDesigned for high amplification efficiencyMicrobiological testing/measurementLibrary creationHigh homologyFlora
The invention discloses a method for detecting nasal flora and a detection kit. The present invention provides a set of primers for the detection of sample flora, including primers for amplifying the V4-V5 region of bacterial 16S rDNA and primers for amplifying the V1-V9 region of bacterial 16S rDNA; the present invention first uses the amplification efficiency The V1‑V9 primers with high specificity and low human homology first enrich the 16S rDNA fragment, and then use nested PCR to amplify the target segment V4V5 double fragment, which overcomes the need to directly amplify the V4V5 segment. The amplification primers have high homology with the human genome sequence. When amplifying nasal swab samples with high human gDNA content, there will be serious non-specific amplification problems.
Owner:CAPITALBIO CORP
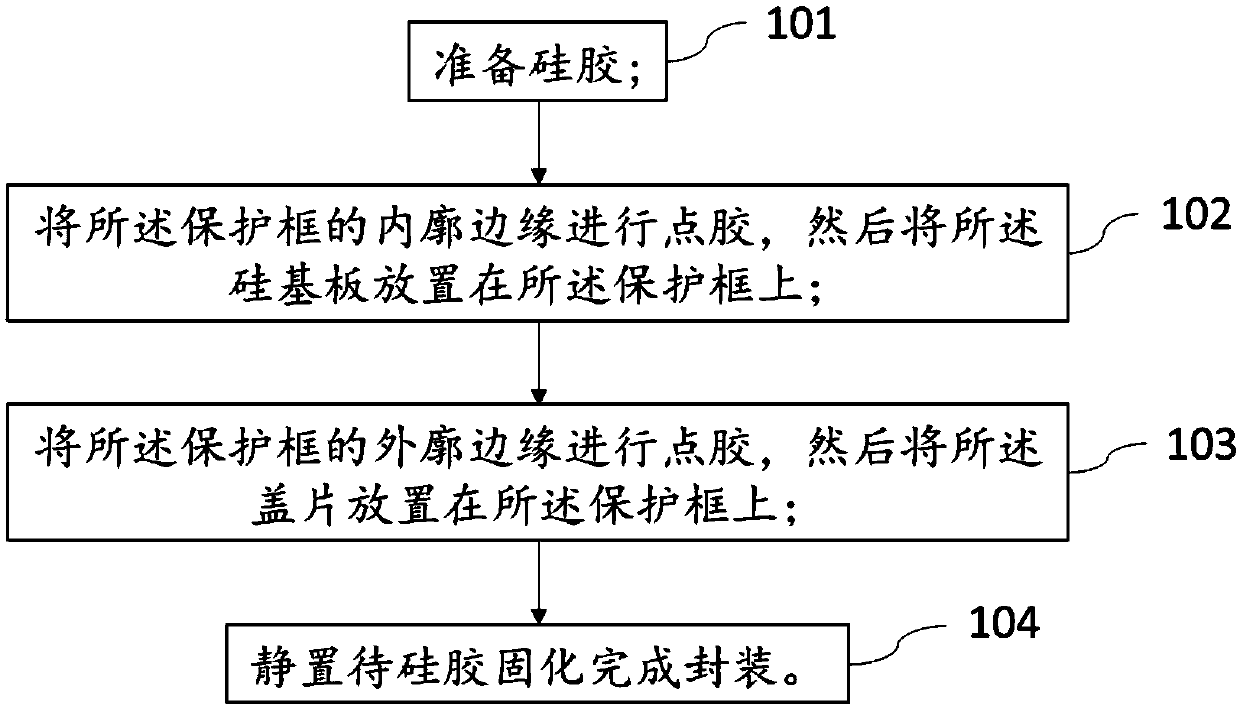
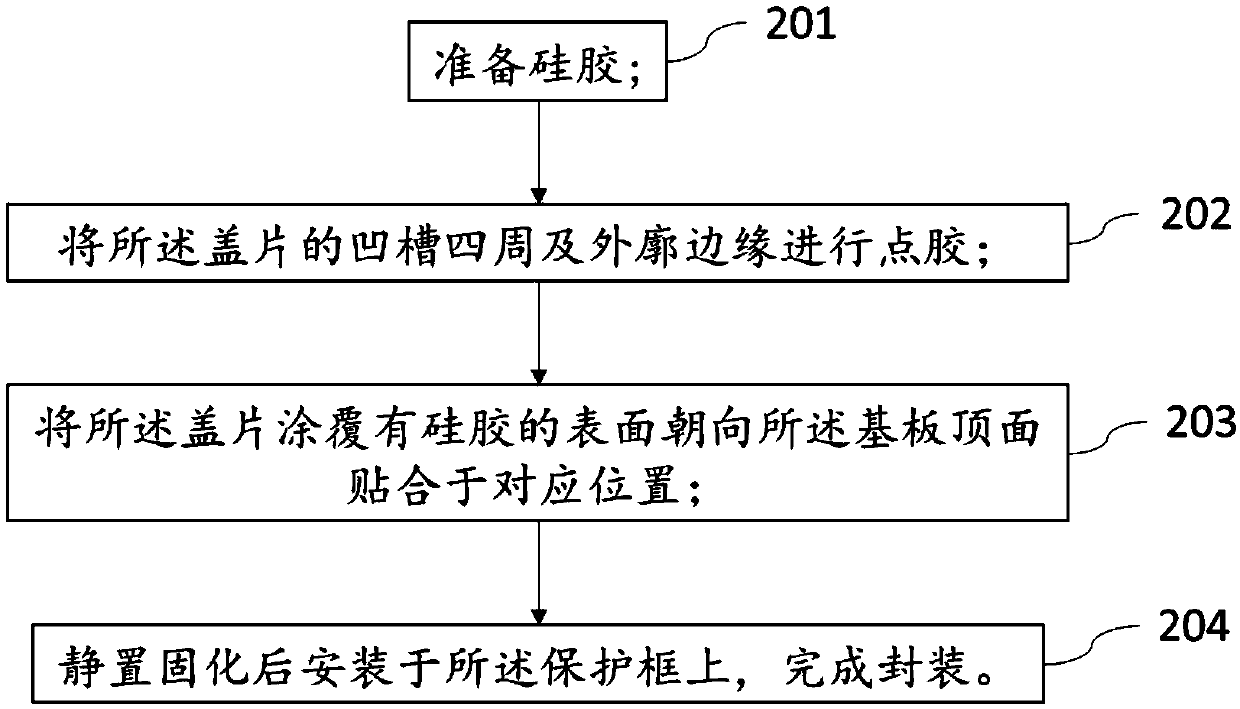
Gene sequencing chip and packaging method thereof
ActiveCN110846390ASolve the problem of contaminating the surface of the chipSolving the problem of contaminating chip surfacesMicrobiological testing/measurementCrystallographyBiotechnology
The invention provides a gene sequencing chip and a packaging method thereof. The chip comprises a protecting frame, an aminated substrate and a cover plate, wherein the aminated substrate is arrangedin the protecting frame and the cover plate stacks on the substrate. The method is characterized in that the contact areas of the protecting frame, the substrate and the cover plate are adhered through the curing reaction of coated silica gel to form a sealed structure, wherein the silica gel comprises one or the combination of dealcoholized silica gel, deaminated silica gel and deamidized silicagel, and the silica gel and the curing reaction byproduct of the silica gel do not have reaction with amino. The method has the advantages that the silica gel series glue which does not have reactionwith the amino is used to perform adhering and sealing, the problem that the glue pollutes the surface of the chip during the packaging of the aminated chip is solved effectively, complete sequencingdata is guaranteed, and sequencing quality is increased.
Owner:SHENZHEN HUADA GENE INST
Solid phase substrates, their processing methods and uses
ActiveCN111100786BImprove performanceIncrease distribution densityBioreactor/fermenter combinationsBiological substance pretreatmentsPhysical chemistrySilanization
A solid phase substrate, its processing method and application, the solid phase substrate has at least one silanized surface, the surface is a surface exposed to a specific environment for a certain period of time, the specific environment is an inert gas environment, and the temperature of the specific environment is selected from 37°C to 60°C, a certain period of time is not less than 2 hours. The invention improves the distribution density and stability of the nucleic acid molecules fixed on the surface of the solid phase substrate, improves the chip quality, and can improve the sequencing quality when used in sequencing.
Owner:GENEMIND BIOSCIENCES CO LTD
Solid-phase substrate as well as preparation method and application thereof
PendingCN114524623AImprove performanceIncrease distribution densityMicrobiological testing/measurementCoatingsPhysical chemistryBiology
The invention relates to the technical field of nucleic acid chips, in particular to a solid-phase substrate and a treatment method and application thereof. The solid-phase substrate is provided with at least one silanized surface, the silanized surface is a second surface formed by exposing a silanized first surface to a specific environment for a certain time, the specific environment is an inert gas environment, the temperature of the specific environment is selected from 37 DEG C to 60 DEG C, and the certain time is not less than 2 hours. The performance of the solid-phase substrate is improved, so that the distribution density and the stability of nucleic acid molecules fixed on the surface of the solid-phase substrate are improved, and the quality of the nucleic acid chip is finally improved.
Owner:GENEMIND BIOSCIENCES CO LTD
Reduced genome library building method for millet
InactiveCN108165646AAvoid two-way complementarityImprove data utilizationMicrobiological testing/measurementLibrary creationEnzyme digestionDNA Endonuclease
The invention relates to a reduced genome library building method for millet. The method includes the steps of: 1. utilizing restrictive endonuclease ApeK I for enzyme digestion on the genome DNA of several millet samples respectively to obtain an enzyme-digested product; 2. connecting a bar code joint F and a universal joint R with the enzyme-digested product; 3. mixing the connection products ofthe several millet samples at an equal volume and performing purification to obtain a purified connection product; 4. taking the purified connection product, using a universal primer F and 1 or morebar code primers R for PCR amplification to obtain a PCR product; 5. purifying the PCR product to obtain a target fragment, thus obtaining a sequencing library; and 6. using a high-throughput sequencing platform for sequencing of the sequencing library. The method provided by the invention has the advantages of high detection flux, low cost, good library building stability, and accurate and reliable sequencing result, and at the same time simplifies the genome sequencing and library building steps.
Owner:GRAIN RES INST HEBEI ACAD OF AGRI & FORESTRY SCI
Genome-wide analysis of APA based on 3t-seq
InactiveCN103911449BAvoid damageHigh yieldMicrobiological testing/measurementPoly-A RNABiotin-streptavidin complex
The invention relates to a method for detecting a PAS (Poly A Site) of an RNA (Ribose Nucleic Acid), and provides a method for analyzing APA (Alternative Polyadenylation) based on 3T-seq (TT-mRNA Terminal Sequencing) within a whole-genome range. The method for analyzing APA comprises the following steps: preparing magnetic beads containing oligo dT, i.e., mixing an oligo dT primer modified by 5' end biotin and the magnetic beads with streptavidin; mixing the magnetic beads with total RNA and screening RNA containing Poly A; performing reverse transcription, synthesizing the second strand of a double-stranded cDNA and breaking the double-stranded cDNA; removing the Poly A structure; after terminal modification, connecting a sequencing linker; and recovering libraries. According to the method for analyzing APA, RNA horizontal operation is reduced, a Poly A sequence is horizontally removed from the DNA, and the influence of the Poly structure on sequencing is eliminated; double-stranded DNA breaking enzyme is selected for treatment, and on the premise that the quality of the libraries is not affected, the DNA is broken horizontally; the experiment process is simplified, the experiment difficulty and cost are lowered and the method for analyzing APA is wide in application range.
Owner:SHANGHAI JIAOTONG UNIV
High-throughput sequencing-based library construction method and reagents for large sample volume mixed library construction of PCR products
ActiveCN108504651BReduce the cost of building a libraryIncrease profitMicrobiological testing/measurementLibrary creationPhosphorylationLigation
The invention discloses a library construction method and reagent for pooling of large-sample-size PCR products based on high-throughput sequencing. The library construction method comprises the following steps: separately subjecting each of a plurality of PCR products derived from DNAs of different samples to annealing with a random primer having a tag sequence and carrying out an isothermal amplification reaction under the action of an isothermal amplification enzyme; mixing isothermal amplification products derived from different samples and carrying out piece selection on the mixed products; subjecting the products of the piece selection to repairing of 5' phosphorylated blunt ends and A addition reaction of 3' ends; connecting the products with linker sequences so as to obtain ligation products capable of distinguishing library information; and subjecting the ligation products to PCR amplification to obtain a library applicable to high-throughput sequencing. The method provided bythe invention realizes pooling without dependence on an ultrasonic breaking instrument, is lowered in library construction cost and complexity, reduces data waste, and improves the base randomness and coverage depth uniformity of sequencing data, thereby reducing the sequencing cost of a single sample.
Owner:CHEERLAND BIOTECH CO LTD
A method for multiple amplification library construction for trace dna and its special kit
ActiveCN108929901BGood uniformity of amplificationEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMutation detection
The invention discloses a method for multiple amplification library building of trace DNA and a special kit thereof. The multiple amplification library construction method of the present invention can realize multiple amplification sequencing using trace DNA or even DNA at the single cell level as a template, and the entire reaction process can be realized in a single tube. It is proved by experiments that the library construction method of the present invention achieves up to 207 multiple amplifications of one-tube amplicons at the level of single cells, and the fidelity is higher than that of existing commercial kits, which not only has the advantages of convenient operation , low cost, high sequencing quality and high sensitivity, and can also meet the sequencing needs of the two sequencing platforms. It is suitable for the field of high-throughput low-frequency mutation detection, such as ctDNA mutation detection, and the field of germline mutation detection with low input DNA, such as single-cell mutation detection and screening.
Owner:CAPITALBIO CORP
Method for rapid quantification of high-throughput sequencing library of MGI platform and kit
ActiveCN113186262AHigh actual molarityGuaranteed accuracyMicrobiological testing/measurementDNA/RNA fragmentationHigh throughput sequenceBioinformatics
The invention provides a method for rapid quantification of a high-throughput sequencing library of an MGI platform and a kit. The method comprises the following step of carrying out fluorescent quantitative PCR detection on a to-be-detected high-throughput sequencing library by using a composition containing a primer and a probe; wherein the composition containing the primers and the probes comprises a first primer and a first probe which are used for detecting an MGI library, and a second primer and a second probe which are used for detecting an arabidopsis PHYA gene, the first primer comprises sequences shown as SEQ ID NO: 1 and SEQ ID NO: 2, and the first probe comprises a sequence shown as SEQ ID NO: 3. The kit comprises the first primer and the first probe, and further comprises the second primer and the second probe, the second primer comprises sequences as shown in SEQ ID NO: 4 and SEQ ID NO: 5, and the second probe comprises a sequence as shown in SEQ ID NO: 6. The provided method or kit can be used for rapid quantification of the high-throughput sequencing library.
Owner:SHANGHAI REALBIO TECH CO LTD
Lysate for extracting plant cell nucleus
PendingCN114277094AImprove sequencing qualityExtract impactMicrobiological testing/measurementBiotechnologyGenomics
The invention provides a lysate which is used for plant cell nucleus extraction and ATAC-seq experiments based on a lysate of 10x Genomics Company. Under the condition that the basic components of the original formula are not changed as far as possible, the proper concentration of each component is found, and cell nucleuses of different tissue parts, development periods and stress states of plants can be extracted. It is found for the first time that the fixed time needs to be adjusted according to species, tissue parts, plant development periods and environmental states (such as stress of heat, cold, salt, drought and the like). It is found for the first time that extraction is carried out after plant materials are fixed, and beta-mercaptoethanol is added into original lysate components, so that plant cell nucleus extraction and plant ATAC-seq experiments can be carried out. Experiments prove that when the lysate provided by the invention is used for extracting the plant cell nucleus, the nuclear membrane of the obtained cell nucleus is complete, and the sequencing quality is high; the method is suitable for different tissue parts, different development periods and different environment states of different plant species.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
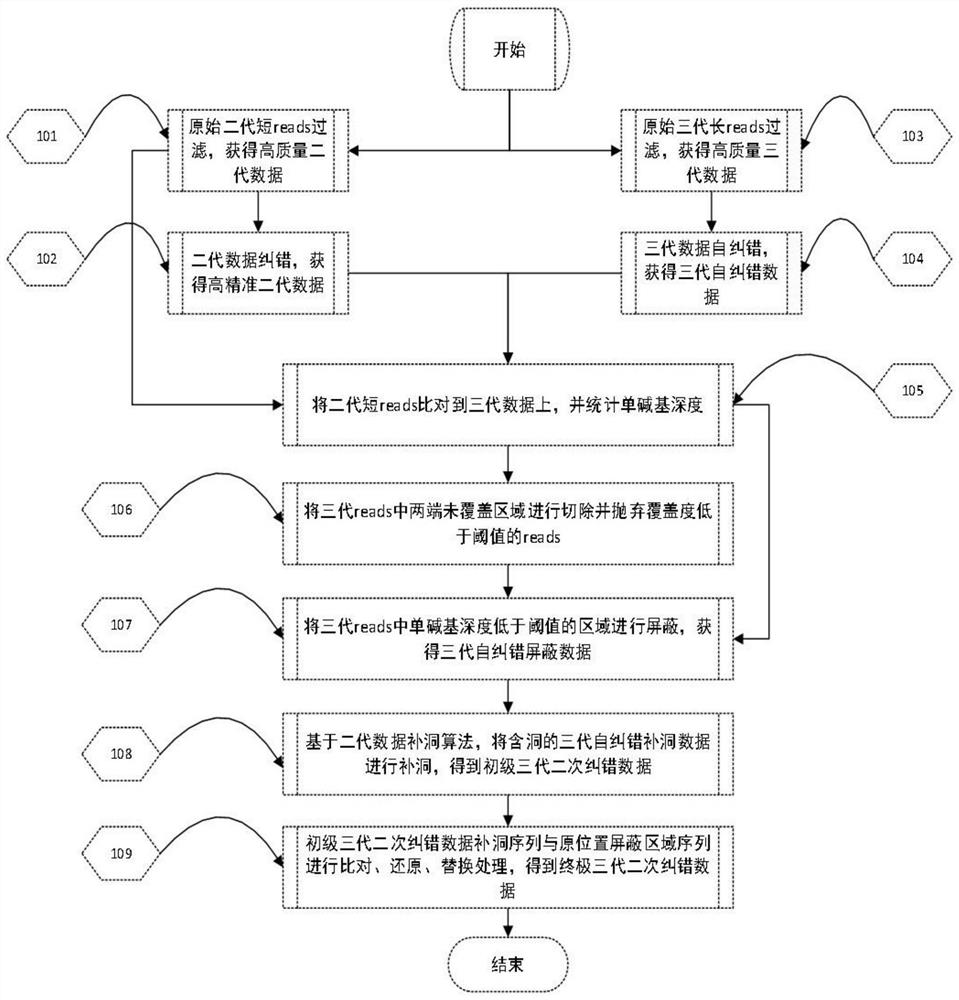
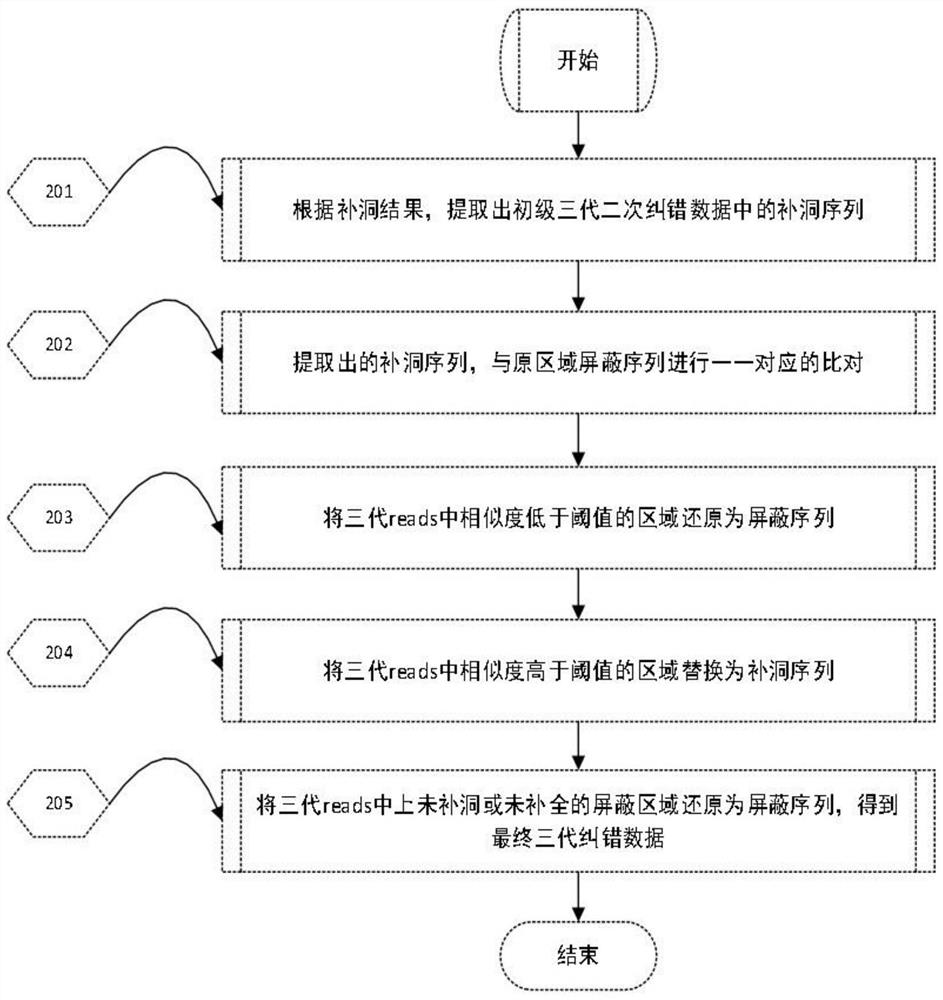
A method for processing raw data of nucleic acid third-generation sequencing and its application
ActiveCN108573127BReduce error rateImprove sequencing qualityProteomicsGenomicsThird generation sequencingBioinformatics
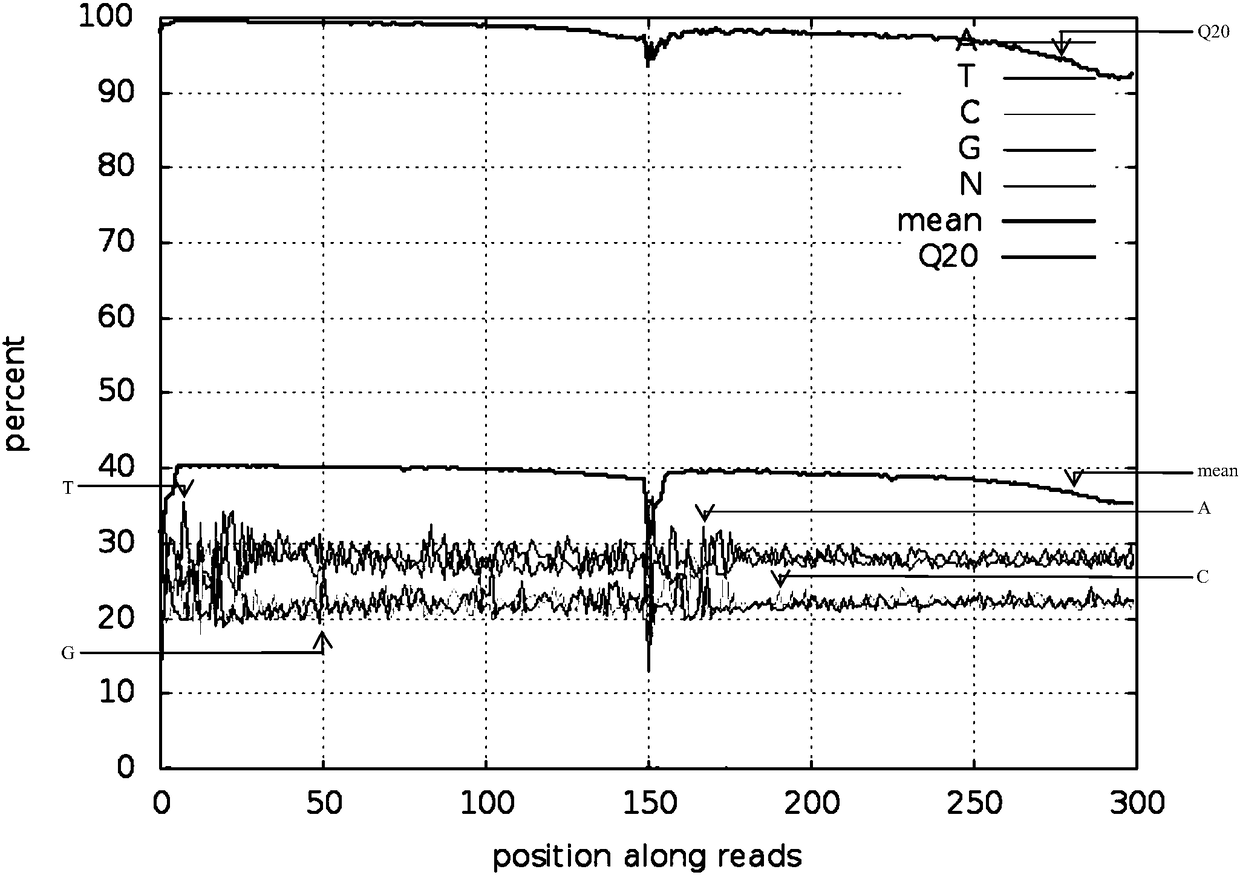
The present application discloses a method for processing raw data of nucleic acid third-generation sequencing and its application. The method for processing the raw data of the third-generation nucleic acid sequencing of the present application includes comparing the second-generation short-sequence data to the third-generation self-error-correcting data, and counting the single bases of the third-generation self-error-correcting data in the comparison results Coverage depth, mask the area with a single base coverage depth lower than the threshold as N, and use the second-generation sequencing hole-filling software to fill holes in the N-shielded area to obtain nucleic acid third-generation sequencing with a low single-base error rate data. The method for processing the raw data of the third-generation nucleic acid sequencing of the present application uses the second-generation short-sequence data to compare with the third-generation long-sequence data, and uses the hole-filling software of the second-generation sequencing to compare the single base coverage in the comparison results The low-depth N-shielding region is complemented, which effectively reduces the single-base error rate in the third-generation sequencing data and improves the sequencing quality.
Owner:BGI TECH SOLUTIONS
Template polynucleotide paired terminal sequencing method and kit
PendingCN114672546AImprove stabilityImprove sequencing qualityMicrobiological testing/measurementBinding sitePolynucleotide
The invention relates to the technical field of gene sequencing, in particular to a template polynucleotide paired terminal sequencing method and a kit. According to the invention, an extension primer for generating a complementary strand is specially designed to form an extension primer structure at least formed by connecting a first extension primer and a second extension primer into a whole, and a binding site of the first extension primer on template polynucleotide is designed to be located at the upstream of a binding site of the second extension primer. In an environment with strand displacement activity, the extension strand of the upstream extension primer displaces the extension strand of the downstream extension primer, and the displaced extension strand of the downstream extension primer is connected to the first extension primer through the second extension primer and then is combined to a template polynucleotide strand through the extension strand of the first extension primer, so that the template polynucleotide is obtained. The template polynucleotide chain is fixed on a solid support, so that the stability of the complementary chain in a cyclic sequencing environment is improved, and the sequencing quality is improved.
Owner:郑州思昆生物工程有限公司
Primer composition for analyzing intestinal flora and application thereof
ActiveUS20200165664A1Increased complexityReduce the ratioMicrobiological testing/measurementPreparing sample for investigationMicrobiologyIntestino-intestinal
Owner:SUZHOU PRECISION BIOTECH CO LTD
Y-shaped linker, kit and method for constructing RRBS library
PendingCN111945232AImprove sequencing qualityHigh outputMicrobiological testing/measurementLibrary creationEnzyme digestionGenetics
The invention provides a Y-shaped linker, a kit and a method for constructing an RRBS library. The Y-shaped linker comprises a methylation linker 1, namely 5'-acactctttccctacacgacgctcttccgatctn-3', and a methylation linker 2, namely 5'-n2n1' agatcggaagagcacacgtctgaactccagtcac-3', wherein the methylation linker 1 and the methylation linker 2 are annealed to form the Y-shaped linker; the bases c inthe methylation linker 1 and the methylation linker 2 are methylationmodified c; n1 and n1' are complementary paired protective base sequences; the number of bases of n1 or n1' is in a range of 6-12;n2 is a cohesive terminal base of an enzyme cutting site; and the number of bases of n2 is in a range of 1-5. The linker is helpful for improving sequencing data output, and also improves the evaluation and accuracy of enzyme digestion efficiency.
Owner:天津诺禾医学检验所有限公司
Highly efficient and rapid homogenized full-length cDNA library construction method
InactiveCN108193284AEasy to operateHigh detection throughputLibrary creationOligonucleotidePolymerase chain reaction
The invention discloses a highly efficient and rapid homogenized full-length cDNA library construction method, which comprises the following steps: under the guide of PCR (Polymerase Chain Reaction) primers and oligonucleotide primrs, mDNA is reversely transcribed by a reverse transcriptase, so that first strand cDNA can be synthesized; when the purification of the first strand cDNA needs to be finished, the first strand cDNA is put onto ice to terminate reaction, the first strand cDNA is then put into a test tube, the test tube is then put onto a preheated PCR instrument, the first strand cDNA utilizes the PCR primers and anchor primers again, an improved Cap-trapper method is utilized to synthesize and amplify second strand cDNA in a PCR instrument and the second strand cDNA is stayed overnight under the condition of 14 DEG C to 20 DEG C, Lambda bacteriophage packaging reaction is respectively carried out on the second strand cDNA, and the unamplified second strand cDNA library is stored under the condition of 1 DEG C to 4 DEG C for 12 to 18 days. When constructing a bacterial cDNA library, the method can utilize characteristics to carry out reverse transcription to provide a theoretical basis, the recombination rate is high, and the requirement of current biological development is met.
Owner:WUHAN IGENEBOOK BIOTECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com