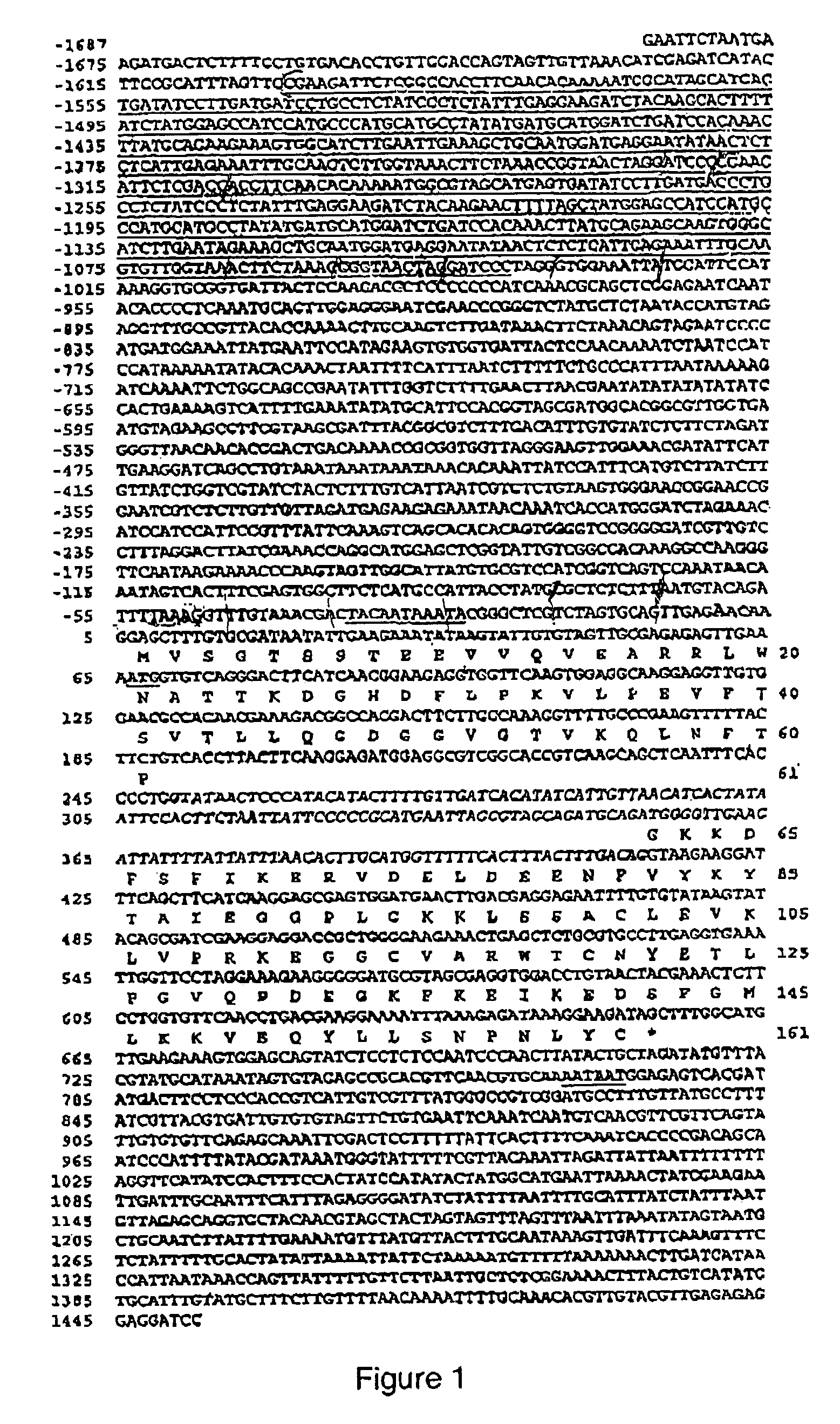
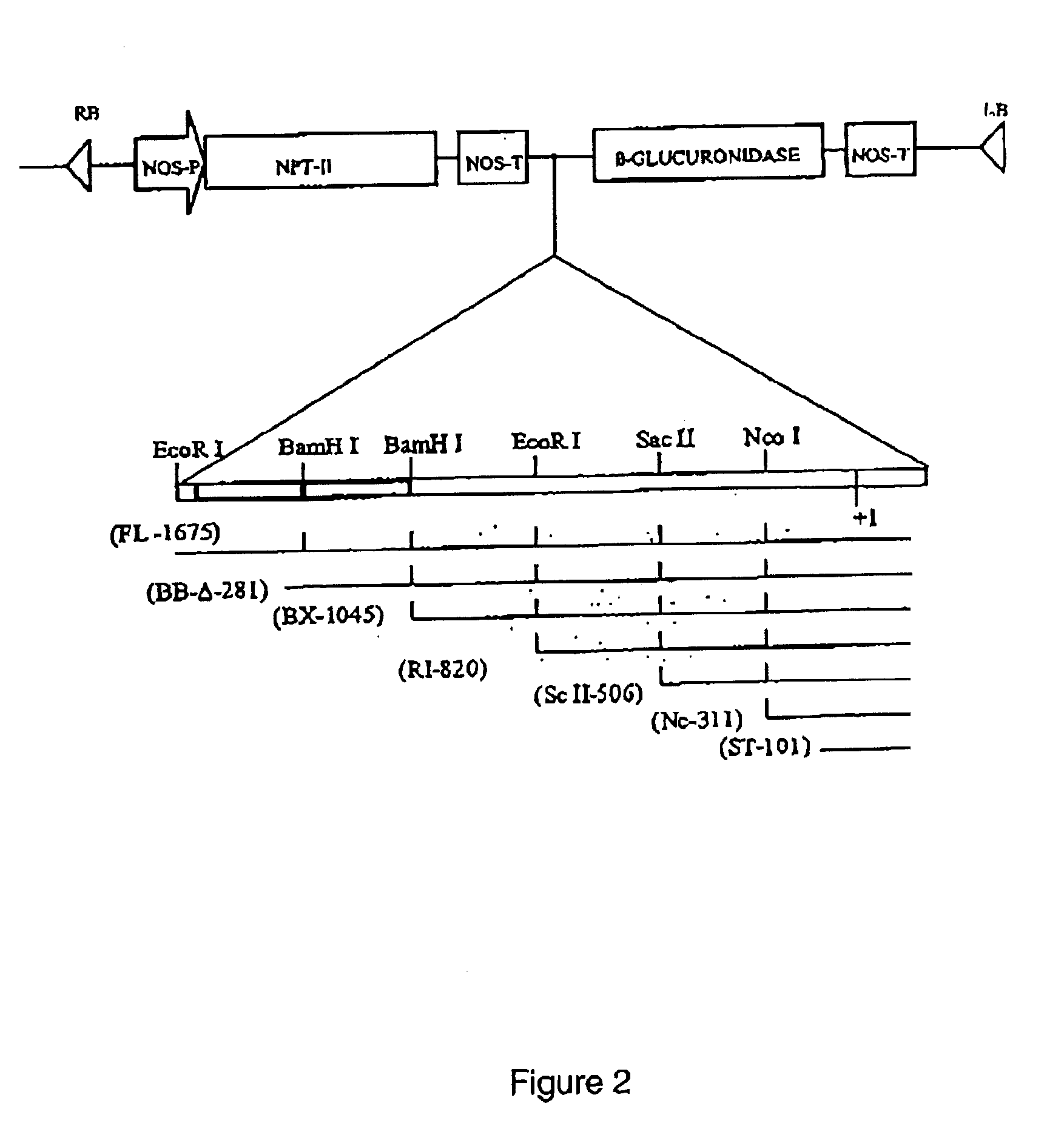
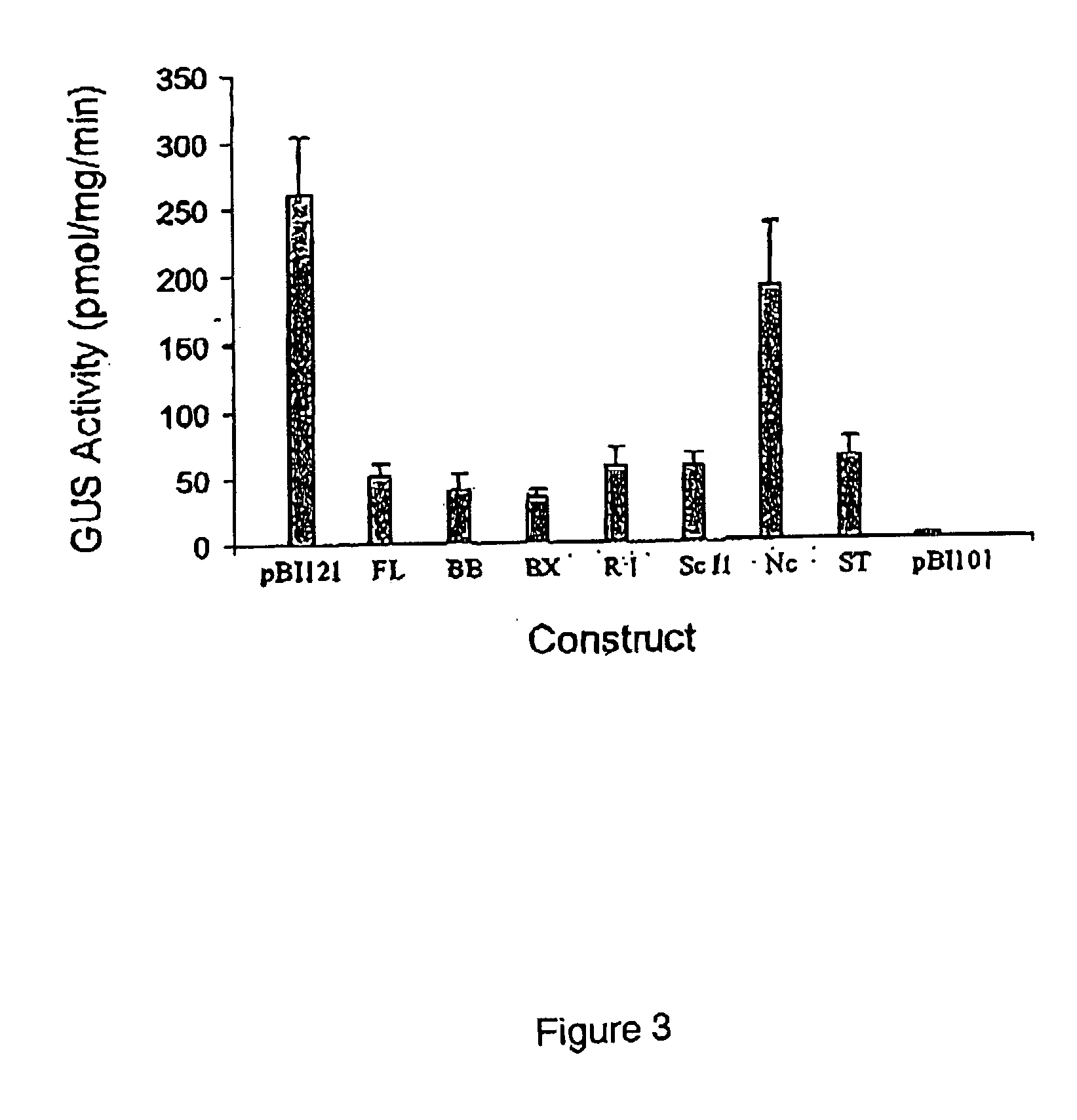
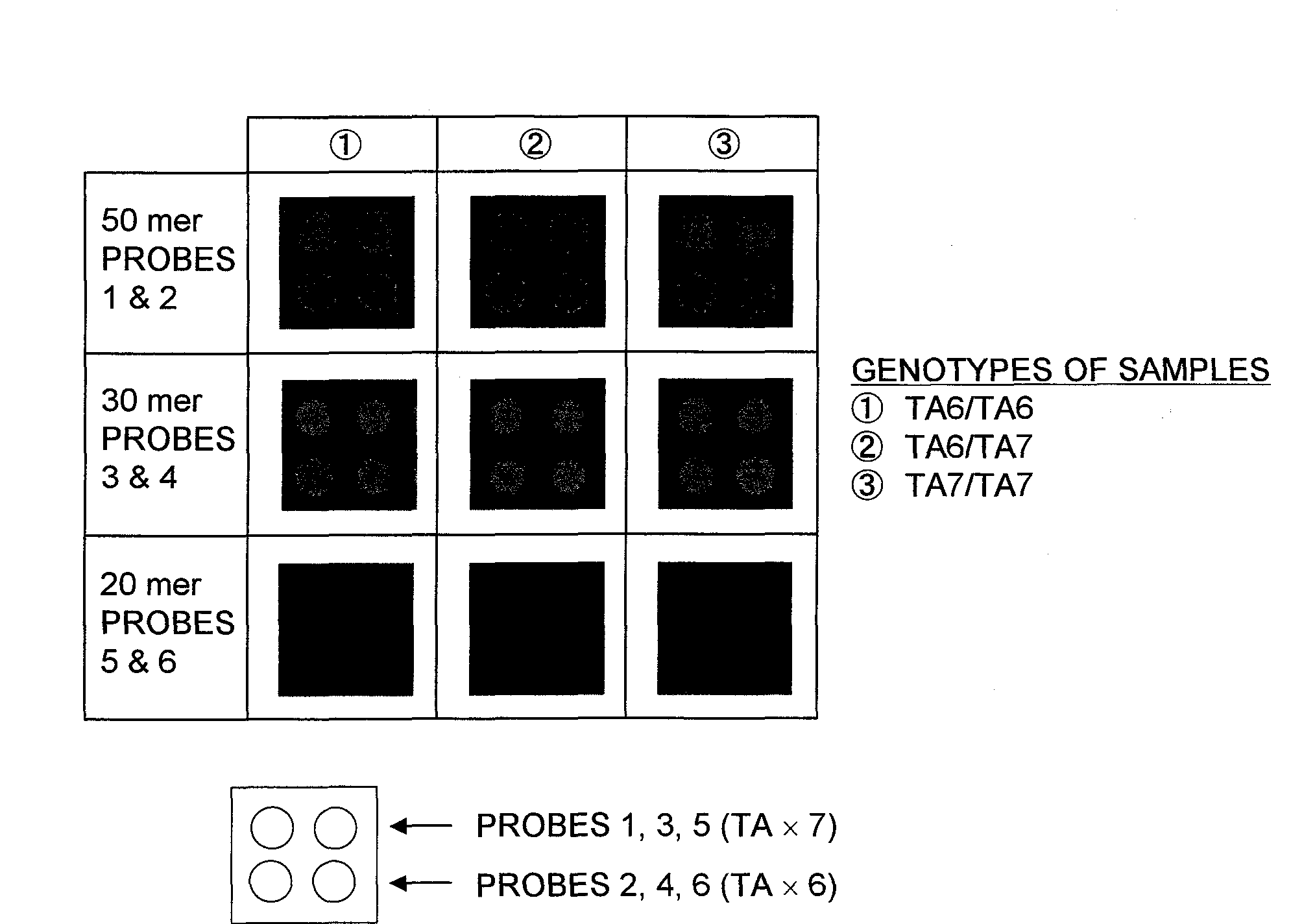
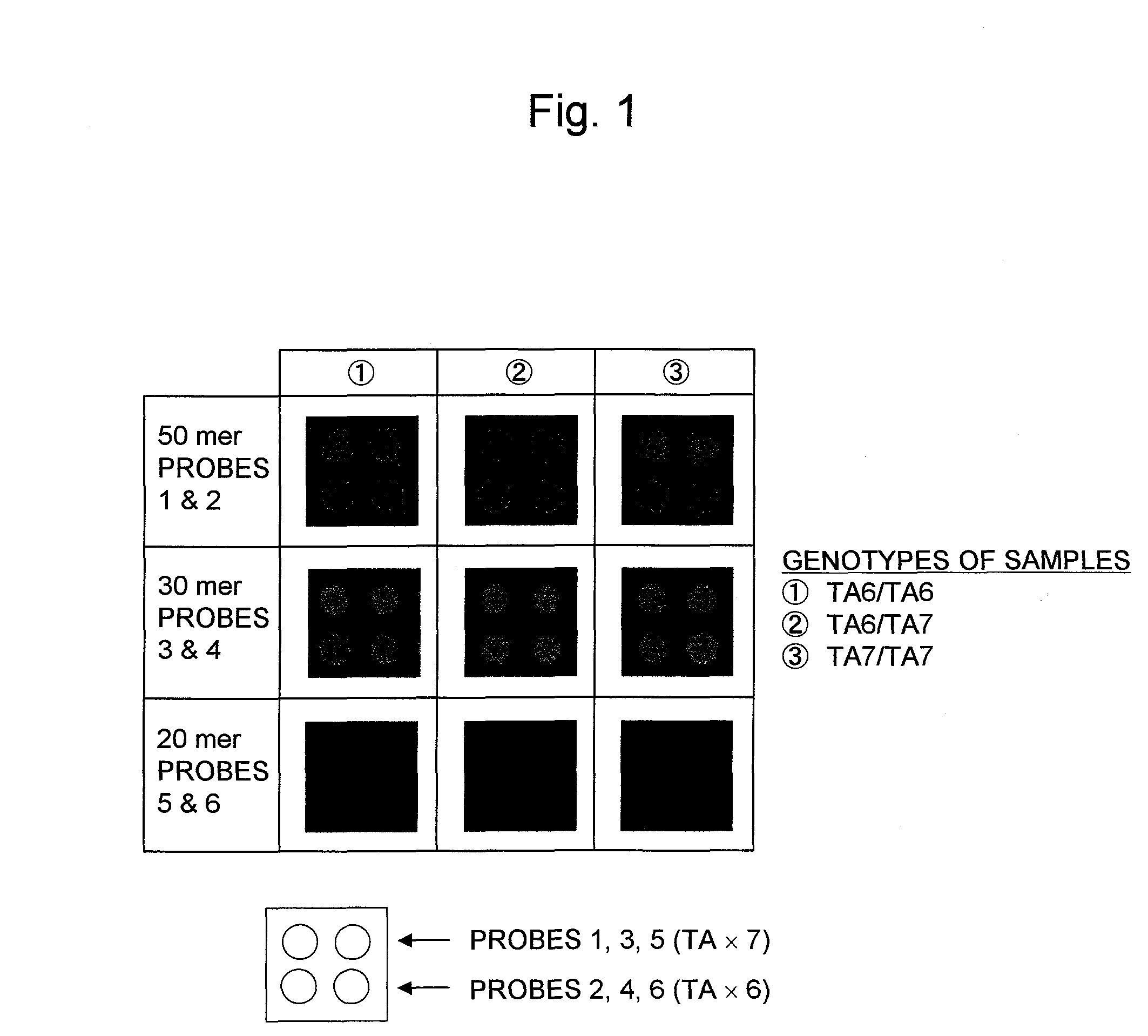
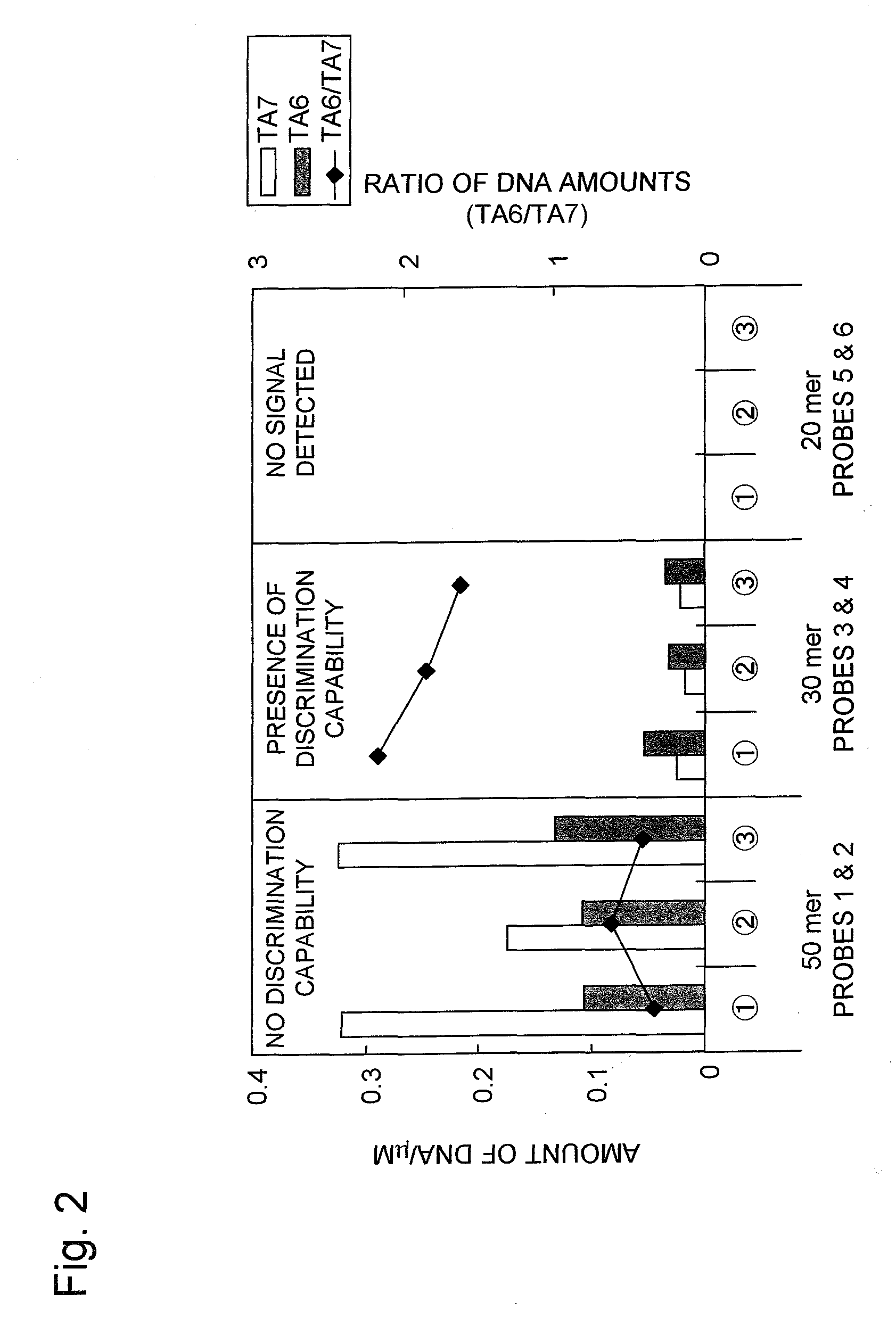
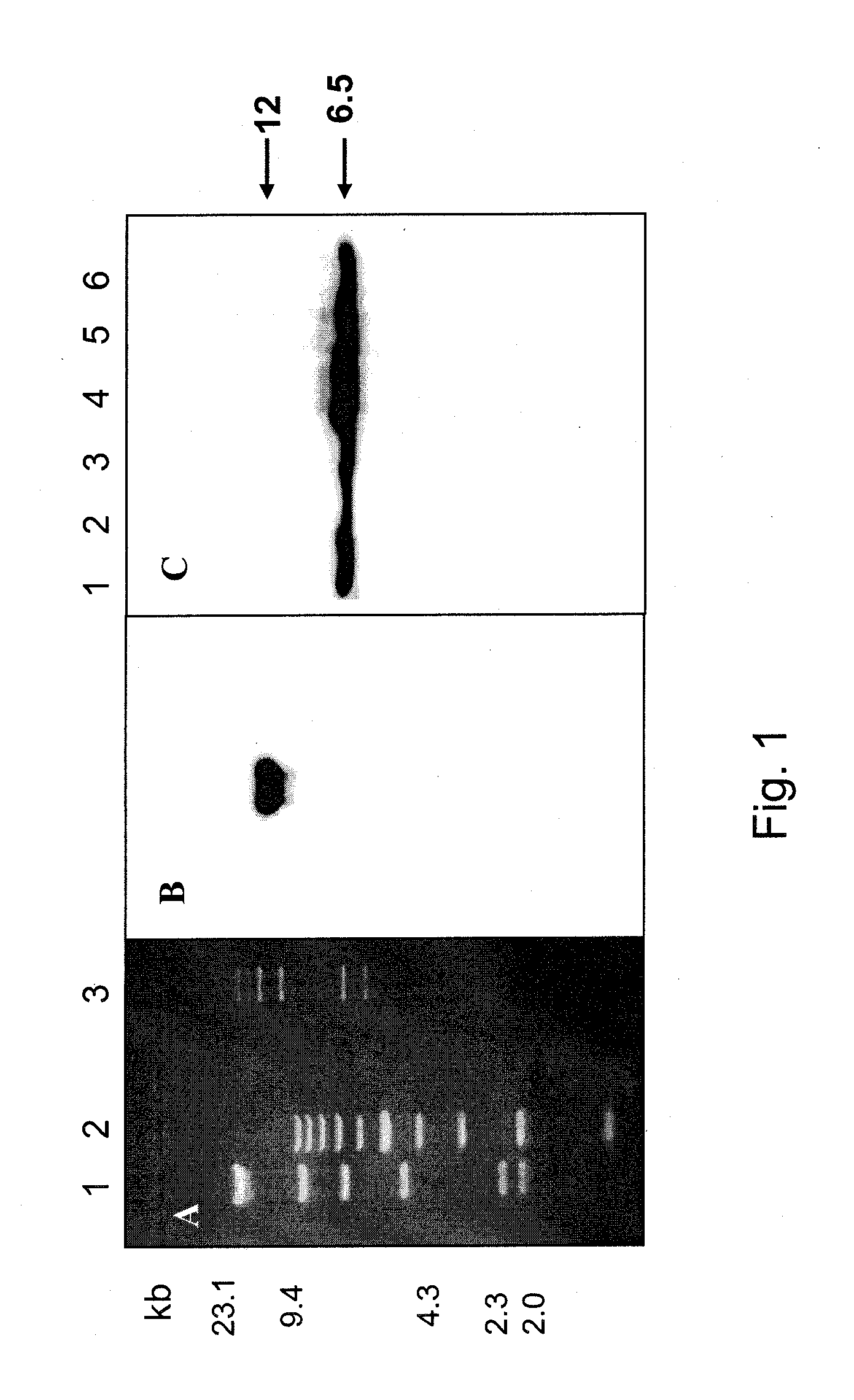
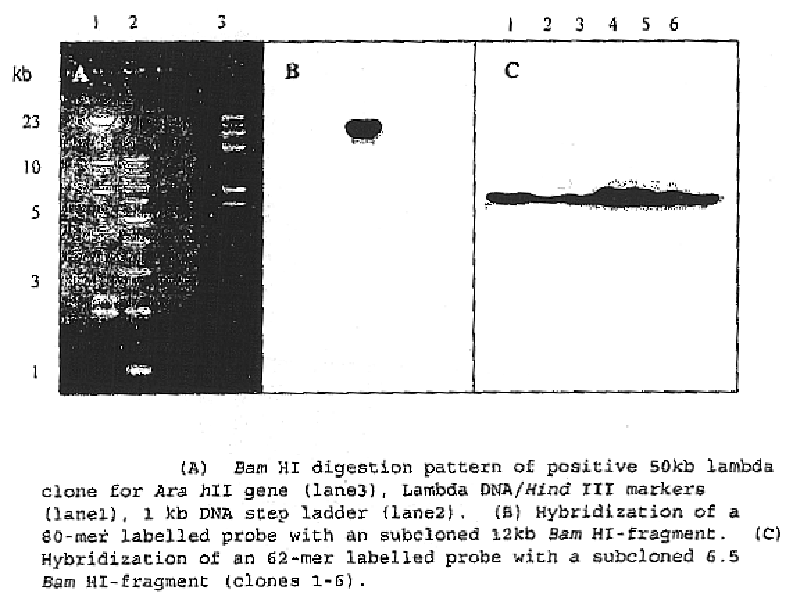
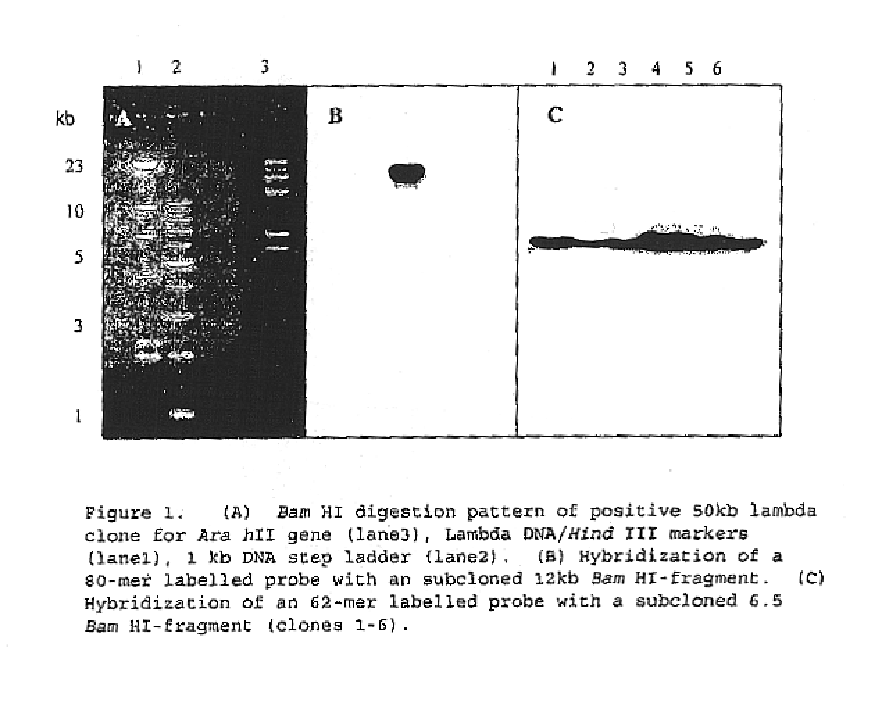
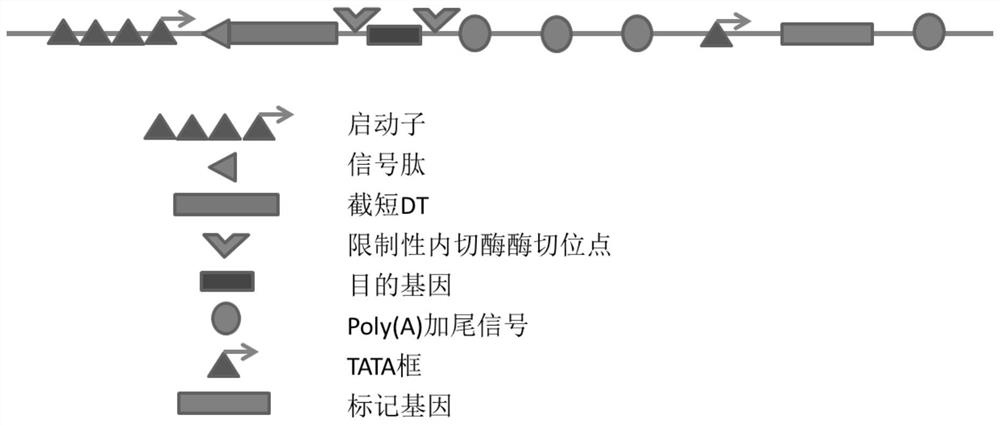
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
37 results about "TATA box" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In molecular biology, the TATA box (also called the Goldberg-Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The prokaryotic homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence.
Artificial promoter libraries for selected organisms and promoters derived from such libraries
InactiveUS7199233B1Strong impactNone is suitable for purposeSugar derivativesMicrobiological testing/measurementNucleotideNucleobase
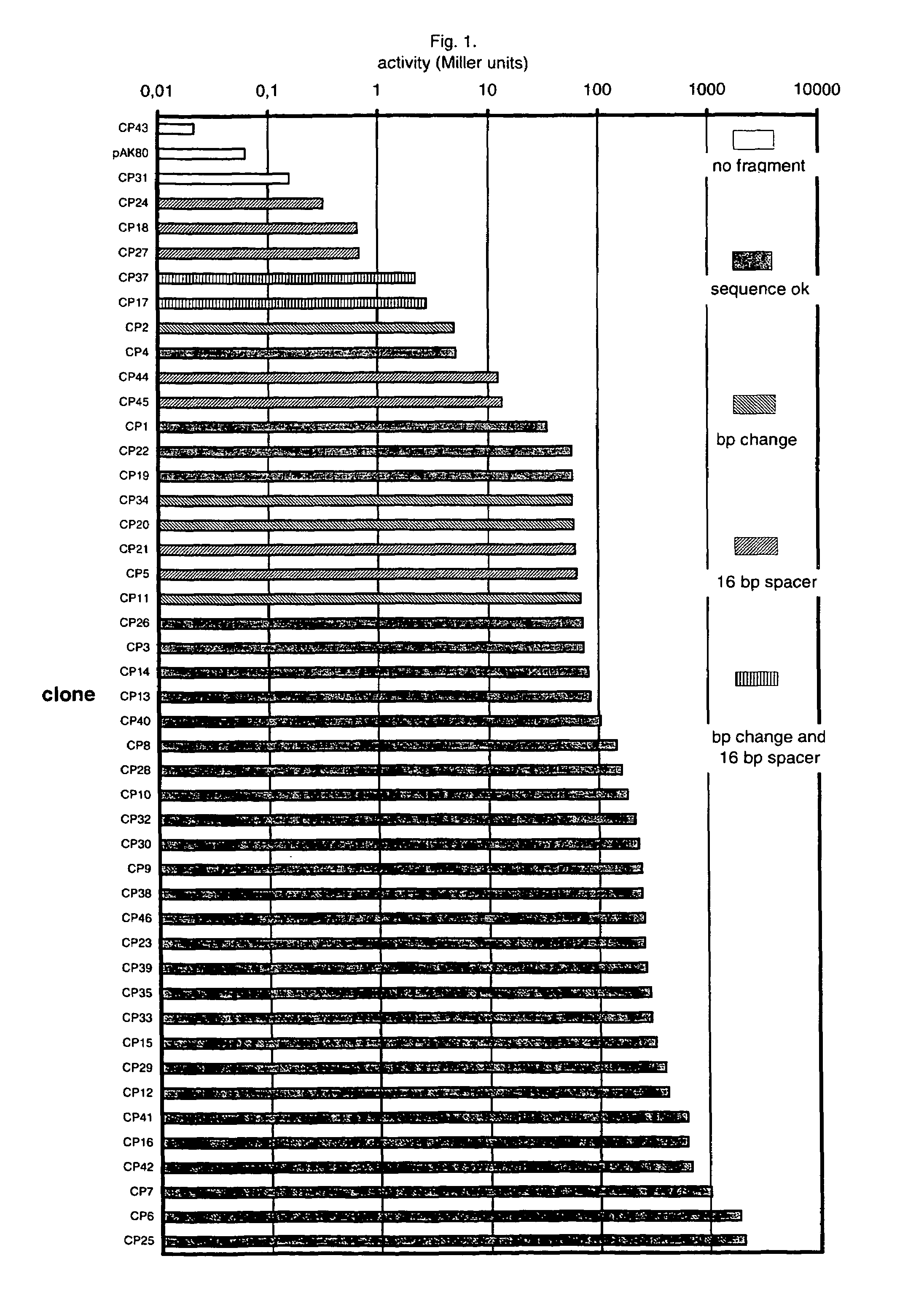
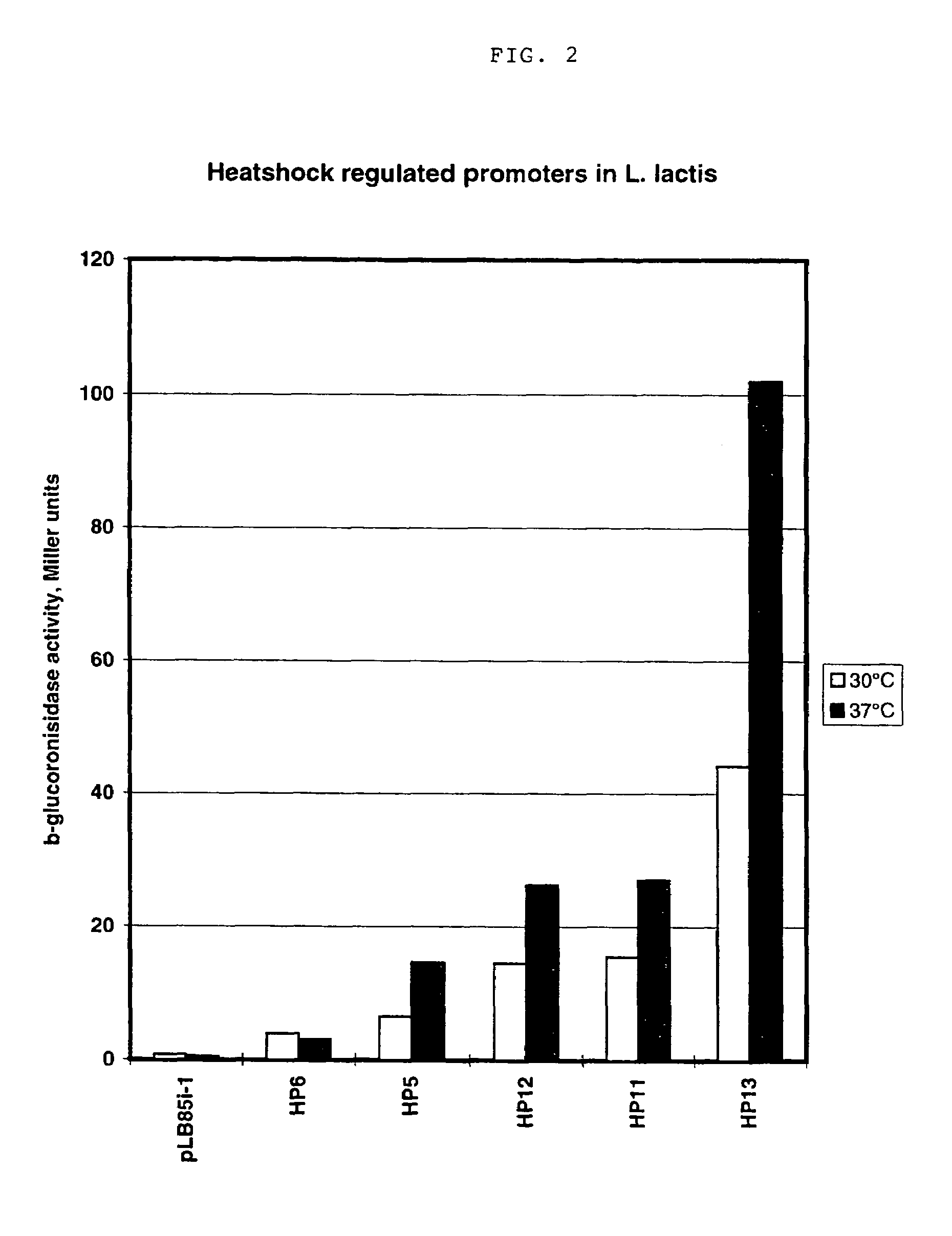
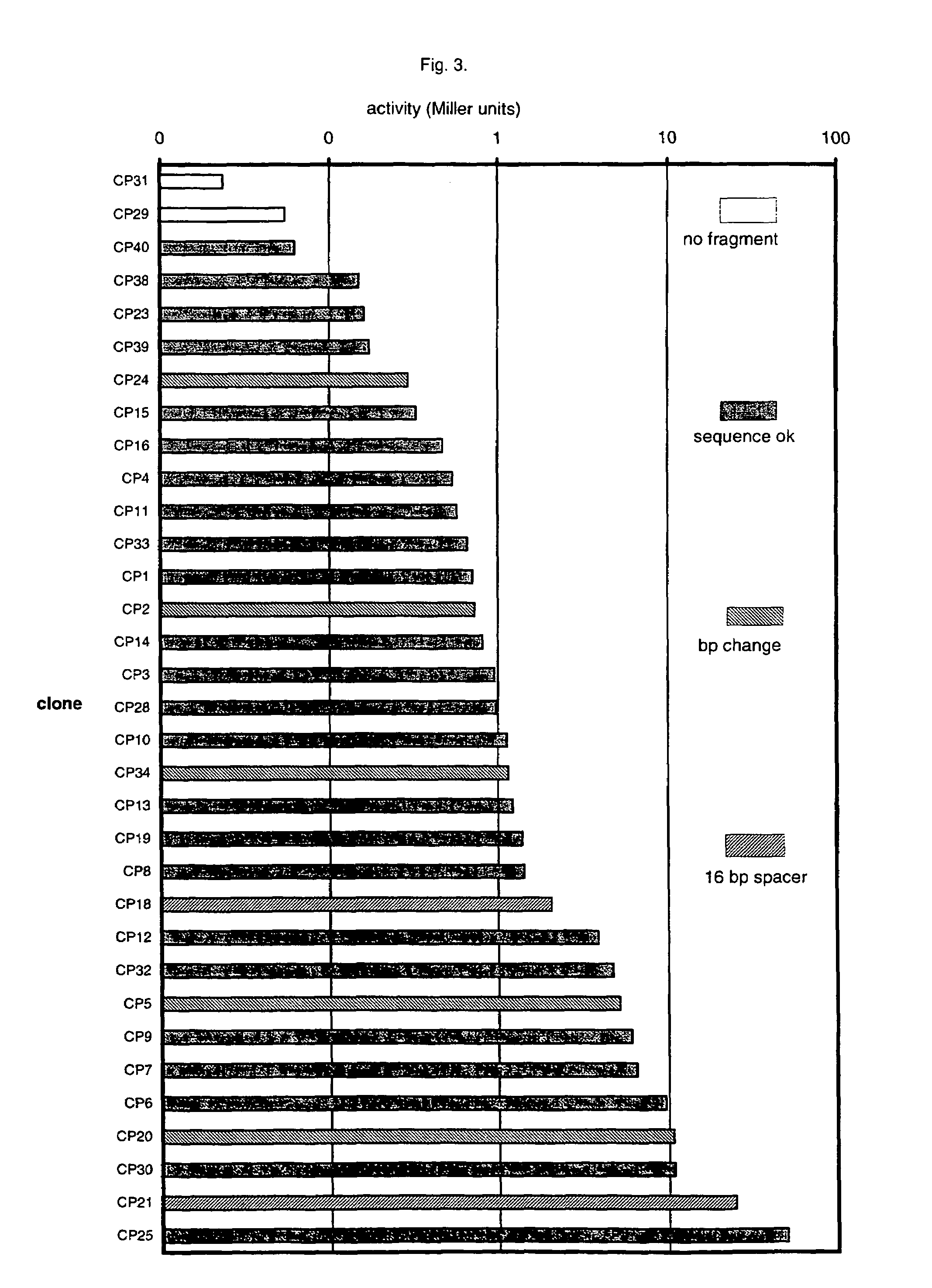
An artificial promoter library (or a set of promoter sequences) for a selected organism or group of organisms is constructed as a mixture of double stranded DNA fragments, the sense strands of which comprise at least two consensus sequences of efficient promoters from said organism or group of organisms, or parts thereof comprising at least half of each, and surrounding intermediate nucleotide sequences (spacers) of variable length in which at least 7 nucleotides are selected randomly among the nucleobases A, T, C and G. The sense strands of the double stranded DNA fragments may also include a regulatory DNA sequence imparting a specific regulatory feature, such as activation by a change in the growth conditions, to the promoters of the library. Further, they may have a sequence comprising one or more recognition sites for restriction endonucleases added to one or both of their ends. The selected organism or group of organisms may be selected from prokaryotes and from eukaryotes; and in prokaryotes the consensus sequences to be retained most often will comprise the −35 signal (−35 to −30): TTGACA and the −10 signal (−12 to −7): TATAAT or parts of both comprising at least 3 conserved nucleotides of each, while in eukaryotes said consensus sequences should comprise a TATA box and at least one upstream activation sequence (UAS). Such artificial promoter libraries can be used, e.g., for optimizing the expression of specific genes in various selected organisms.
Owner:JENSEN PETER RUHDAL +1
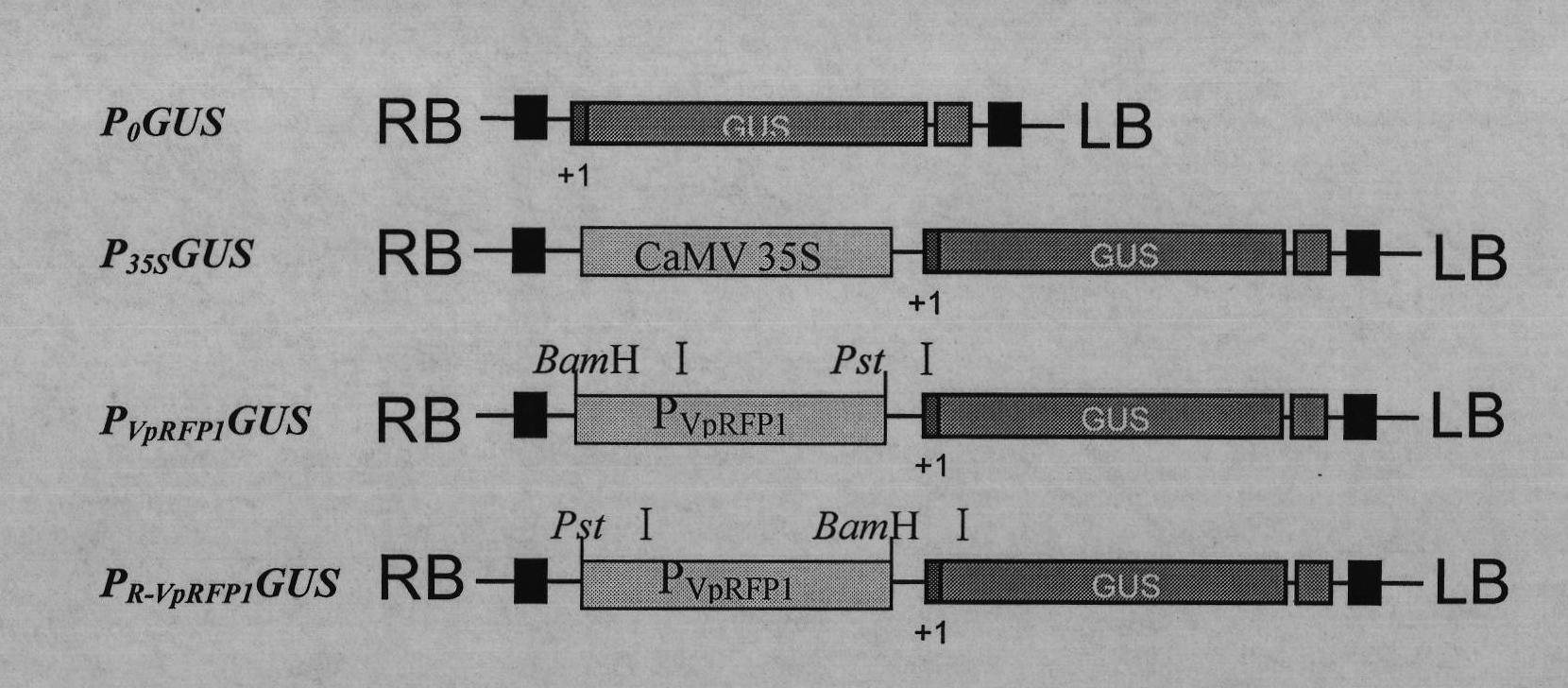
Grapevine powdery mildew resistance transcription factor gene VpRFP1 promoter sequence and application thereof
InactiveCN102121005AMicrobiological testing/measurementVector-based foreign material introductionPlant hormoneDisease
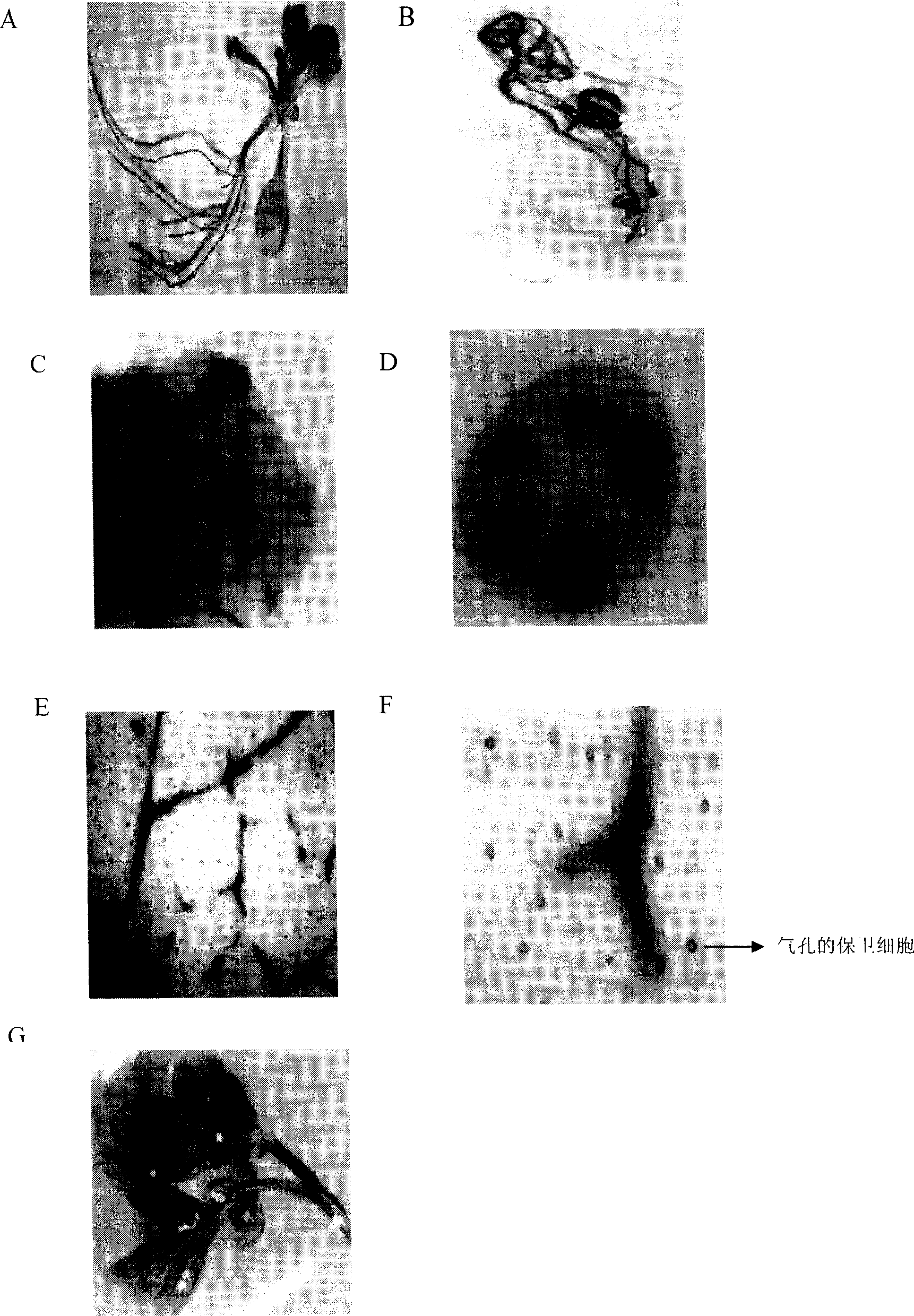
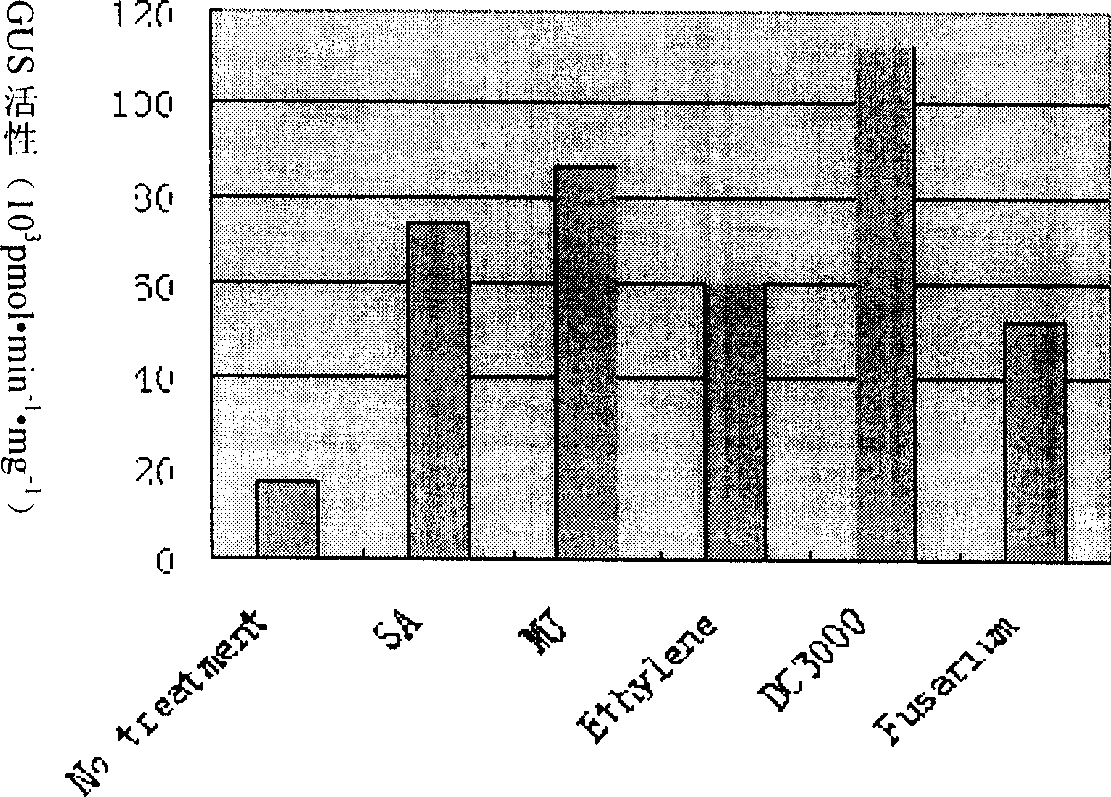
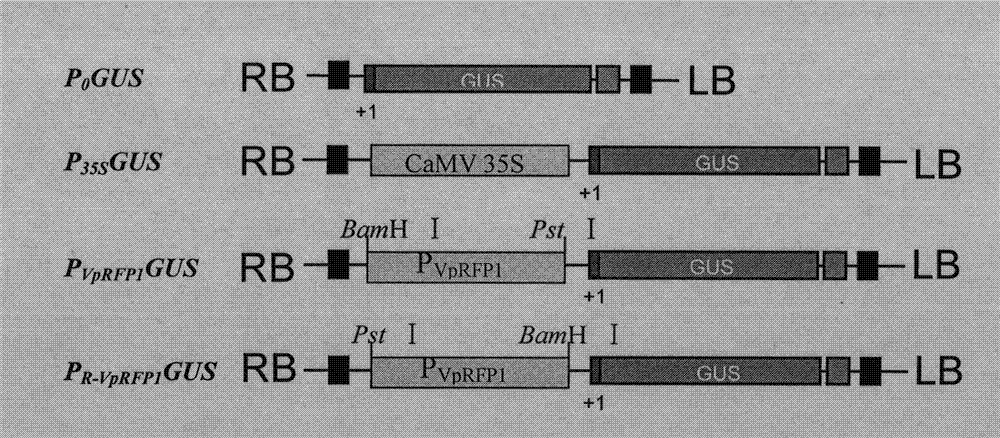
The invention relates to a grapevine powdery mildew resistance transcription factor gene VpRFP1 promoter sequence and application thereof. In the sequence, a full-length 1249bp promoter sequence of a Chinese wild vitis pseudoreticulata powdery mildew transcription factor gene VpRFP1 is cloned by adopting chromosomal walking in combination with a nested polymerase chain reaction (PCR) technology, wherein the sequence comprises 103 TATA-boxes and 27 CAAT-boxes, has 8 elements related to plant defense reaction, 14 photoreaction elements, 5 plant hormone response elements, and 14 other unknown elements. By adopting an agrobacterium tumefaciens-mediated instantaneously converted nicotiana benthamiana leaves analyzing method, the function of the Chinese wild vitis pseudoreticulata powdery mildew transcription factor gene VpRFP1 promoter proves that: the promoter has stronger promotion activity, can actively respond to pathogen invasion and disease resistant signal substances, and can improve the disease resistance and high temperature resistance of wheat, rice, potato, corn, soybean, rape and other plants through a transgenic method.
Owner:NORTHWEST A & F UNIV
Cotton tissue specific and pathogenic bacterium inducing promoter and its use
InactiveCN1900281AUnderstand interactionReduce outputFermentationVector-based foreign material introductionBudGuard cell
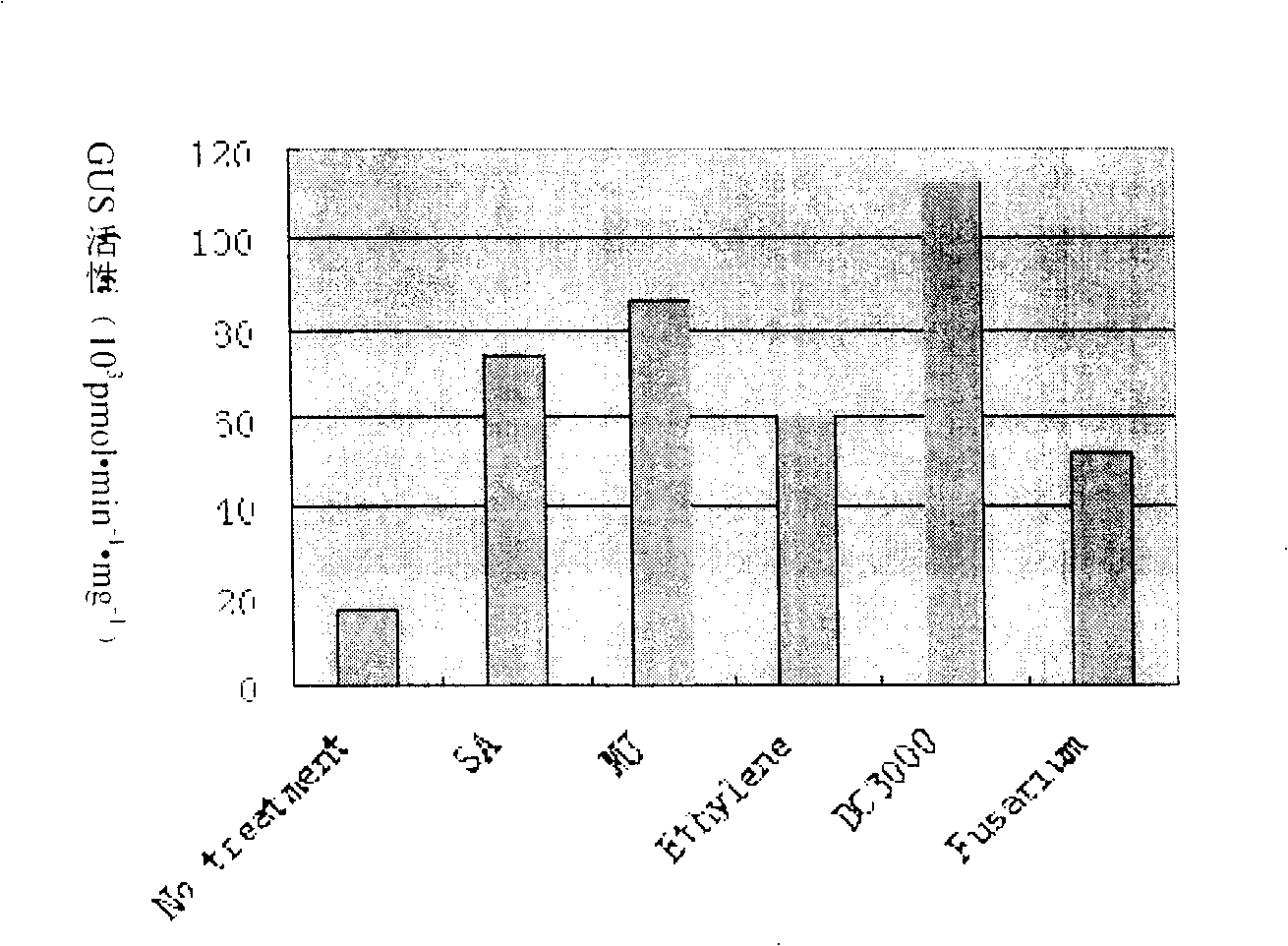
The present invention relates to cotton tissue specific and pathogenic bacterium inducing promoter and their application, and belongs to the field of biotechnology. The promoter contains CAAT-box, TATA-box, ethylene, methyl jasmonate, abscisic acid response element, pathogenic bacterium inducer response elements W boxes, GT-1, MYB, MYBST1, MYB1LEPR, etc. There are also specific root expressing elements to enhance the expression of GUS gene, which expresses specifically in the phloem of root, bud and stem, the vein of foliage and the guard cell of air pore. Paraffin section observation shows that GHNBS promoter expresses in phloem companion cell. After treatment with SA, MeJA, Ethylene, pathogenic bacteria of blight and pseudomonas syringae DC3000, GUS will have obviously raised activity, so that it is one organ specific and pathogenic bacterium relevant promoter.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Root-specific conifer gene promoter and its use
There is provided an isolated polynucleotide sequence useful as a promoter in plant root tissue. The sequence is derived from the promoter for the Western white pine gene PR10. The sequence comprises at least 45 nucleotidcs, and has at least two Dof elements which individually may be in a forward or inverted orientation. Each Dof element has a sequence NNNWAAAGNNN, wherein N is A, T, G, or, C, and, W is A or T. The Dof elements are separated from each other by between 10 and 20 nucleotides. The sequence also has a TATA box located between 15 and 25 nucleotides in the 3′ direction of the closest Dof element. In some instances, the polynucleotide has one Dof element with the sequence NNNWAAAGNNN and another with the sequence NNNCTTTWNNN. In some instances the polynucleotide sequence is one where the 5′ Dof element sequence is CTCTCTTTAAT, and another Dof element (which may be the next Dof element) sequence is TTTTAAAGGTT and the TATA box sequence is TACAATAAATA or TATAWAWA, wherein W is A or T. Also provided are sequences useful in modulating gene expression in stem and leaf tissue.
Owner:HER MAJESTY THE QUEEN & RIGHT OF CANADA REPRESENTED BY THE MIN OF NATURAL RESOURCES
Cre-lox based method for conditional RNA interference
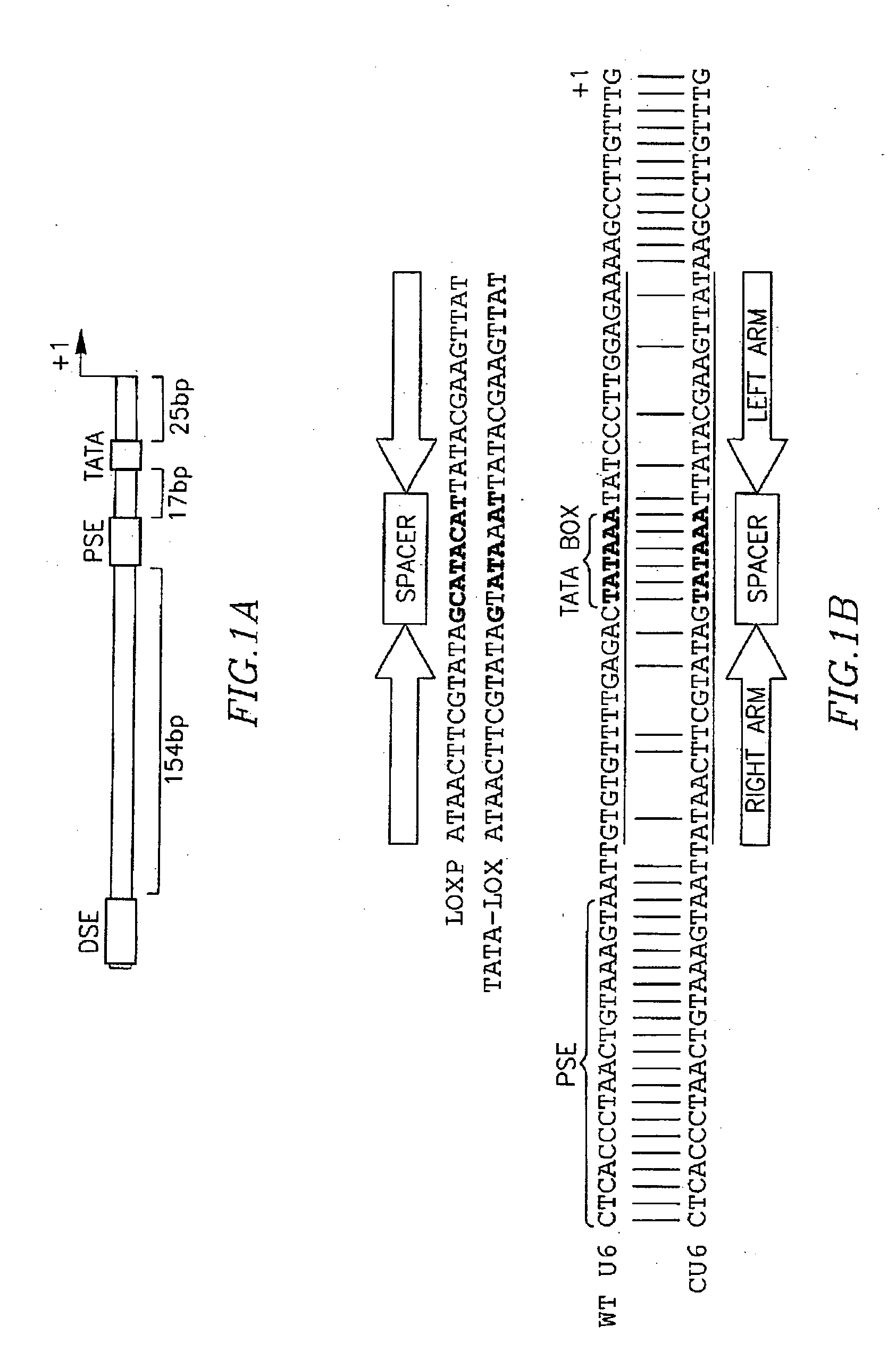
The present invention relates to vectors, compositions and methods for conditional, Cre-lox regulated, RNA interference. Vectors for use in conditional expression of a coding sequence based on a strategy in which the mouse U6 promoter is modified to include a hybrid between a LoxP site and a TATA box, and their use in conditional expression in transgenic mice are disclosed. The vectors allow for spatial and temporal control of miRNA expression in vivo.
Owner:MASSACHUSETTS INST OF TECH
Method for determining risk of adverse effect of irinotecan and kit for it
ActiveUS20080153093A1Sure easySimple and efficient to determineMicrobiological testing/measurementFermentationNucleic Acid ProbesIrinotecan
A method for determining the risk of adverse effects of irinotecan (CPT-11), a synthetic anticancer drug, by detecting polymorphisms in the TATA box within the promoter region of the UDP-glucuronosyl transferase gene. A kit for detecting the adverse effects of irinotecan containing at least one pair of nucleic acid probes.
Owner:TOYO KOHAN CO LTD +1
Down-regulation and silencing of allergen genes in transgenic peanut plants
InactiveUS20050114924A1Reduced and undetectable allergen protein contentReduced protein contentOther foreign material introduction processesPlant peptidesTransgeneAmino acid
An allergen-free transgenic peanut seed is produced by recombinant methods. Peanut plants are transformed with multiple copies of each of the allergen genes, or fragments thereof, to suppress gene expression and allergen protein production. Alternatively, peanut plants are transformed with peanut allergen antisense genes introduced into the peanut genome as antisense fragments, sense fragments, or combinations of both antisense and sense fragments. Peanut transgenes are under the control of the 35S promoter, or the promoter of the Ara h2 gene to produce antisense RNAs, sense RNAs, and double-stranded RNAs for suppressing allergen protein production in peanut plants. A full length genomic clone for allergen Ara h2 is isolated and sequenced. The ORF is 622 nucleotides long. The predicted encoded protein is 207 amino acids long and includes a putative transit peptide of 21 residues. One polyadenilation signal is identified at position 951. Six additional stop codons are observed. A promoter region was revealed containing a putative TATA box located at position−72. Homologous regions were identified between Ara h2, h6, and h7, and between Ara h3 and h4, and between Ara h1P41B and Ara h1P17. The homologous regions will be used for the screening of peanut genomic library to isolate all peanut allergen genes and for down-regulation and silencing of multiple peanut allergen genes.
Owner:DODO HORTENSE +3
Bidirectional promoters for small RNA expression
InactiveUS20060171924A1Efficient and multi-gene targeted RNA silencingPromote disseminationBiocideVectorsRNA SequenceTATA box
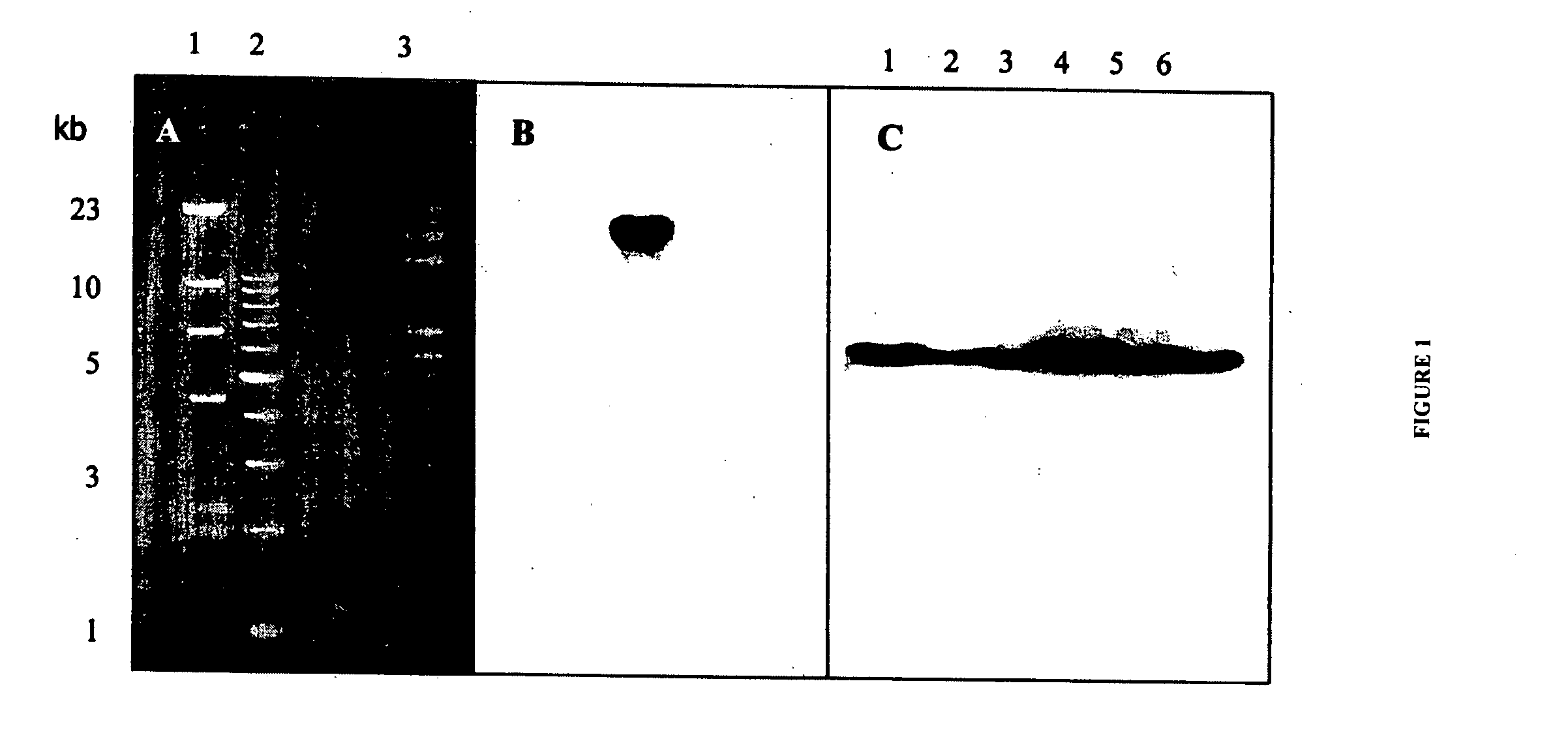
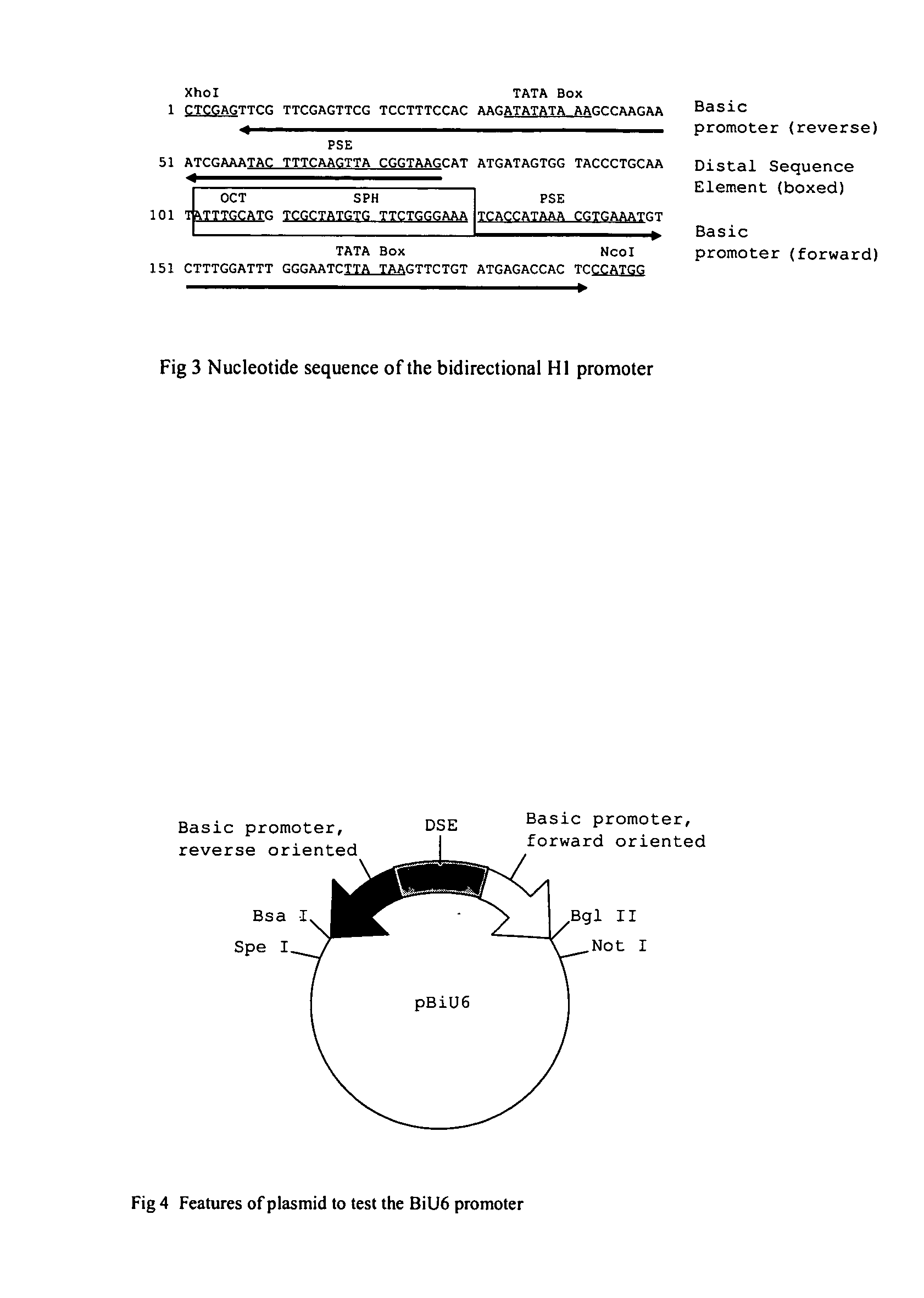
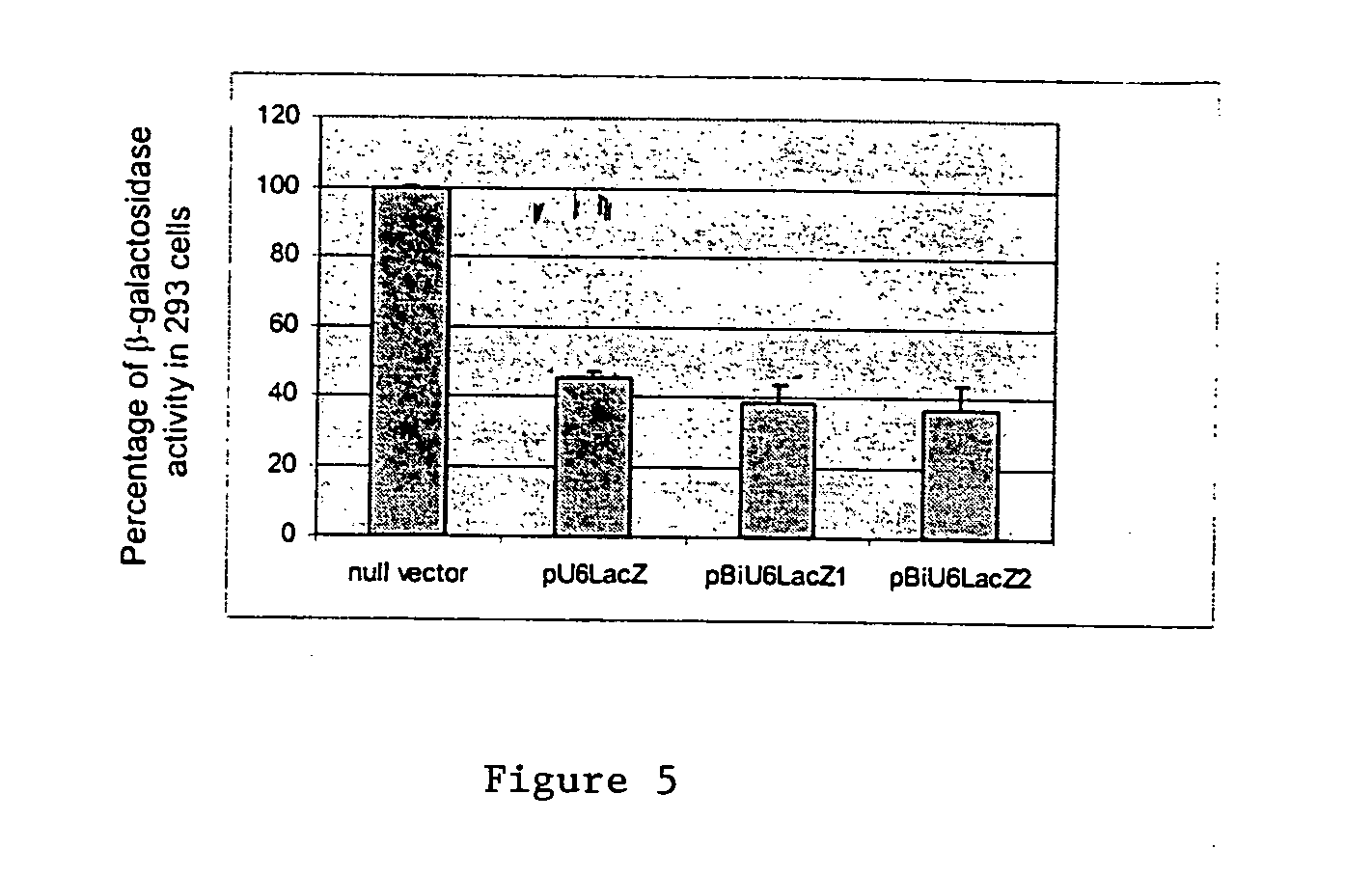
The invention provides bidirectional promoters for expressing two or more short RNA sequences from a single promoter. A particular embodiment of the bidirectional promoters of the invention include: 1) a Pol III promoter that contains a TATA box, a PSE and a DSE; and 2) a Pol III promoter that includes a PSE and a TATA box fused to the 5′ end of said DSE in reverse orientation. Vector embodiments are also disclosed comprising the novel bidirectional promoters of the invention, as well as methods of making and using these promoters.
Owner:LUO KE +2
Promoter batch capture method
ActiveCN107091929AEfficient separationEfficient acquisitionMicrobiological testing/measurementBiological testingConserved sequenceTATA box
The invention provides a promoter batch capture method. According to the promoter batch capture method, when a plant gene is transcribed, a complex of a 'DNA-protein factor' formed in a promoter area is utilized, an improved chromatin immunoprecipitation technology is applied, and DNA containing an unknown plant gene promoter fragment is sorted out. The promoter batch capture method is a technology for specifically capturing an unknown plant gene promoter fragment by utilizing the property that protein factors of OsTBP2 and OsTFIIB are tightly combined with promotion and transcribe of TATA box short-chain conserved sequence which on the promoter and through the generation of a specific antibody of the two factors and through the combination of the chromatin immunoprecipitation technology (ChIP). In the technology, by utilizing the advantages that most promoters contain one TATA box short-chain conserved sequence, and through the combination of technology that the unknown plant gene promoter fragment is separated through the newly developed chromatin immunoprecipitation technology (ChIP), large-scale capture of the plant promoter is easily achieved, and the technology has high commercial value.
Owner:RICE RES ISTITUTE ANHUI ACAD OF AGRI SCI
Promoter for expressing specificity of plant tissue and later development and application thereof
InactiveCN101831424AMitigating the impact of agronomic traitsIncrease concentrationBacteriaMicroorganism based processesPlant tissueOrgan Specificity
The invention relates to a promoter for the specificity expression and the later development expression of a plant tissue and application thereof, belonging to the biotechnology field. In the invention, a promoter sequence of a cotton cyclopropane fatty acid synthase with the length of 2.6kb is obtained. Place analysis proves that the promoter sequence contains a CAAT box, a TATA box, a cupric ion induced expression element, a pathogenicbacteria induced expression element, a dry response element, a cold stress response element, and the like as well as some root specificity expression elements and floral organ specificity elements. After GUS (beta-Glucuronidase) is dyed, a traditional gene is specially expressed at a later development stage by the promoter, and the expression parts are only restricted at a stem base part, a root and an anther.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Lilium chalcone synthase genes (chs) promoter as well as preparation method and use thereof
InactiveCN101230347AFermentationVector-based foreign material introductionHot start PCRChalcone synthase
The invention discloses a newly found DNA sequence of the promoter of the chalcone synthase (chs) gene and the manufacturing method and use for the same. The promoter is used for PCR with a joint from sorbonne, and clones 899bp of the upstream regulation sequence of chs gene by combining the characteristics of the nested PCR, TD PCR, hot-started PCR and two-step PCR. The analysis in the promoter database Plant CARE shows that, the sequence has a basically conservative region TACPyAT which expresses specifically in flowers, and has the main cis-acting elements with which the primary promote is provided such as a transcription initiation related TATA box, a transcription auxiliary CAAT box, a G box and CCAAT etc. The promoter can regulate the specific expression of the target gene in the plant, and can provide an effective promoter element for the constructing the plant expression vector, increase the expression of foreign gene, and minimum the biological energy consumption.
Owner:NORTHWEST A & F UNIV
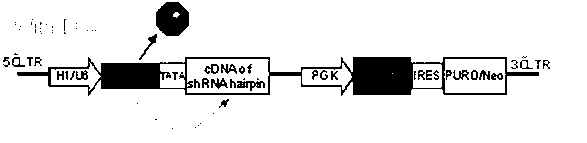
Inducible shRNA (short hairpin ribonucleic acid) lentiviral expression vector and construction method and application thereof
The invention provides an inducible shRNA (short hairpin ribonucleic acid) lentiviral expression vector and a construction method and application thereof. The inducible shRNA lentiviral expression vector comprises an shRNA expression frame and a tetracycline repressor protein expression frame, wherein the shRNA expression frame comprises a first promoter, a tetracycline response element being combined with tetracycline repressor protein, a TATA box and an shRNA coding sequence, and the tetracycline repressor protein expression frame comprises a second promoter, a tetracycline repressor protein expression gene, an internal ribosome entry site (IRES) DNA (deoxyribonucleic acid) sequence and a marker gene. Compared with an existing inducible shRNA expression vector, the inducible shRNA lentiviral expression vector has the advantages of being capable of rapidly establishing inducible shRNA expressing a silent gene in various cell strains by using a single vector, having no need of clone selection of cells, and being applicable to in vitro culture and in vivo study of cells.
Owner:傅开屏 +1
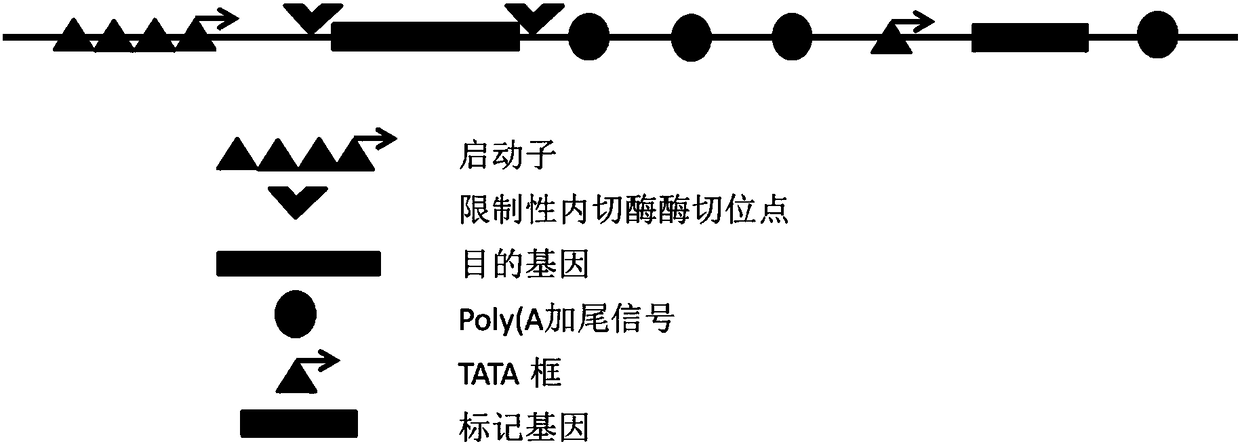
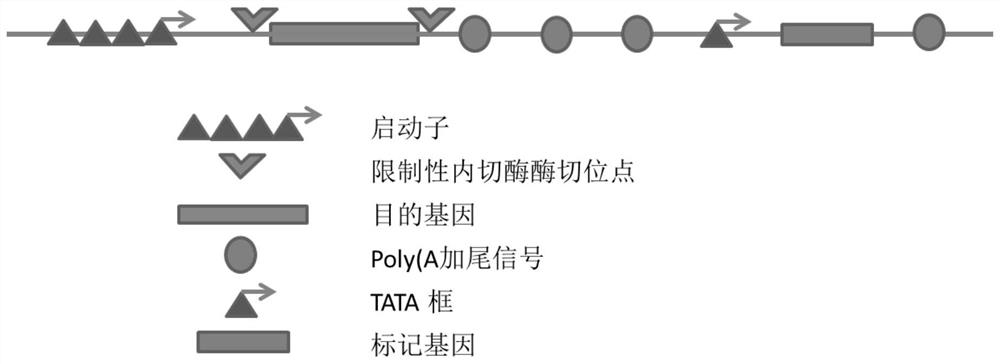
Expression vector of mammalian cell for industrial production
ActiveCN108315351ARead through to avoidVector-based foreign material introductionEnzyme digestionTATA box
The invention provides an expression vector for expressing protein / polypeptide by a mammalian host cell expression protein / polypeptide. The expression vector contains a marker gene and a restriction-endonuclease enzyme digestion site, wherein the restriction-endonuclease enzyme digestion site is used for being inserted into insertion of a target gene; the quantity of Poly(A) tailing signals behindthe target gene is 2 or more than 2; a starting component of the marker gene is only a TATA box. The upstream end of the selectable marker gene of the expression vector provided by the invention contains two or more Poly(A) tailing signals; the interference, to the activity of the downstream TATA box, of an upstream strong promoter is minimized, so as to drive the expression of the selectable marker gene. The expression vector provided by the invention can be used for generating stable and further high-yield cell lines for the expression of recombinant protein within a short time; moreover, the gene amplification does not need drug selection / medication; the selection can be directly carried out in the condition that any no drug induced amplification does not exists, and these cell lines can be used for stably expressing high-level recombinant protein for a long time, and is quite applicable to the industrialized production of the protein or the polypeptide.
Owner:济南海湾生物工程有限公司
AaPDR2 gene promoter as well as functional verification method and application thereof
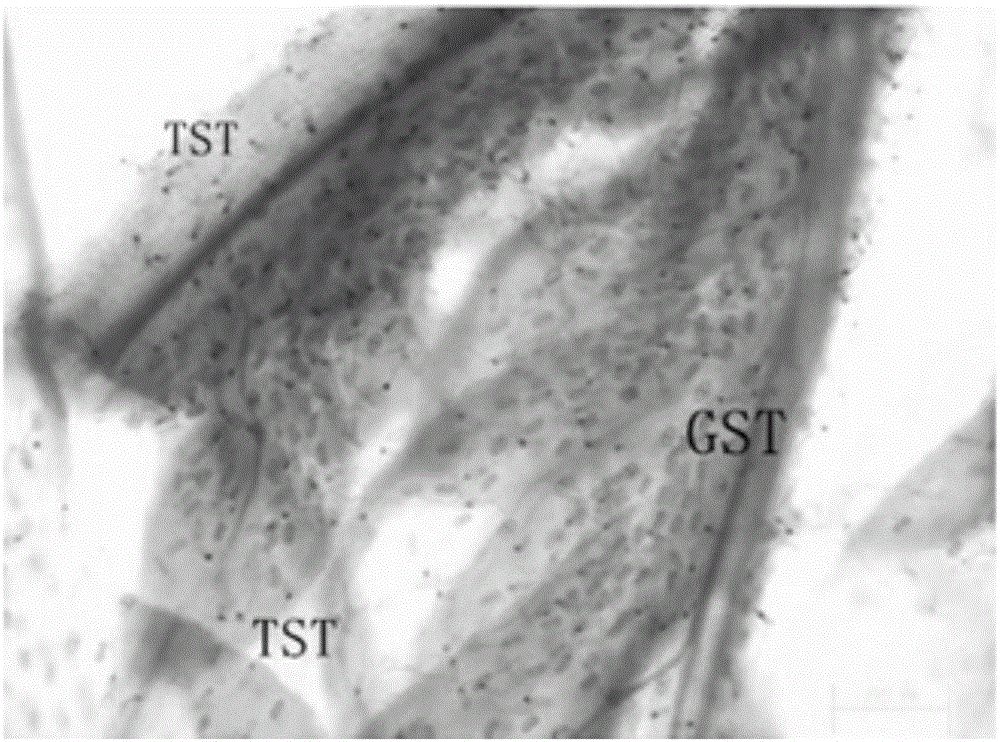
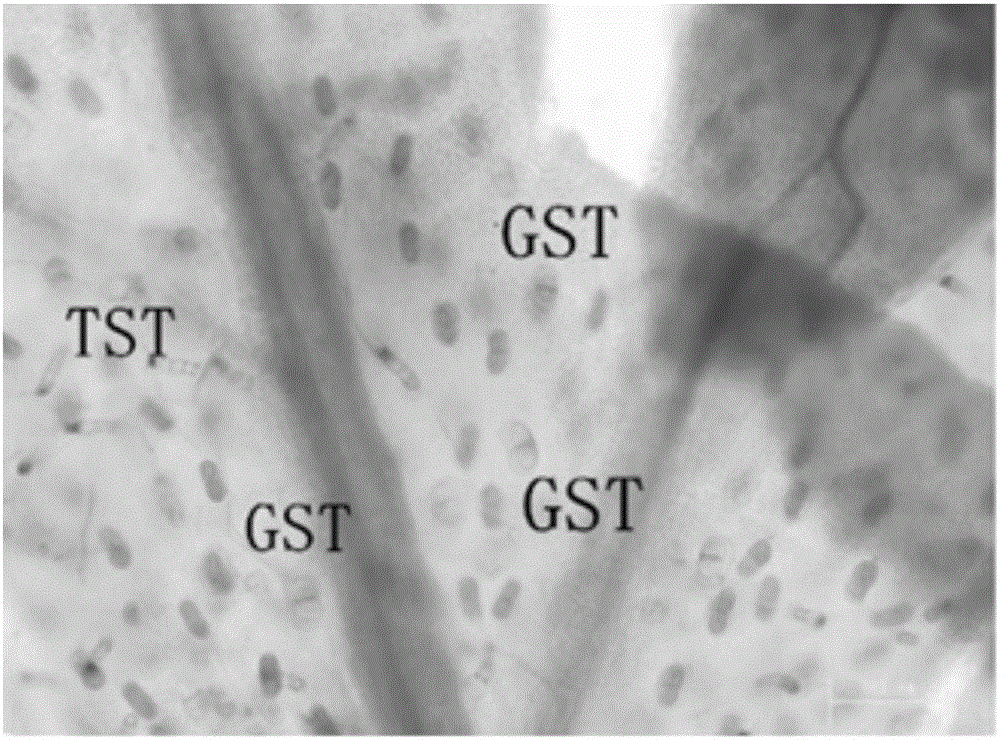
The invention relates to an AaPDR2 gene promoter as well as a functional verification method and application thereof. The AaPDR2 gene promoter comprises the following cis-acting elements: ABRE, BoxI, CAAT-box, CCAAT-box, CGTCA-motif, G-Box, G-box, GA-motif and TATA-box. Further, the DNA (Deoxyribonucleic Acid) sequence of the AaPDR2 gene promoter is shown in SEQ ID NO.1. The AaPDR2 gene promoter has a function of predominantly expressing genes guided by the AaPDR2 gene promoter in glandular secretory trichome and T shape trichome of young leaves of Artemisia annua L., and the function is confirmed by a GUS reporter gene. Therefore, the AaPDR2 gene promoter disclosed by the invention has important significance for genetic engineering breeding for expressing and generating metabolites by using plant glandular trichome tissues.
Owner:SHANGHAI JIAO TONG UNIV
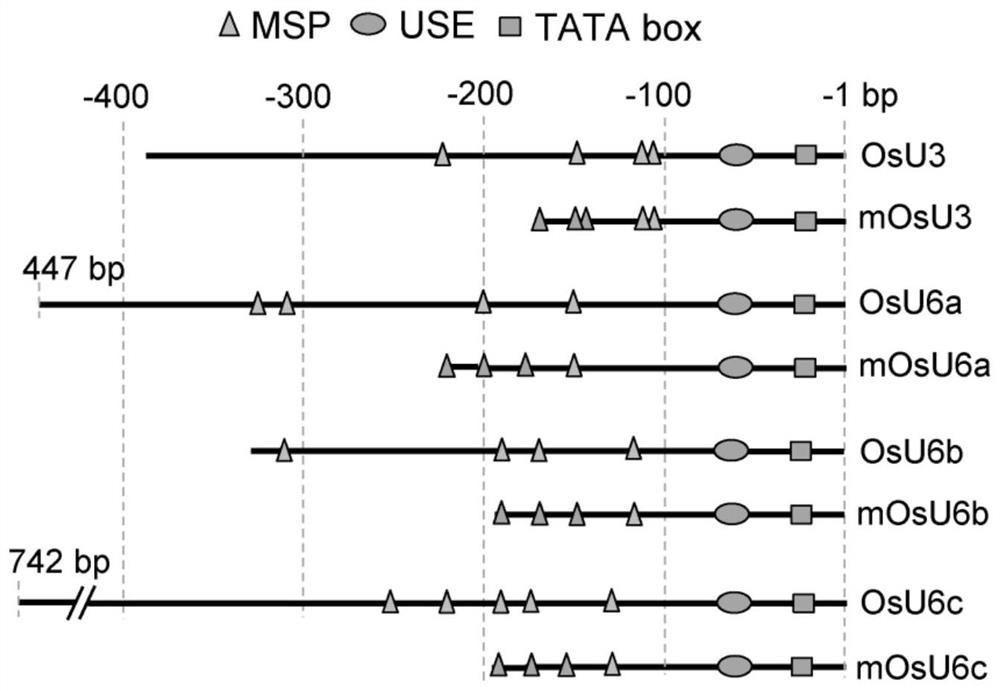
Short small-nuclear RNA promoter and construction method and application thereof in genome editing
ActiveCN111019946AImprove editing efficiencyShorten the lengthVector-based foreign material introductionAngiosperms/flowering plantsWild typeNuclear gene
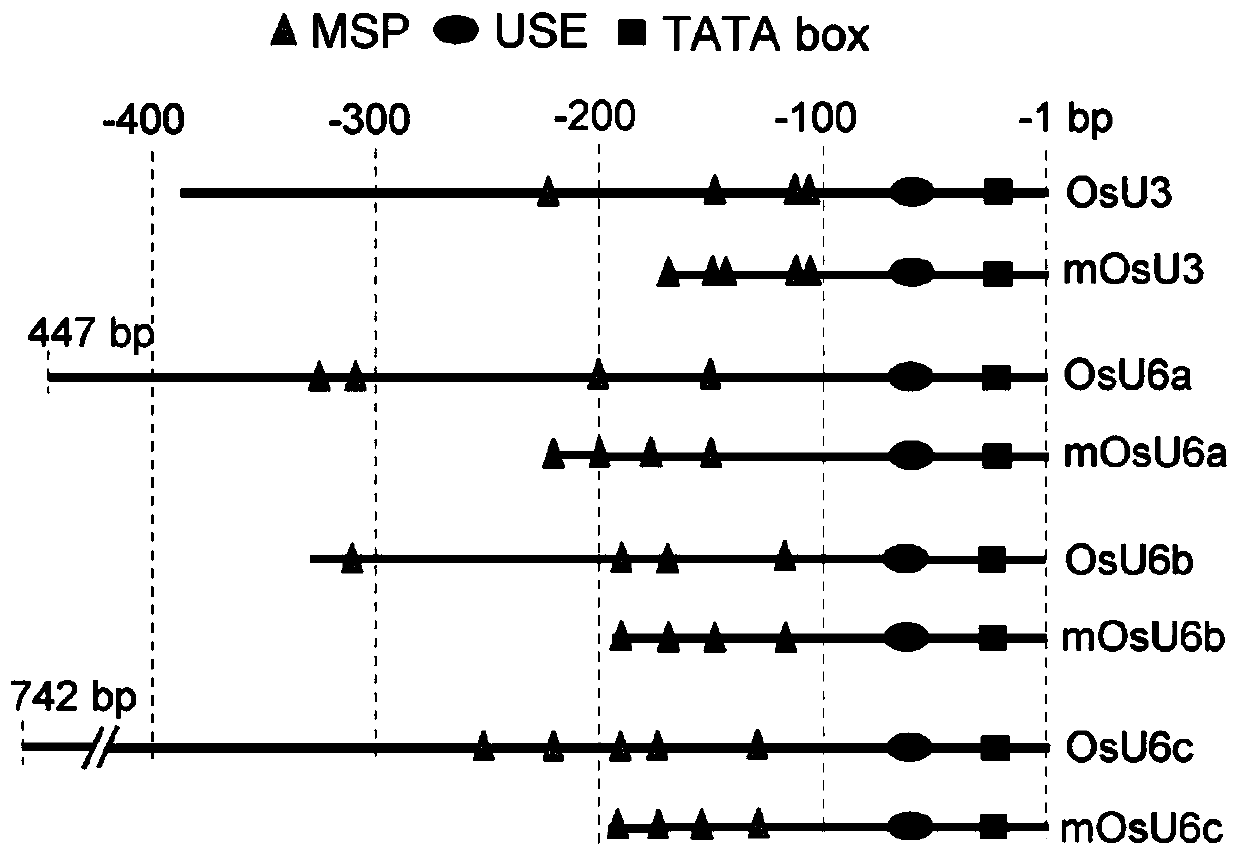
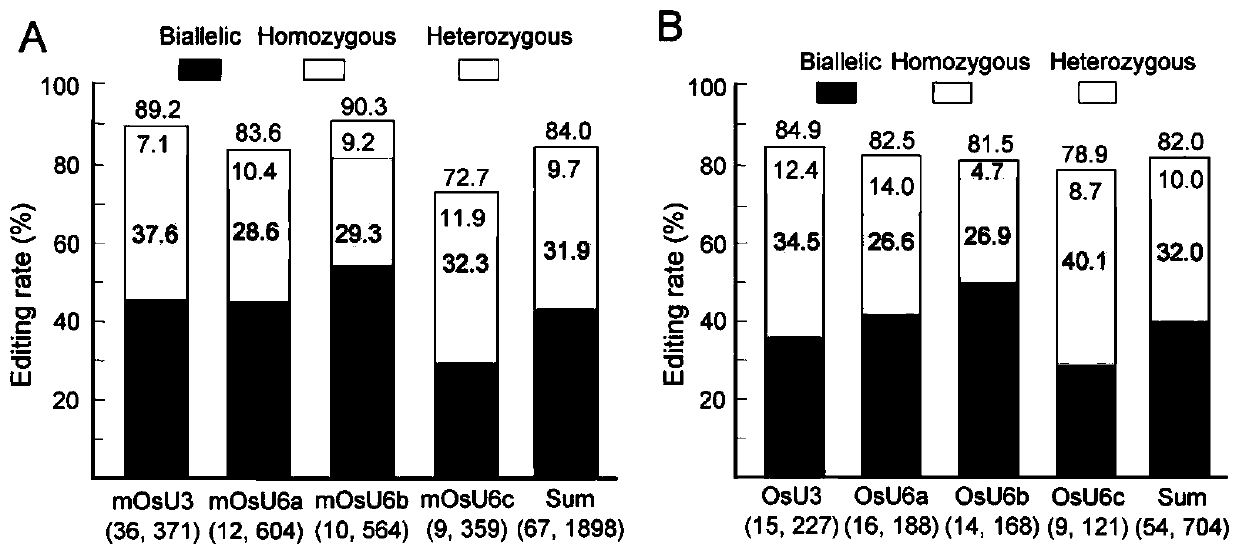
The invention provides a short small-nuclear RNA promoter as well as a construction method and application thereof. The length of the short small-nuclear RNA promoter is 150-300bp, and the short small-nuclear RNA promoter sequentially contains one TATA box conservative element, one USE conservative element and at least one MSP conservative element in the direction from an end 3' to an end 5'. Compared with an existing wild type small-nuclear RNA gene promoter, the short small-nuclear RNA promoter constructed in the invention has the advantages that the promoter has stronger transcriptional activity and obviously higher editing efficiency of driving the sgRNA and is shorter, the length of each sgRNA expression cassette can be reduced, the efficiency of constructing a vector by multi-targetsgRNA cloning is improved, the simultaneous editing of multiple targets and multiple genes is facilitated, and the promoter has a good application prospect in the aspect of genome editing.
Owner:SOUTH CHINA AGRI UNIV
Down-regulation and silencing of allergen genes in transgenic peanut seeds
InactiveUS20110004959A1Sugar derivativesOther foreign material introduction processesBiotechnologyNucleotide
An allergen-free transgenic peanut seed is produced by recombinant methods. Peanut plants are transformed with multiple copies of each of the allergen genes, or fragments thereof, to suppress gene expression and allergen protein production. Alternatively, peanut plants are transformed with peanut allergen antisense genes introduced into the peanut genome as antisense fragments, sense fragments, or combinations of both antisense and sense fragments. Peanut transgenes are under the control of the 35S promoter, or the promoter of the Ara h2 gene to produce antisense RNAs, sense RNAs, and double-stranded RNAs for suppressing allergen protein production in peanut plants. A full length genomic clone for allergen Ara h2 is isolated and sequenced. The ORF is 622 nucleotides long. The predicted encoded protein is 207 amino acids long and includes a putative transit peptide of 21 residues. One polyadenilation signal is identified at position 951. Six additional stop codons are observed. A promoter region was revealed containing a putative TATA box located at position −72. Homologous regions were identified between Ara h2, h6, and h7, and between Ara h3 and h4, and between Ara h1P41B and Ara h1P17. The homologous regions will be used for the screening of peanut genomic library to isolate all peanut allergen genes and for down-regulation and silencing of multiple peanut allergen genes.
Owner:NGATEGEN INC +1
Down-regulation and silencing of allergen genes in transgenic peanut seeds
Owner:NGATEGEN INC +1
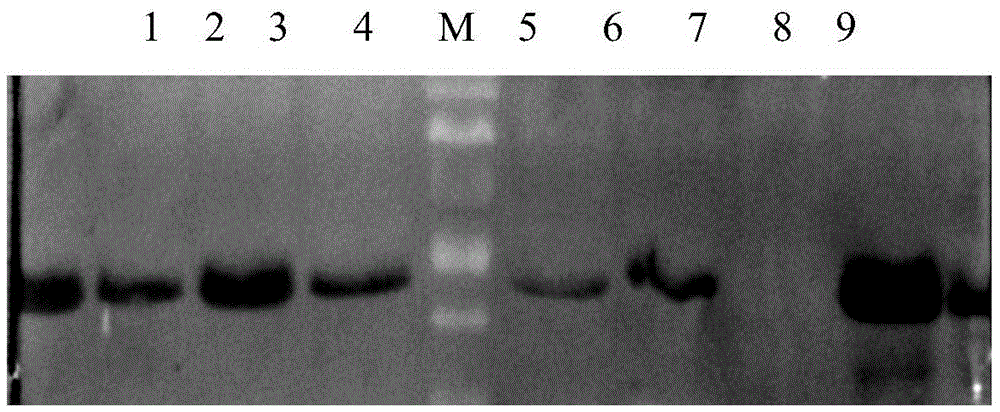
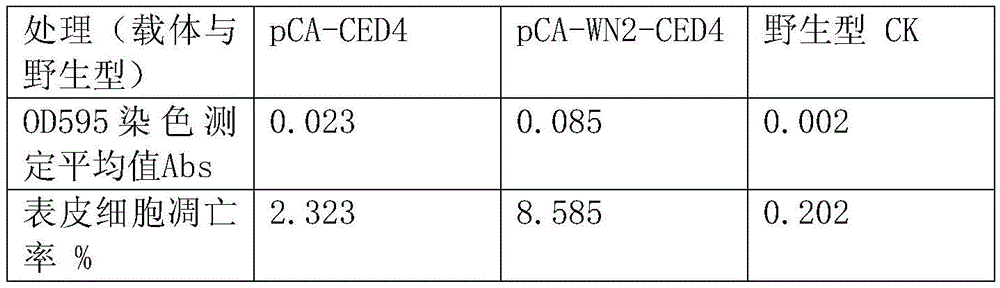
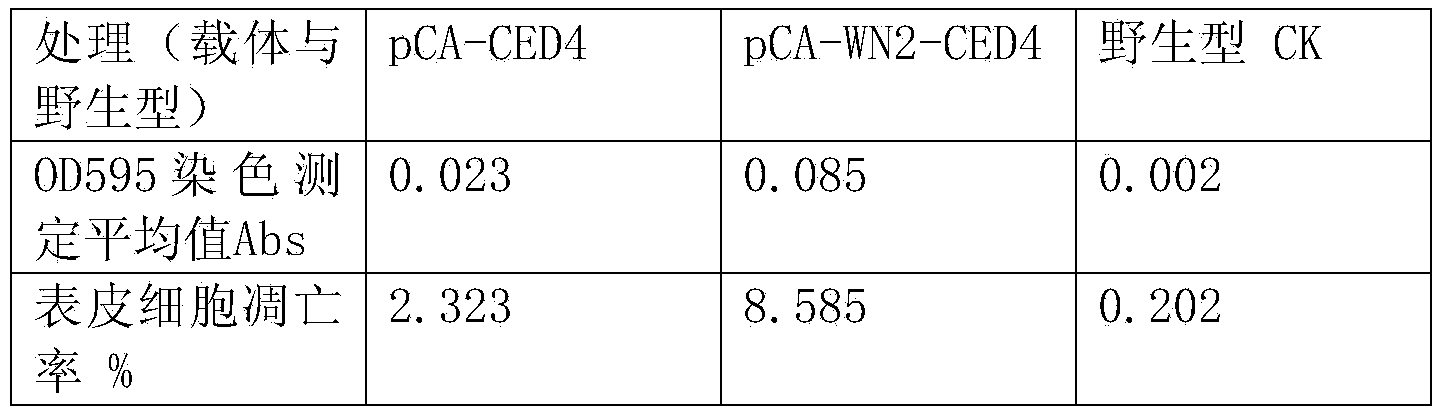
A nematode apoptosis regulation gene ced-4 inducible promoter, rice expression vector and preparation method thereof
ActiveCN103882019BGood effectVector-based foreign material introductionDNA/RNA fragmentationAgricultural scienceNematode
Owner:YICHUN UNIVERSITY
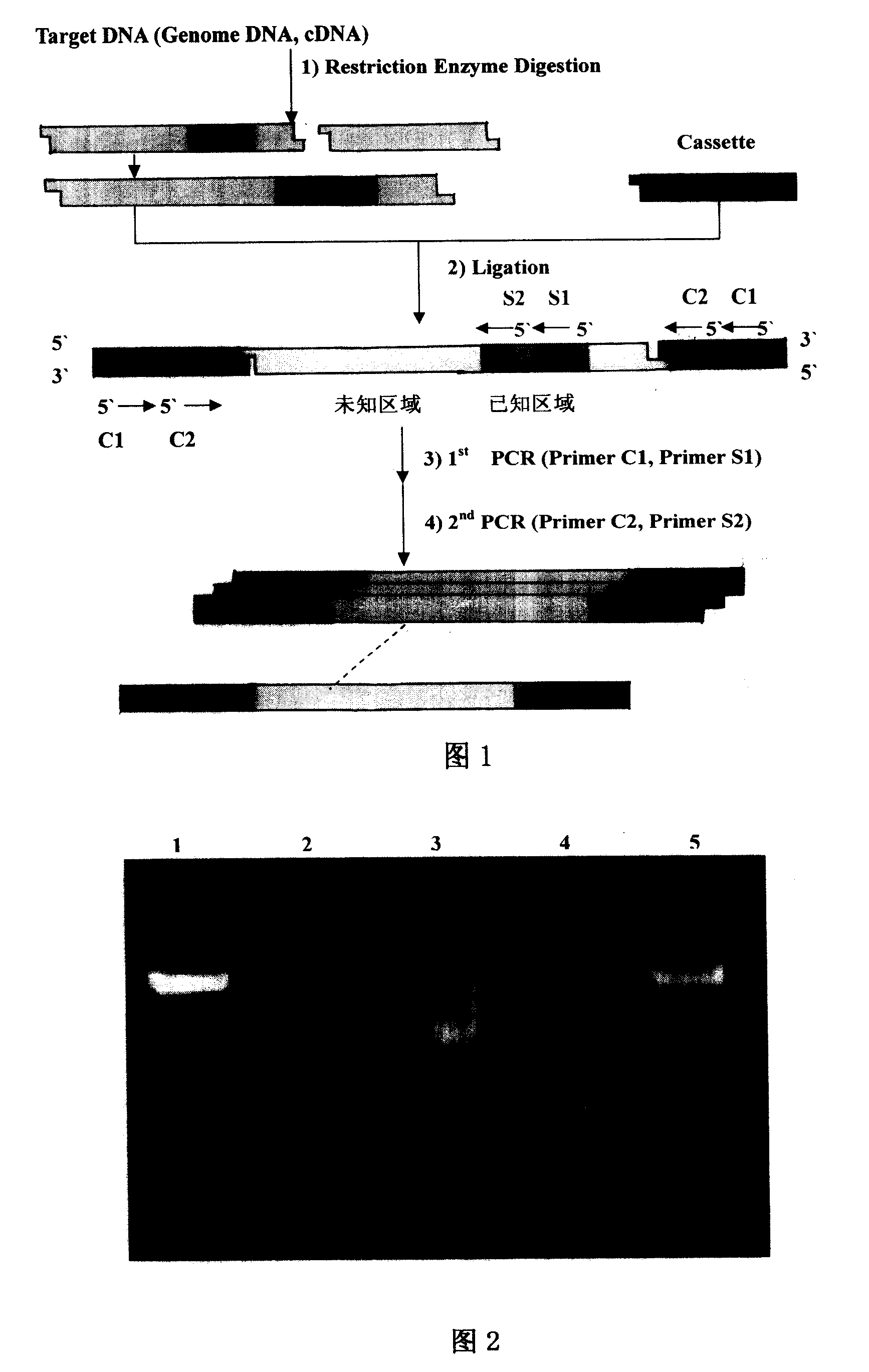
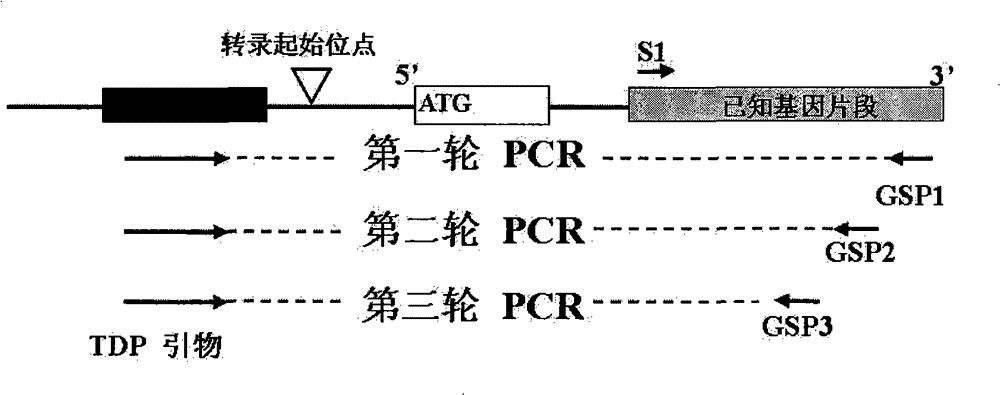
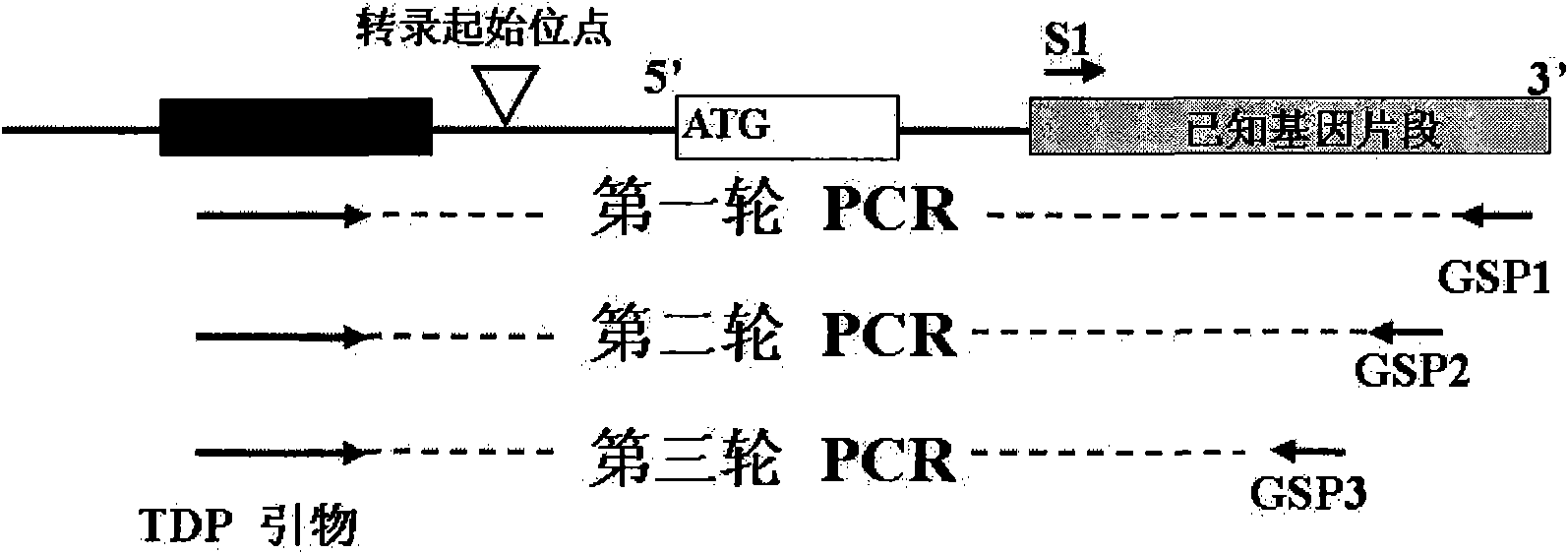
Method for cloning end of target gene 5' and special kit thereof
InactiveCN101824448BPrevent extractionSimplify the experimental stepsFermentationRNA extractionConserved sequence
The invention discloses a method for cloning the end of target gene 5'and a special kit thereof. The method comprises the following steps of: respectively combining a degenerate primer TDP which is designed according to a TATA-box conserved sequence with gene specific primers GSP1, GSP2 and GSP3 into 3 pairs of primers; and amplifying the end of the gene 5' by means of semi-nested PCR with the 3 pairs of primers, wherein the Tm values of the gene specific primers GSP1, GSP2 and GSP3 are higher than the Tm value of the TDP degenerate primer at 3-5 DEG C, and the combination positions between the gene specific primers GSP1, GSP2 and GSP3 and the gene are sequentially near to the end of the gene 5'. The method for cloning the end of target gene 5' does not need to use expensive RNA extraction reagent, RACE extraction reagent and the like, thereby reducing the experiment cost.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
A mammalian cell expression vector for industrial production
The present invention provides an expression vector for expressing proteins / polypeptides in mammalian host cells, comprising a marker gene and a restriction endonuclease cleavage site for inserting a target gene, and a Poly(A) tail after the target gene The number of signals is two or more, and the promoter element of the marker gene is only the TATA box. The upstream of the selectable marker gene of the expression vector provided by the present invention contains two or more Poly(A) tailing signals to minimize the interference of the strong upstream promoter on the activity of the downstream TATA box to drive the expression of the selectable marker gene. The expression vector of the present invention can generate stable and high-yielding cell lines for recombinant protein expression in a short time, and gene amplification does not require drug selection / mediation, and can be directly selected without any drug-induced amplification, These cell lines can stably express high levels of recombinant proteins for a long time and are very suitable for industrial production of proteins or polypeptides.
Owner:济南海湾生物工程有限公司
A method for batch capture of promoters
ActiveCN107091929BEfficient separationEfficient acquisitionMicrobiological testing/measurementBiological testingConserved sequenceTATA box
The invention provides a method for capturing promoters in batches. The method uses the complex of "DNA-protein factors" formed in the promoter region when plant genes are transcribed, and applies the improved chromosomal immunoprecipitation technique to capture the unknown plant DNA sorting out of gene promoter fragments. The present invention utilizes the property that the protein factors OsTBP2 and OsTFIIB tightly bind to the TATA box short-chain conserved sequence on the promoter to initiate transcription, and generates specific antibodies for these two factors, combined with chromatin immunoprecipitation (ChIP), to specifically target A technique for capturing promoter fragments of unknown plant genes. This technology utilizes the characteristic of a short-chain conserved sequence of the TATA box contained in most promoters, combined with the newly developed chromatin immunoprecipitation technique (ChIP) in recent years to isolate unknown plant gene promoter fragments, it is relatively easy Achieving large-scale capture of plant promoters has high commercial value.
Owner:RICE RES ISTITUTE ANHUI ACAD OF AGRI SCI
Activated siRNA able to enhance gene expression
ActiveCN103937794AHigh expressionGenetic material ingredientsGene therapyTarget expressionNucleotide
The invention discloses an activated siRNA able to specifically enhance gene expression through a gene promoter with a TATA-box motif in the target. The activated siRNA has the characteristics that: (1) the activated siRNA is complementary with a position taking the TATA-box motif as the center; (2) the first two base groups of an antisense strand adopt UA; (3) the length is 23 nucleotides; (4) methylation modification is conducted on the 3' end of the antisense strand; and (5) mismatch of the 3' end of the antisense strand is avoided. The activated siRNA provided by the invention can significantly enhance the target expression for a long time, and has great application value and broad application prospects in biotechnology, gene therapy drug development and other fields.
Owner:GUANGZHOU QIANYANG BIO-TECH PHARM CO LTD
RNA polymerase III promoter, process for producing the same and method of using the same
InactiveUS7504492B2Without impairing functionEasily substitutedSugar derivativesHydrolasesCre recombinaseTATA box
The present invention provides an RNA polymerase III promoter characterized in that a part of its sequence is substituted with a sequence capable of undergoing specific DNA recombination by Cre recombinase in such a manner functions of PSE and a TATA box are not impaired. This RNA polymerase III promoter can be used in a RNAi induction method with the condition that RNAi molecules to be used in analyzing gene function can be induced at an arbitrary point of time or only in a specific tissue (organ).
Owner:UENO HIROYUKI +2
Cotton tissue specific and pathogenic bacterium inducing promoter and its use
InactiveCN100415887CUnderstand interactionReduce outputFermentationVector-based foreign material introductionBudBlight
The present invention relates to cotton tissue specific and pathogenic bacterium inducing promoter and their application, and belongs to the field of biotechnology. The promoter contains CAAT-box, TATA-box, ethylene, methyl jasmonate, abscisic acid response element, pathogenic bacterium inducer response elements W boxes, GT-1, MYB, MYBST1, MYB1LEPR, etc. There are also specific root expressing elements to enhance the expression of GUS gene, which expresses specifically in the phloem of root, bud and stem, the vein of foliage and the guard cell of air pore. Paraffin section observation shows that GHNBS promoter expresses in phloem companion cell. After treatment with SA, MeJA, Ethylene, pathogenic bacteria of blight and pseudomonas syringae DC3000, GUS will have obviously raised activity, so that it is one organ specific and pathogenic bacterium relevant promoter.
Owner:JIANGSU ACAD OF AGRI SCI
A kind of aapdr2 gene promoter and its function verification method and application
Owner:SHANGHAI JIAO TONG UNIV
Grapevine powdery mildew resistance transcription factor gene VpRFP1 promoter sequence and application thereof
InactiveCN102121005BMicrobiological testing/measurementVector-based foreign material introductionDiseasePlant hormone
The invention relates to a grapevine powdery mildew resistance transcription factor gene VpRFP1 promoter sequence and application thereof. In the sequence, a full-length 1249bp promoter sequence of a Chinese wild vitis pseudoreticulata powdery mildew transcription factor gene VpRFP1 is cloned by adopting chromosomal walking in combination with a nested polymerase chain reaction (PCR) technology, wherein the sequence comprises 103 TATA-boxes and 27 CAAT-boxes, has 8 elements related to plant defense reaction, 14 photoreaction elements, 5 plant hormone response elements, and 14 other unknown elements. By adopting an agrobacterium tumefaciens-mediated instantaneously converted nicotiana benthamiana leaves analyzing method, the function of the Chinese wild vitis pseudoreticulata powdery mildew transcription factor gene VpRFP1 promoter proves that: the promoter has stronger promotion activity, can actively respond to pathogen invasion and disease resistant signal substances, and can improve the disease resistance and high temperature resistance of wheat, rice, potato, corn, soybean, rape and other plants through a transgenic method.
Owner:NORTHWEST A & F UNIV
A short small small nuclear RNA promoter and its construction method and application in genome editing
ActiveCN111019946BShorten the lengthIncrease the lengthVector-based foreign material introductionAngiosperms/flowering plantsWild typeNuclear gene
Owner:SOUTH CHINA AGRI UNIV
Method for cloning end of target gene 5' and special kit thereof
InactiveCN101824448APrevent extractionSimplify the experimental stepsFermentationRNA extractionConserved sequence
The invention discloses a method for cloning the end of target gene 5'and a special kit thereof. The method comprises the following steps of: respectively combining a degenerate primer TDP which is designed according to a TATA-box conserved sequence with gene specific primers GSP1, GSP2 and GSP3 into 3 pairs of primers; and amplifying the end of the gene 5' by means of semi-nested PCR with the 3 pairs of primers, wherein the Tm values of the gene specific primers GSP1, GSP2 and GSP3 are higher than the Tm value of the TDP degenerate primer at 3-5 DEG C, and the combination positions between the gene specific primers GSP1, GSP2 and GSP3 and the gene are sequentially near to the end of the gene 5'. The method for cloning the end of target gene 5' does not need to use expensive RNA extraction reagent, RACE extraction reagent and the like, thereby reducing the experiment cost.
Owner:INST OF BOTANY CHINESE ACAD OF SCI
Application of IRT1 promoter insertion element in apple
The invention discloses an application of an IRT1 promoter insertion element in an apple. The invention discloses a method for identification or auxiliary identification of the iron-deficiency-resistant degree of a plant. The method comprises detecting a genome gene sequence of an IRT1 protein of a to-be-detected plant. The iron-deficiency-resistant performance of a to-be-detected plant having the genome gene sequence, the promoter area of which has a 5'-ATTATAA-3' sequence, of the IRT1 protein is larger or selectively larger than that of a to-be-detected plant having the genome gene sequence, the promoter area of which doesn't have a 5'-ATTATAA-3' sequence, of the IRT1 protein. According to deficiency and existence of an element TATA-box of the IRT1 promoter, descendant iron-deficiency-resistant plants can be quickly screened. The application makes a significant sense in the field of the iron-deficiency-resistant performance of a plant.
Owner:CHINA AGRI UNIV
Nematode apoptosis regulatory gene ced-4 inducible promoter, rice expression vector and preparation method thereof
ActiveCN103882019AGood effectVector-based foreign material introductionDNA/RNA fragmentationAgricultural scienceNematode
The invention discloses a nematode apoptosis regulatory gene ced-4 inducible promoter wn-2, a rice expression vector pCA-Wn-2-CED-4 and a preparation method of the pCA-Wn-2-CED-4 expression vector. According to the invention, a TATA-box core sequence shared by animal and plant genes is added into the reported core sequence having the best inductive effect in a W-box series of the promoter, therefore, a novel wn-2 sequence of a nematode apoptosis gene in rice expression is innovatively designed; the wn-2 and the inherent promoter of the rice expression vector are fused, so that the novel pCA-Wn-2-CED-4 expression vector of the rice expression vector is constructed; the pCA-Wn-2-CED-4 expression vector has the superior effect of inducing rice expression of the target gene ced-4; and the pCA-Wn-2-CED-4 expression vector is screened and connected to correct vector clone and successfully converted into rice varieties, such as Nipponbare and Lijiang Xintuan dark rice.
Owner:YICHUN UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com