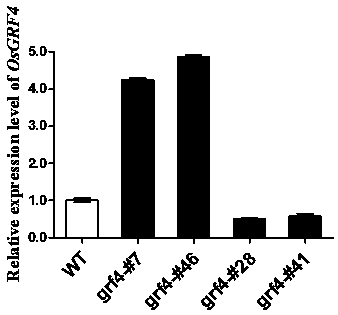
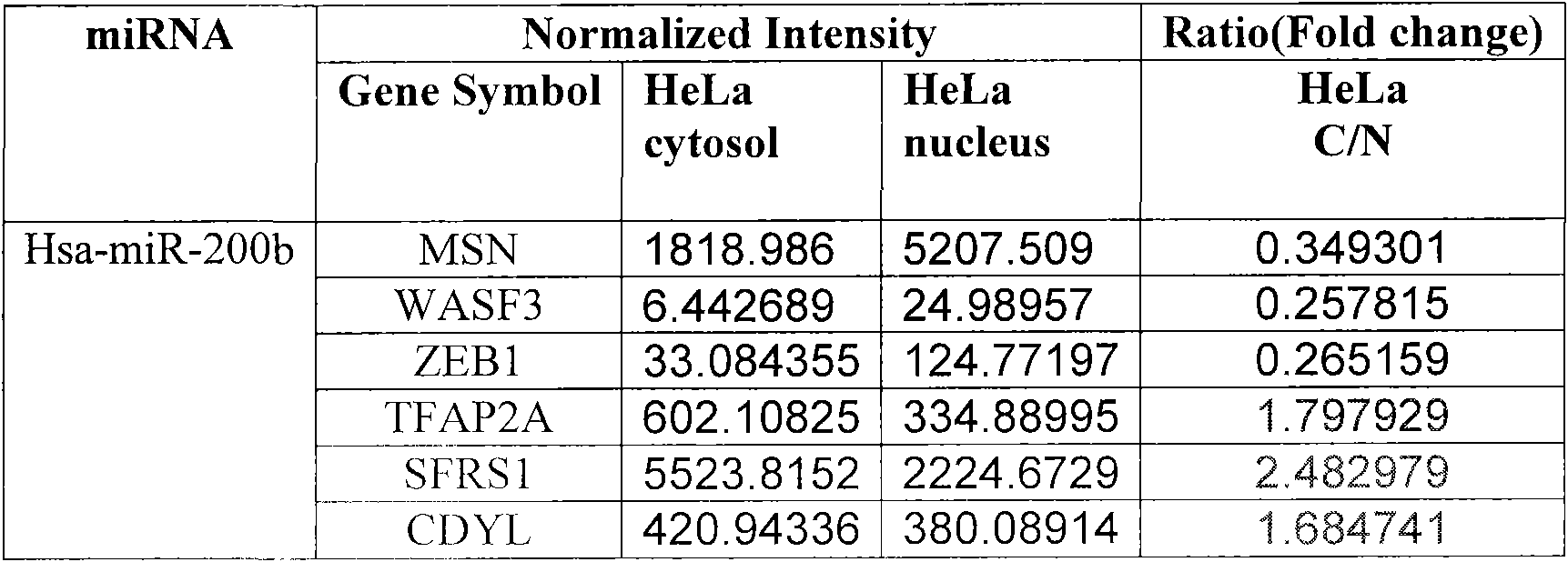
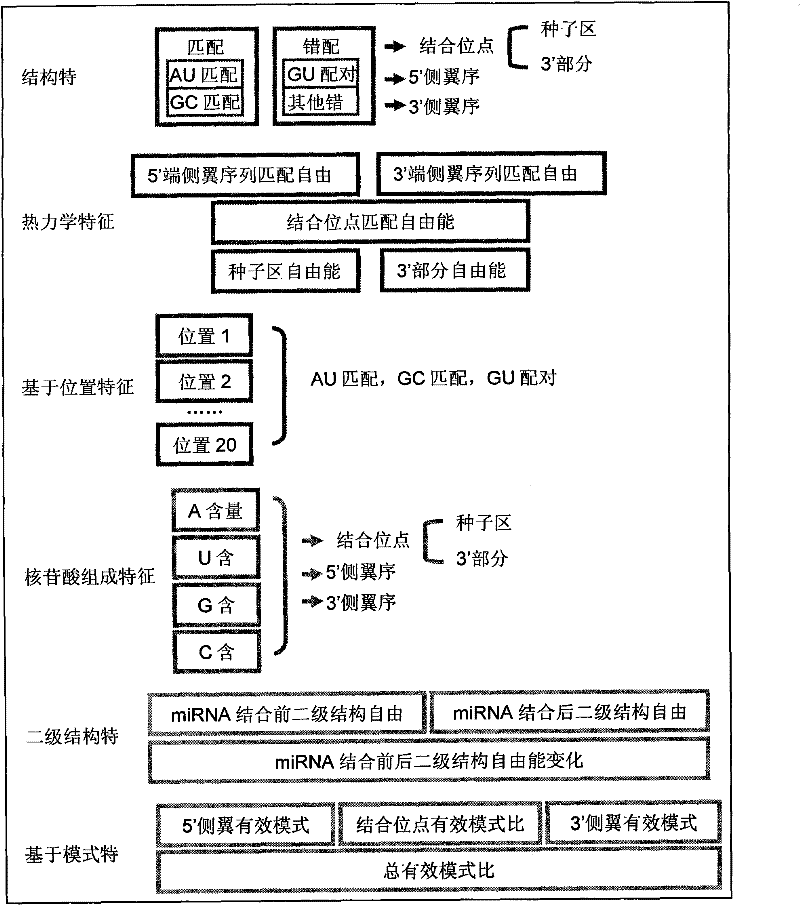
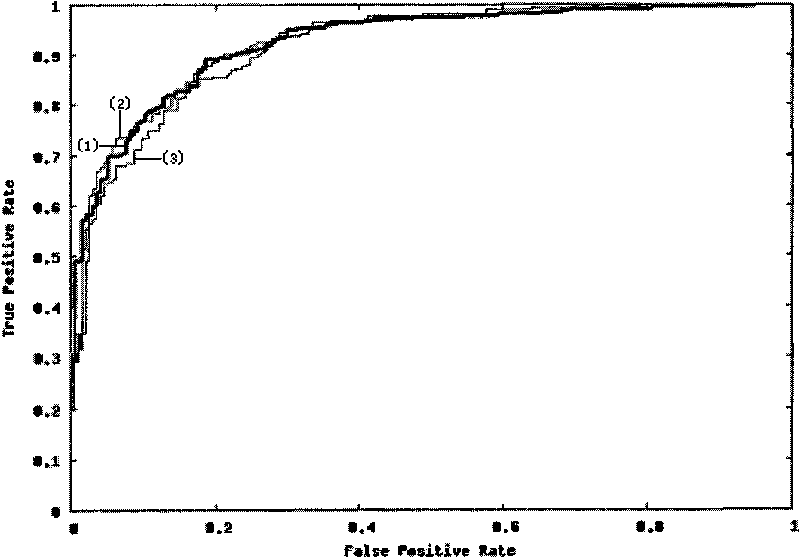
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
34 results about "Mirna target gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Sequence characteristic analysis method for forecasting miRNA target gene
ActiveCN106599615AMeasuring Binding PossibilitiesBalance differenceBiostatisticsSequence analysisData setRelevant feature
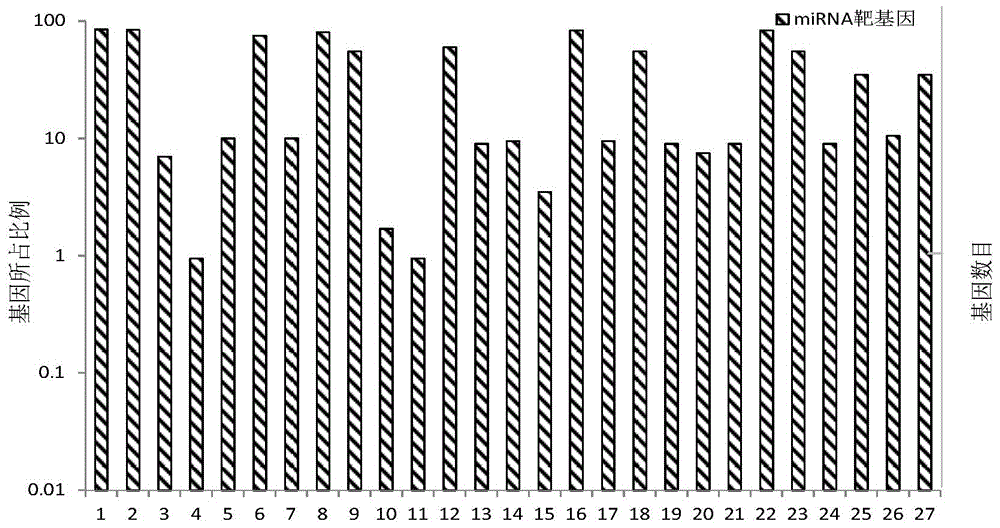
The invention discloses a sequence characteristic analysis method for forecasting a miRNA target gene. The method comprises the steps of constructing related characteristics of 27 miRNA-target point pairing sequences on the basis of a CLASH experiment data set, and forming a characteristic set comprising 84 characteristic values by combining traditional characteristic; and performing machine learning by using a random forest model, and constructing a miRNA target gene forecast model to perform miRNA target gene recognition. The model constructed according to the method has the advantages of high accuracy, sensitivity, specificity and precision, and the miRNA target gene can be relatively and accurately forecasted.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
Recommendation model based miRNA target gene prediction method
ActiveCN105808976AImprove accuracyIncreased sensitivityHybridisationSpecial data processing applicationsExperimental validationRecommendation model
The present invention discloses a recommendation model based miRNA target gene prediction method (miRTRS). The method comprises: constructing a bipartite graph of an miRNA and a gene by using experimentally verified miRNA target gene data; on this basis, calculating a possibility that one gene is an miRNA target gene by using a bipartite graph based recommendation algorithm, and introducing biological data, i.e. sequence similarity between miRNAs into the recommendation algorithm; and finally, sorting recommendation values in a descending order, and taking what is ranked at the front as an miRNA target gene relation. The method disclosed by the present invention is simple and easy for use; and compared with the existing miRNA target gene prediction method, the method provided by the present invention is significantly improved on the aspects of accuracy, sensitivity and specificity of prediction, and provides valuable reference information for scientists to perform experiments and further study of miRNA target gene discovery.
Owner:CENT SOUTH UNIV
Method for researching oryza sativa and pathogen interaction mode
ActiveCN104911261AQuick and effective screeningStrong disease resistanceComponent separationMicrobiological testing/measurementDiseaseTotal rna
The invention discloses a method for researching an oryza sativa and pathogen interaction mode, and belongs to the field of plant biotechnology. The method for researching the interaction mode of oryza sativa in response to pathogen infection through integrated application of a transtriptome, a proteome and a miRNA group comprises the concrete steps: A, respectively taking oryza sativa leaf blade total RNAs, Small RNAs and total proteins of a disease-resistant material and a susceptible material at different time points before and after pathogen inoculation; B, respectively carrying out differential-expression spectrum analysis, differential proteomics analysis and miRNA identification and target gene analysis; and C, integrating and associating differential genes, proteins and miRNA target genes identified by the three ways, to reveal disease-resistant and susceptible reaction paths activated or inhibited in the oryza sativa and pathogen interaction process, so as to relatively systematically illuminate an oryza sativa-magnaporthe oryzae interaction mechanism. The method can quickly and effectively screen candidate genes associated with oryza sativa disease resistance.
Owner:SOUTH CHINA AGRI UNIV
Method of constructing degradome sequencing library
InactiveCN103334159AEasy to operateBreak free from the constraints of bioinformatics predictionsLibrary creationMirna target geneBioinformatics
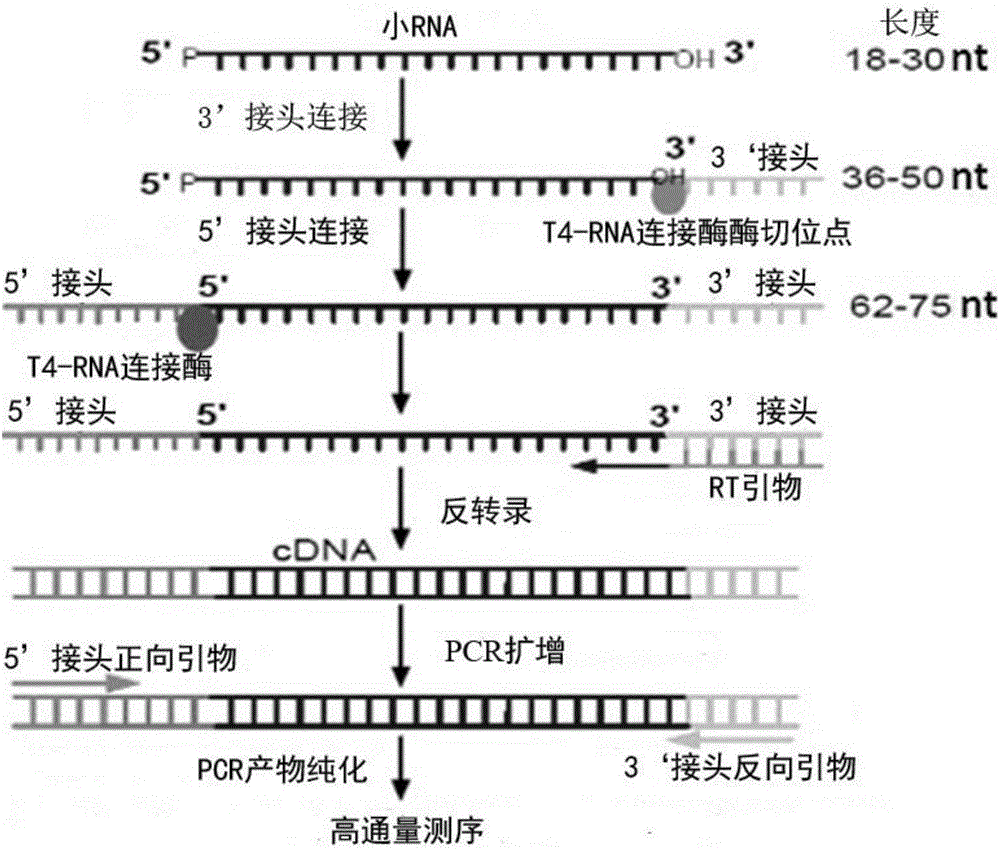
The invention discloses a method of constructing degradome sequencing library. The method comprises following steps: sorting mRNA sections of miRNA target gene, connecting connectors of 5' terminals of mRNA sections of miRNA target gene, and amplifying cDNA of miRAN target gene. The technical scheme facilitates the operation, enables the specific loci where the animal and plant miRNAs act on the target gene to be located accurately, breaks through the bioinformatics prediction limitation, finds the target gene where miRNA acts on, and has the characteristics of efficiency, rapidness, intuition and accuracy.
Owner:HANGZHOU LC BIOTECH
Method for screening MicroRNAs of nilaparvata lugens with effect on oryza sativa resistance adaptability
ActiveCN108504748AMicrobiological testing/measurementSpecial data processing applicationsBiologyMirna target gene
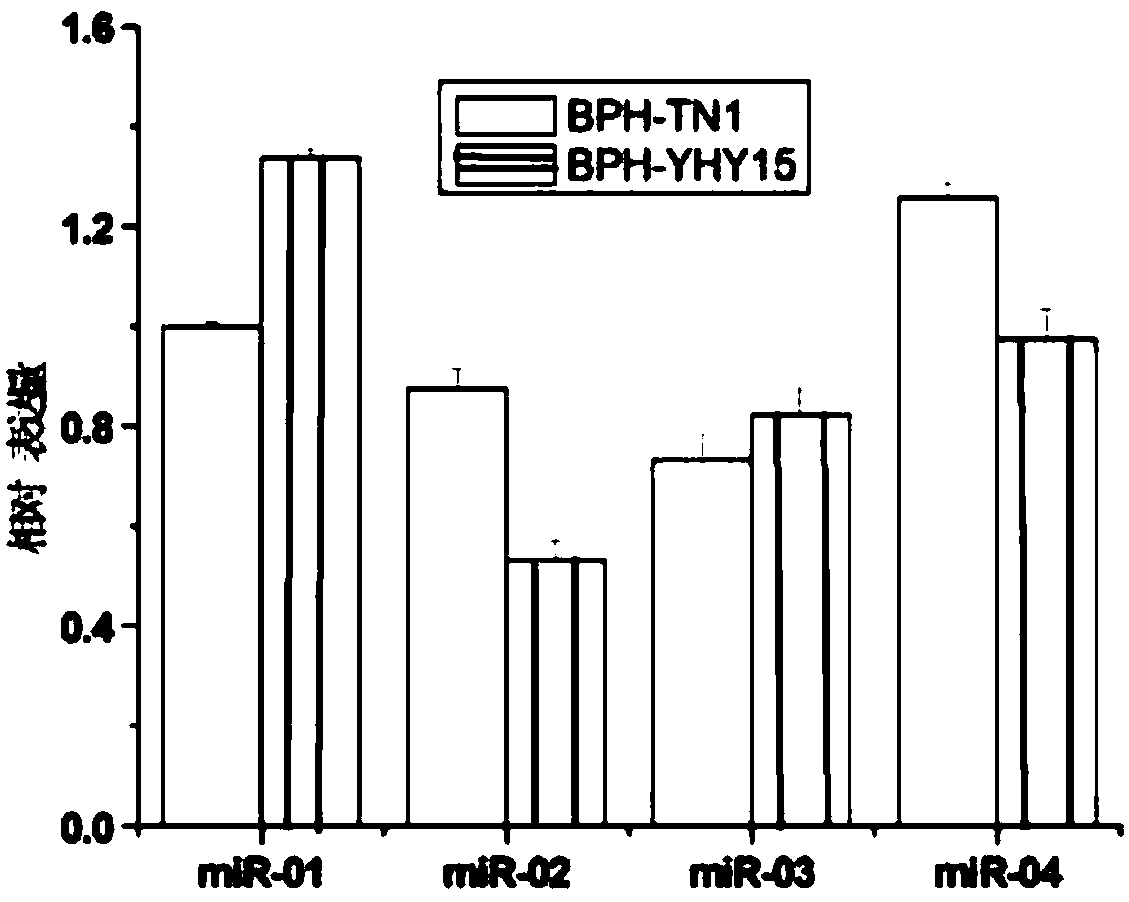
The invention discloses a method for screening MicroRNAs of nilaparvata lugens with effect on oryza sativa resistance adaptability, and relates to the bioengineering technology. The method comprises the following steps: (1) sample collection; (2) sequencing of small RNA; (3) pretreatment of sequencing raw data; (4) sequence alignment; (5) new miRNA prediction; (6) analysis of miRNA differential expression; (7) prediction of differential miRNA target genes; (8) synthesis of miRNA mimic, miRNA inhibitor and control chain; (9) microinjection and artificial feeding of miRNA mimic and miRNA inhibitor to nilaparvata lugens and verification of the function of miRNAs; (10) preliminary study on oryza sativa resistance adaptability change of the nilaparvata lugens after microinjection and artificialfeeding. Small RNAs of two nilaparvata lugens groups are deeply sequenced with a high-throughput sequencing technology, and are analyzed, identified and predicted according to subsequent bioinformatics, differences between the two nilaparvata lugens groups on insect-susceptible oryza sativa TN1 and insect-resistant oryza sativa YHY 15 are compared, the miRNA associated with host resistance adaptability is analyzed, screened and discovered, and the new method is provided for oryza sativa insect-resistant breeding.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI
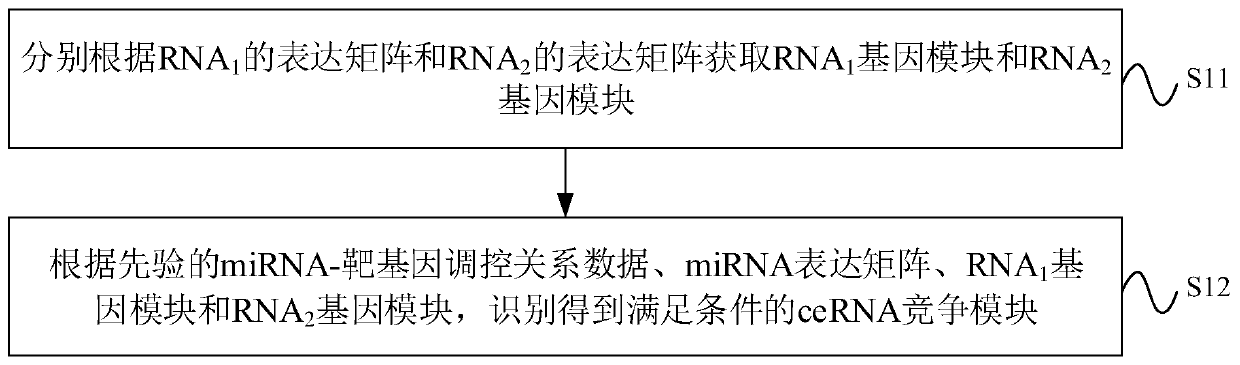
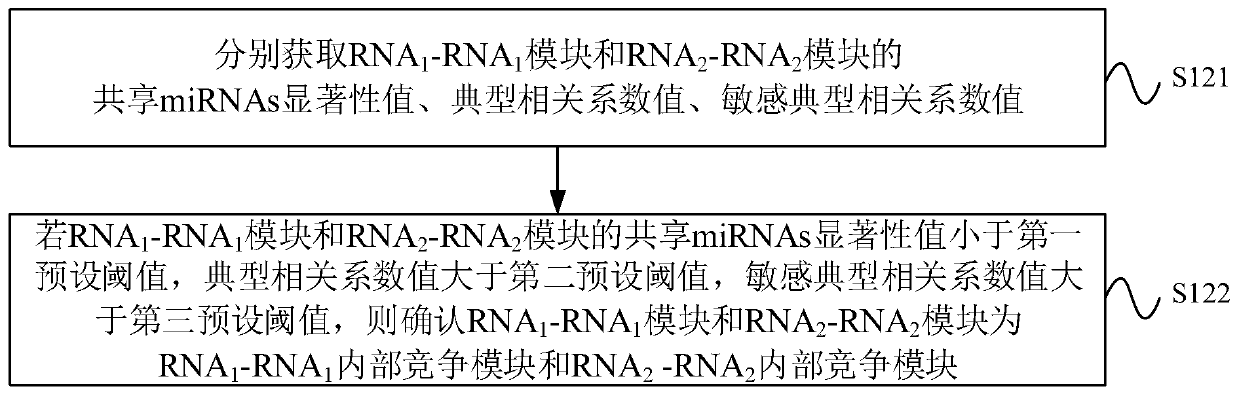
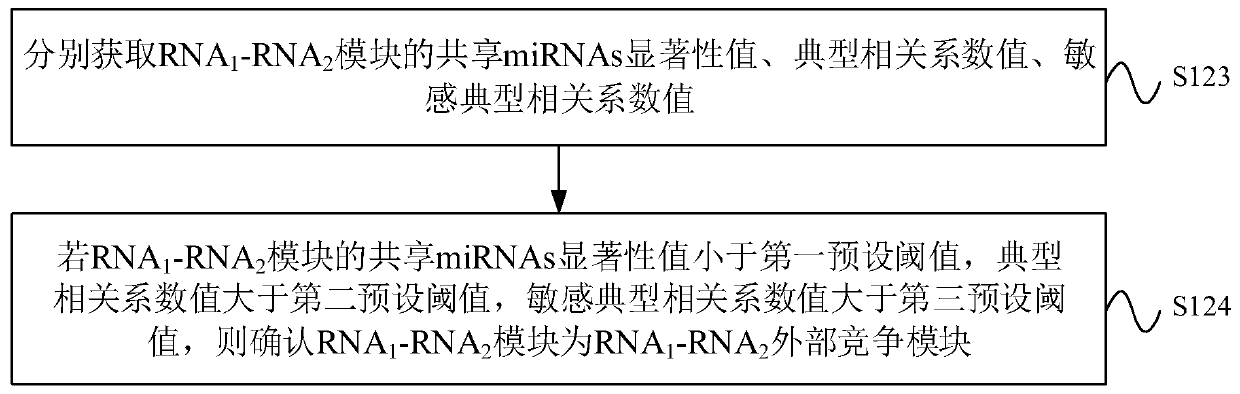
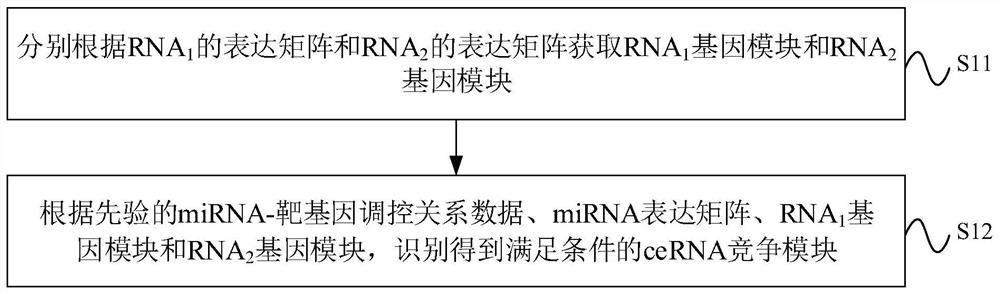
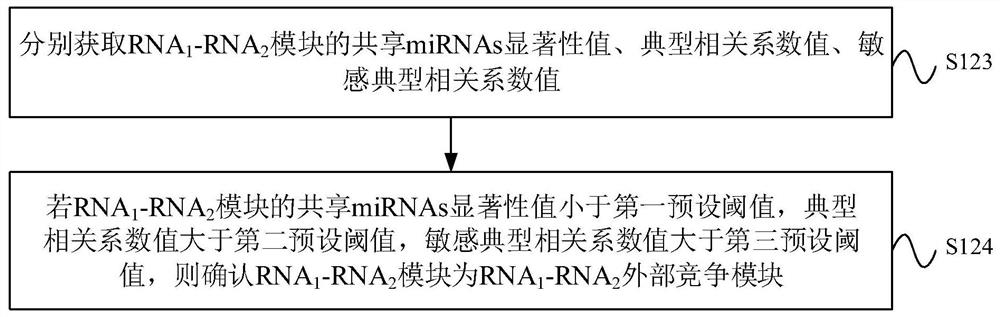
CeRNA competition model identification method and device, electronic equipment and storage medium
ActiveCN111383709AAccurate identificationCharacter and pattern recognitionProteomicsAlgorithmEngineering
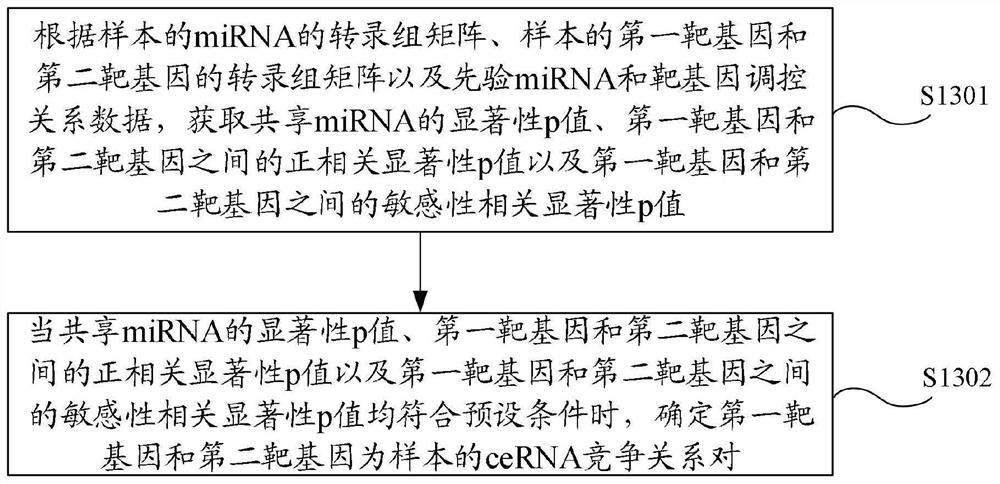
The invention provides a ceRNA competition model identification method and device, electronic equipment and a storage medium, and relates to the field of gene identification. The ceRNA competition model identification method comprises the steps of obtaining an RNA1 gene module and an RNA2 gene module according to the expression matrix of the RNA1 and the expression matrix of the RNA2, wherein theRNA1 and the RNA2 are miRNA target genes; regulating relation data, a miRNA expression matrix, an RNA1 gene module and an RNA2 gene module according to the prior miRNA-target gene, and performing identification for obtaining the ceRNA competition module which satisfies a condition, wherein the ceRNA competition module comprises an inner competition module and an outer competition module. Because the inner and outer competition conditions of the ceRNA are considered, the inner and outer competition strength of the ceRNA and the influence of the miRNA to the inner and outer competition strengthsof the ceRNA are measured in a module level. Therefore, the method provided by the invention can accurately identify the ceRNA competition module.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA +1
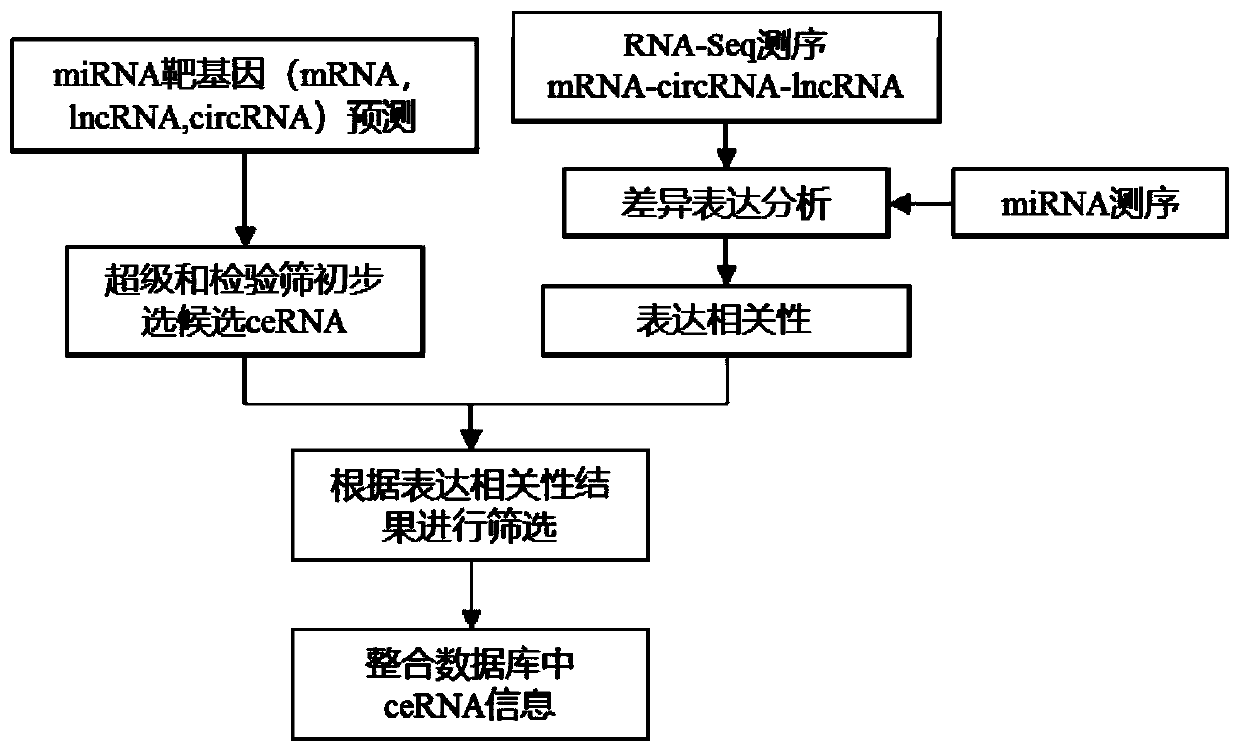
Method for carrying out ceRNA prediction based on miRNA target gene prediction and related expression analysis
The invention discloses a method for carrying out ceRNA prediction based on miRNA target gene prediction and related expression analysis. The method is characterized by comprising the following steps:carrying out correlation analysis on a ceRNA action pair; screening potential ceRNA relationship pairs according to the number of common MERs; evaluating the regulation and control function of the miRNA on a pair of competitive lncRNA-mRNA by adopting two scoring modes according to the expression correlation of the miRNA and the ceRNA to the three; and obtaining results of statistical test significance, and sorting the results according to scores. The method for carrying out ceRNA prediction based on miRNA target gene prediction and related expression analysis has the advantages that after the ceRNA is preliminarily analyzed, the inter-gene expression correlation is analyzed according to an actual sequencing result, and the result is screened; different target gene prediction methods areadopted for animals and plants, so that ceRNA prediction analysis can be performed; and information in a common database is integrated, and the reliability of an analysis result is improved.
Owner:南京派森诺基因科技有限公司
Screening and identifying method of miRNAs (micro Ribonucleic Acids) of fetal fibroblasts of Saanen dairy goats
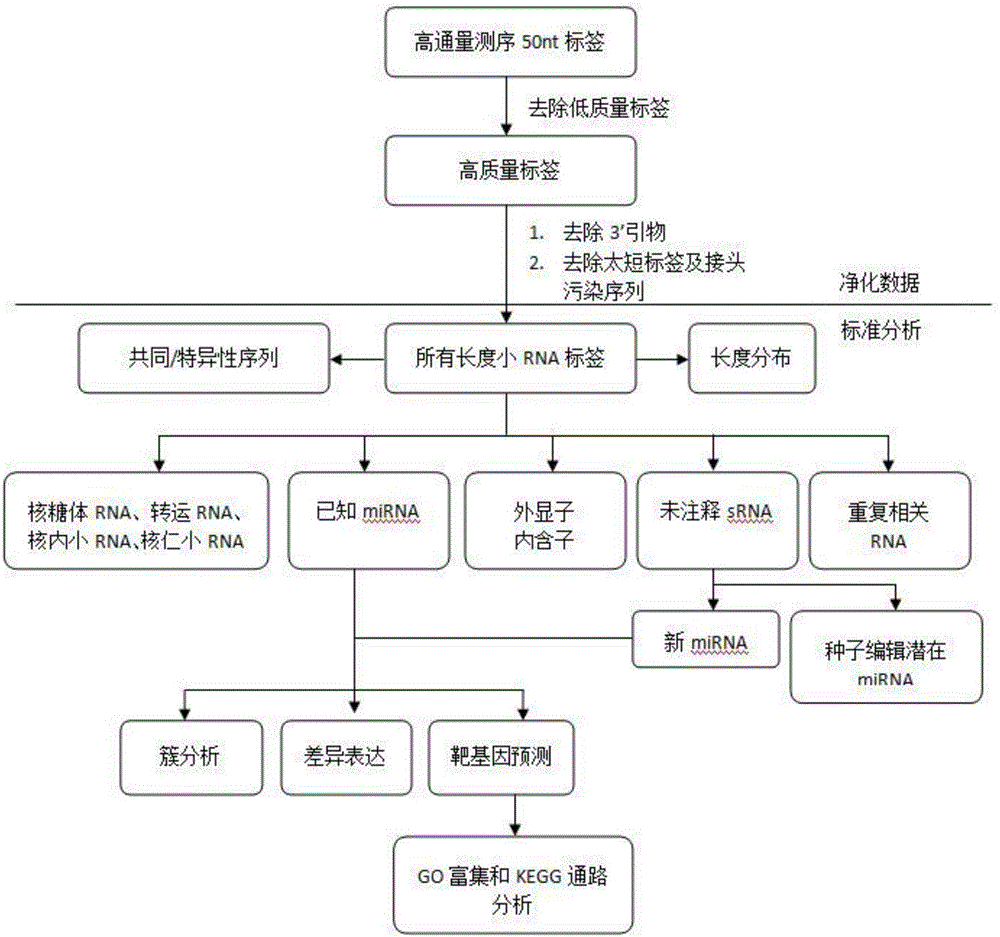
The invention discloses a screening and identifying method of miRNAs (micro Ribonucleic Acids) of fetal fibroblasts of Saanen dairy goats. The method comprises the following steps: obtaining the fetal fibroblasts of the Saanen dairy goats; extracting RNAs; constructing a library and carrying out high-throughput sequencing; and carrying out data processing. By adopting the high-throughput sequencing, 16395039total_reads are obtained, and redundant data is taken out to obtain clean_reads 16150181 containing Unique sRNAS 205857. Bioinformatic analysis is carried out to obtain 44 known miRNAs and 247 candidate new miRNAs. The quantity of miRNA target genes is 5401, and the quantity of sites of the target genes is 6069; and the quantity of candidate miRNA target genes is 8401, and the quantity of sites of the target genes is 10832.
Owner:TARIM UNIV
Predictive analysis method of micro ribonucleic acid (miRNA) target genes of cattle
The invention relates to a predictive analysis method of micro ribonucleic acid (miRNA) target genes of the cattle and is suitable for prediction of the miRNA target genes of the cattle. The predictive analysis method of the miRNA target genes of cattle comprises the following steps: (1) establishing a 3'-untranslated region (UTR) data base, (2) obtaining a sequence of the miRNA of the cattle for species without 3'-UTR sequences, (3) obtaining a protein sequence of the cattle, (4) deducting a sequence coding for amino acids in protein (CDS) from the miRNA by writing a practical extraction and reporting language (PERL) script in order to obtain 5'-UTRs and 5'-UTR sequences of the genes, wherein the 3'- UTR sequences are gathered together, and the 3'-UTR data base is established, (5) downloading relevant data for species with an open 3'-UTR, (6) carrying out target genes prediction respectively by a plurality of software, and referring to miRGator software with regard to some miRNA which does not correspond and (7) carrying out statistic analysis and counting the target genes which are obtained through prediction in order to determine a most possible target gene. Generally, in step (6), intersection of a plurality of software prediction results is adopted, namely an overlap part.
Owner:SHANGHAI CLUSTER BIOTECH
Method for constructing degradome sequencing library
PendingCN108570712AReduce the starting amountEasy to operateLibrary creationMirna target geneComputational biology
The invention discloses a method for constructing a degradome sequencing library. The method comprises the following steps: the sorting of mRNA fragments of a miRNA target gene; the ligation of 5' terminal adapters with the mRNA fragments of the miRNA target gene; reverse transcription with random primers; and the amplification of the cDNA of the miRNA target gene. According to a technical schemeof the invention, ligation of specific 3' terminal adapters is not needed, so operation is simple, little time is needed, and requirements on a sample size are greatly reduced; the method can accurately find out specific sites where animal and plant miRNAs act on target genes, get rid of the limitation of bioinformatics prediction, and actually find out target genes on which miRNA acts through experiments; and the method is characterized in that the method is efficient, fast, visual and accurate.
Owner:杭州联川基因诊断技术有限公司
Method for relieving miRNA inhibition function to promote target gene expression
ActiveCN110157730AEasy to operateLow running costVector-based foreign material introductionGenome editingRecognition sequence
The invention discloses a strategy for relieving a miRNA inhibition function to promote target gene expression, and provides a specific implementation method based on a genome editing technology. A gene editing technology is used for editing the recognition sequence of a miRNA target gene, base substitution mutation and amino acid deletion mutation with normal target gene functions are screened, the recognition sequence destroys mutants not inhibited by upstream miRNA, and the simple and efficient method is provided for promoting the functional research and functional application of the targetgene.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
mRNA nucleoplasmic ratio variation-based method for identifying miRNA target gene and application thereof
The invention belongs to the technical field of biology, and relates to a rapid and accuracy message Ribonucleic Acid (mRNA) nucleoplasmic ratio variation-based method for identifying a mirna Ribonucleic Acid (miRNA) target gene, and to application of the method in the biomedical field. The method is realized through the following technical scheme of: predicting possible target genes of a certain miRNA through the conventional miRNA target prediction bioinformatics software, such as Targetscan; measuring the mRNA nucleoplasmic ratio variation of the possible target genes in a cell by utilizing a biochip or real-time quantitative Polymerase Chain Reaction (PCR); searching the possible target genes with an mRNA nucleoplasmic ratio of more than 1; and further confirming in the protein level through Western blot.
Owner:INST OF BASIC MEDICAL SCI ACAD OF MILITARY MEDICAL SCI OF PLA
Kit for identifying miRNA (micro-ribonucleic acid) target genes and applications thereof
ActiveCN102168133AMicrobiological testing/measurementVector-based foreign material introductionMultiple cloning siteCloning Site
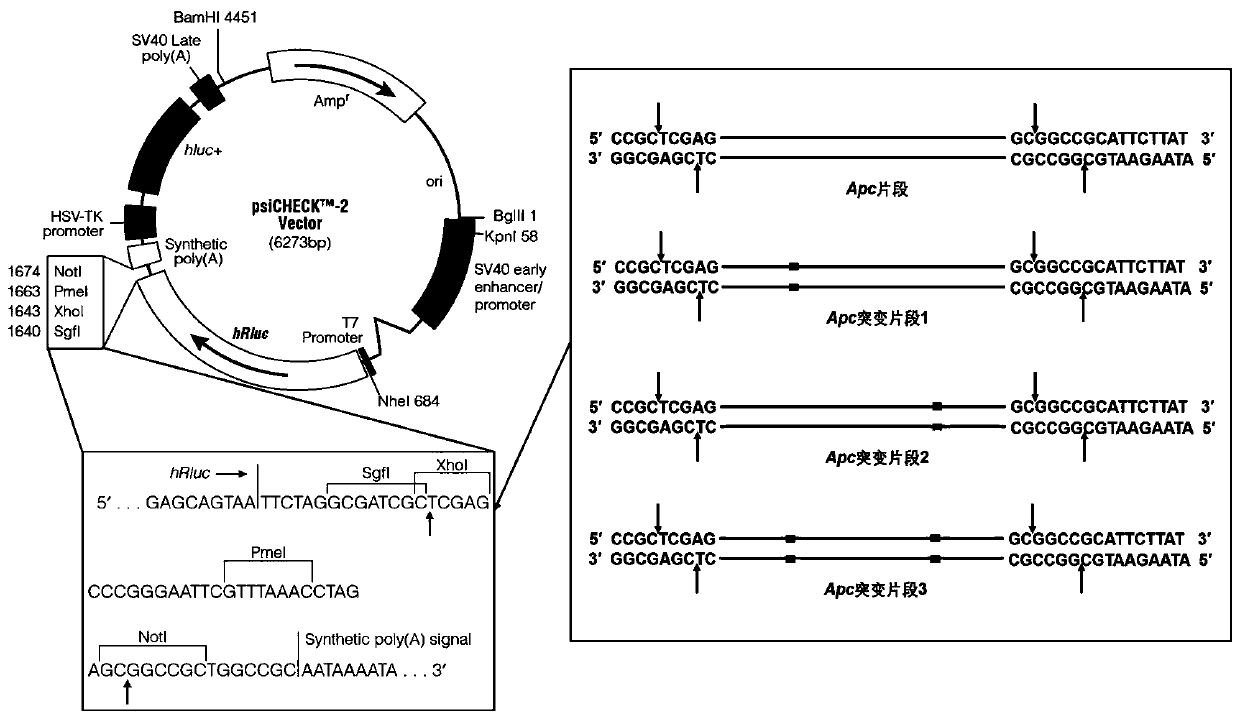
The invention discloses a kit for identifying miRNA (micro-ribonucleic acid) target genes and applications thereof, namely the application of a report vector in identification of miRNA target genes and / or detection of miRNA expression, the application of the kit in the identification of the miRNA target genes and / or the detection of the miRNA expression, and the application of the vector in the identification of the miRNA target genes and / or the detection of the miRNA expression, wherein the report vector comprises report genes, as well as a promoter and multiple cloning sites, which are positioned at the up stream and down stream of the report genes respectively, and the report genes are genes encoded by firefly luciferase. Experiments prove that the research designs a dual-report gene vector system for identification of the miRNA target genes, which is used for regulating and controlling fast identification of the target genes and evaluating the regulation and the control of the miRNA expression in the miRNA research.
Owner:MICROBE EPIDEMIC DISEASE INST OF PLA MILITARY MEDICAL ACAD OF SCI
Analysis and identification method on miRNA key target gene of blood fluke in specific growth period
InactiveCN104404139AQuickly reveal accurate effectsIdentification helpsMicrobiological testing/measurementStudy methodsMirna target gene
The invention an analysis and identification method on the miRNA key target gene of blood fluke in the specific growth period, and belongs to the field of life medicine science. Bioinformatics is adopted to analyze the blood fluke genome so as to find certain miRNA target gene groups in the blood fluke genome. Then Solexa is utilized to analyze the blood fluke expression difference gene groups in the high expression period of miRNA and the low expression period of miRNA. The overlapped part between the blood fluke expression difference gene groups and the miRNA target gene groups is the candidate key gene groups. Then KEGG analysis is used to find the important genes in the candidate key gene groups so as to determine the key target gene group primarily. Then the miRNA mimic is utilized to carry out in-vitro experiments to further screen out the key target genes, and finally the target relationship between the miRNA and the key target genes is further confirmed through a luciferase reporter system. The establishment of the provided method can help the identification of the key target genes of miRNA in the specific period, help the disclosure of the modulation effect of miRNA, and provide an ideal route for miRNA function researches. At the same time valuable references are provided for the similar research methods on other species.
Owner:TONGJI UNIV
Nucleic acid and application thereof
ActiveCN106591311AEffective control of CG contentReduce the difficulty of synthesisGenetically modified cellsNucleic acid vectorBinding siteMirna target gene
The invention provides a separated nucleic acid with a miRNA target gene binding site sequence. By the separated nucleic acid in an embodiment, miRNA reduction can be realized. Especially, as for high-CG-content miRNA with CG content being up to 90%, reduction efficiency of the nucleic acid is remarkably improved as compared with that in the prior art.
Owner:FIELD OPERATION BLOOD TRANSFUSION INST OF PLA SCI ACAD OF MILITARY +1
Application of mir-26a in non-small cell lung cancer
The invention relates to an application of miR-26a in non-small cell lung cancer (NSCLC). MiR-26a can effectively inhibit the proliferation of NSCLC cells, and promote the apoptosis of lung cancer cells. MiRNA target gene prediction software shows that WNK3 is a potential target spot of miR-26a, the 3'-UTR area of WNK3 has two binding sites in complementary pairing with the seed sequence of miR-26a. The test shows that miR-26a can inhibit the translation and expression of target genes in the protein level. A bifluorescence reporter gene system proves the direct targeted relationship of miR-26a and WNK3. The in-depth study shows that miR-26a opens the Caspase path of cell apoptosis by targeting WNK3 protein, and finally plays the role of inducing cell apoptosis, thus providing an important therapeutic target for the accurate treatment. Through the application of miR-26a in NSCLC, an action mechanism of miR-26a is verified, potential miRNA molecules with an anti-cancer function are provided clinically, a theoretical basis is provided for the study on the relevant drug targets, and novel accurate treatment research direction and application guidance are also provided.
Owner:SHANGHAI UNIV
A method and application for regulating mitochondrial gene expression using small RNA
ActiveCN105200056BReduce abundanceLower levelForeign genetic material cellsDNA/RNA fragmentationMitophagySmall RNA
Owner:WUHAN UNIV
cerna competition module identification method, device, electronic equipment and storage medium
ActiveCN111383709BAccurate identificationCharacter and pattern recognitionProteomicsEngineeringMirna target gene
The invention provides a ceRNA competition module identification method, device, electronic equipment and storage medium, and relates to the field of gene identification. The ceRNA competition module recognition method includes: respectively according to the RNA 1 expression matrix and RNA 2 expression matrix to obtain RNA 1 Gene modules and RNA 2 gene module, where RNA 1 and RNA 2 are miRNA target genes. Based on a priori miRNA-target gene regulatory relationship data, miRNA expression matrix, RNA 1 Gene modules and RNA 2 The gene module identifies a ceRNA competition module that satisfies the conditions, wherein the ceRNA competition module includes an internal competition module and an external competition module. Due to the consideration of ceRNA internal and external competition, the strength of ceRNA internal competition and external competition was measured at the module level, as well as the influence of miRNAs on ceRNA internal and external competition. Therefore, the protocol provided by the present invention can accurately identify ceRNA competition modules.
Owner:UNIV OF ELECTRONICS SCI & TECH OF CHINA +1
Kit for identifying miRNA (micro-ribonucleic acid) target genes and applications thereof
ActiveCN102168133BMicrobiological testing/measurementVector-based foreign material introductionMultiple cloning siteCloning Site
The invention discloses a kit for identifying miRNA (micro-ribonucleic acid) target genes and applications thereof, namely the application of a report vector in identification of miRNA target genes and / or detection of miRNA expression, the application of the kit in the identification of the miRNA target genes and / or the detection of the miRNA expression, and the application of the vector in the identification of the miRNA target genes and / or the detection of the miRNA expression, wherein the report vector comprises report genes, as well as a promoter and multiple cloning sites, which are positioned at the up stream and down stream of the report genes respectively, and the report genes are genes encoded by firefly luciferase. Experiments prove that the research designs a dual-report gene vector system for identification of the miRNA target genes, which is used for regulating and controlling fast identification of the target genes and evaluating the regulation and the control of the miRNA expression in the miRNA research.
Owner:MICROBE EPIDEMIC DISEASE INST OF PLA MILITARY MEDICAL ACAD OF SCI
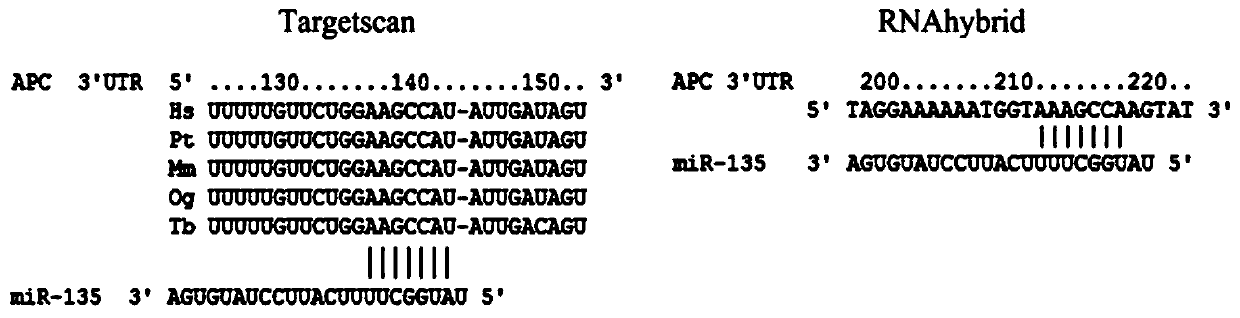
MiRNA target gene double-binding-site fluorescent vector and construction method and application thereof
PendingCN109929879AQuick checkEasy to prepareMicrobiological testing/measurementVector-based foreign material introductionMir 135aBinding site
The invention relates to a miRNA target gene double-binding-site fluorescent carrier and a construction method and application thereof. The miRNA target gene double-binding-site fluorescent carrier comprises two wild-type site fragments bound with a miR-135a-5p seed sequence, a fragment between the first and second wild-type sites, a fragment between first and second wild-type sites, and a fragment between two mutation sites. The invention provides a miRNA target gene double-binding-site fluorescent vector, a construction method and an application thereof. The miRNA target gene double-binding-site fluorescent vector can be used for rapidly and directly detecting the combination of miRNA and a predicted target gene, save the research cost and shorten an experiment period.
Owner:湖南省畜牧兽医研究所
Method for detecting whether mRNa to be tested contains miRNA target site or not and application of method
The invention discloses a method for detecting whether mRNa to be tested contains a miRNA target site or not and an application of the method. The method comprises the steps of 1) providing a target miRNA expression vector and an mRNA expression vector to be tested, wherein the mRNA expression vector to be tested can transfer an mRNA molecule named as an initiation codon region-mRNA-kozak-report gene mRNA to be tested in biologic cells, and the mRNA molecule is formed by connecting an initiation codon region with an mRNA-kozak segment located at the downstream of the initiation codon region and a report gene mRNA located at the downstream of the mRNA-kozak to be tested; 2) constructing restructure biology cells of the miRNA expression vector having transmitting-in purpose and the mRNA expression vector to be tested; and 3) detecting the expression situation of the report gene in the restructured biology cells, and determining whether the mRNA to be tested contains the target miRNA target site or not. The method can be used for screening the miRNA target gene.
Owner:BEIJING UNIV OF AGRI
Method and application thereof for detecting whether mRNA to be detected contains miRNA target site
ActiveCN109868279BVector-based foreign material introductionDNA/RNA fragmentationStart codonMirna target gene
The invention discloses a method for detecting whether the mRNA to be tested contains a miRNA target site and an application thereof. The method includes: 1) providing the target miRNA expression vector and the mRNA expression vector to be tested; the mRNA expression vector to be tested can transcribe the mRNA molecule named as initiation codon region-tested mRNA-kozak-reporter gene mRNA in biological cells , the mRNA molecule is connected by the start codon region, the segment of the mRNA-kozak to be tested downstream of the start codon region and the reporter gene mRNA located at the downstream of the mRNA-kozak to be tested; 2) constructing and introducing the target miRNA expression 3) detecting the expression of the reporter gene in the recombinant biological cells, and determining whether the mRNA to be tested contains the target site of the miRNA of interest. The present invention can be used for screening miRNA target genes.
Owner:BEIJING UNIV OF AGRI
A recommendation model-based method for predicting miRNA target genes
ActiveCN105808976BImprove accuracyIncreased sensitivityHybridisationSpecial data processing applicationsExperimental validationRecommendation model
The present invention discloses a recommendation model based miRNA target gene prediction method (miRTRS). The method comprises: constructing a bipartite graph of an miRNA and a gene by using experimentally verified miRNA target gene data; on this basis, calculating a possibility that one gene is an miRNA target gene by using a bipartite graph based recommendation algorithm, and introducing biological data, i.e. sequence similarity between miRNAs into the recommendation algorithm; and finally, sorting recommendation values in a descending order, and taking what is ranked at the front as an miRNA target gene relation. The method disclosed by the present invention is simple and easy for use; and compared with the existing miRNA target gene prediction method, the method provided by the present invention is significantly improved on the aspects of accuracy, sensitivity and specificity of prediction, and provides valuable reference information for scientists to perform experiments and further study of miRNA target gene discovery.
Owner:CENT SOUTH UNIV
microRNA target position point prediction method based on support vector machine
InactiveCN101710362BMicrobiological testing/measurementSpecial data processing applicationsData setEuclidean vector
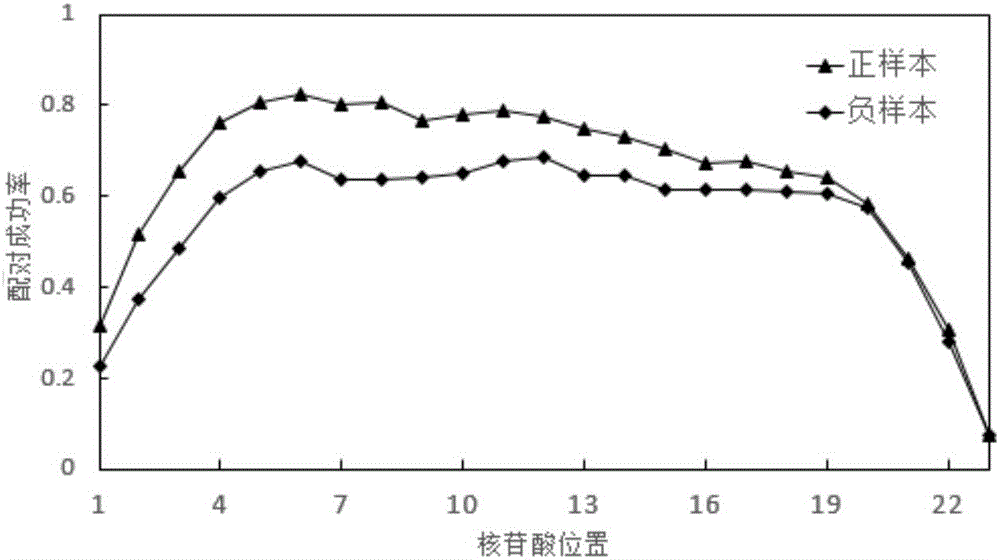
The invention discloses a microRNA target position point prediction method based on a support vector machine, comprising the following steps: 1) building a training data set comprising 278 positive samples and 194 negative samples; 2) building a characteristic set: the sample of each characteristic set is represented by one characteristic vector which contains each aspect information of the miRNA-target position point regulation pair and is divided into six parts, i.e. 128 characteristics; 3) selecting a simplified characteristic set: using a series of characteristic selection algorithms in Weka3 to screen 64 characteristics; 4) result evaluation: comparing the classifying capability of classifiers based on the characteristic set, the simplified characteristic set and the miTarget characteristic set; and 5) function annotation of an miRNA target gene. The invention has the meaning of building a characteristic which is found to be relative with miRNA target position point combination in recent years and developing a set of new miRNA target position point prediction methods; the predictor is optimized by the means of characteristic selection; the detection result comparison shows that the new adopted characteristic can help to predict the miRNA target position point.
Owner:ZHEJIANG UNIV
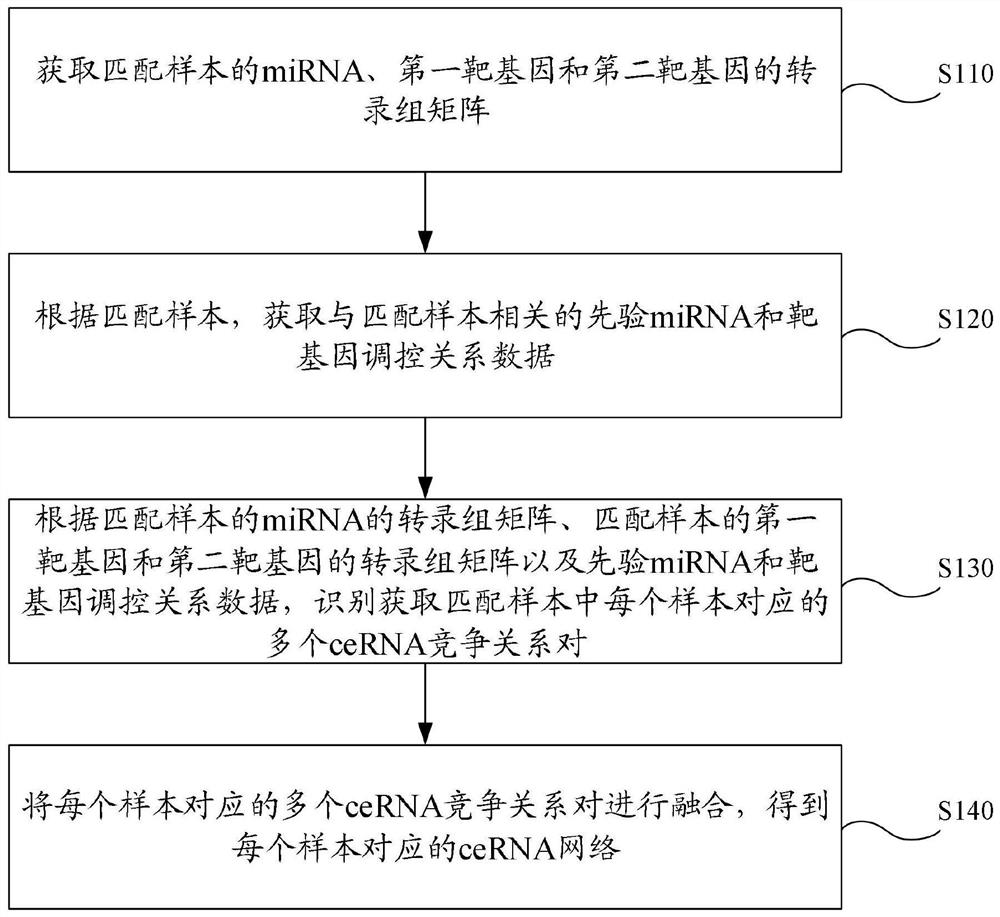
Single-sample ceRNA network identification method and device, electronic equipment and storage medium
The invention provides a single-sample ceRNA network identification method and device, electronic equipment and a storage medium, and relates to the technical field of gene identification. The single-sample ceRNA network identification method comprises the following steps: identifying and acquiring a plurality of ceRNA competition relation pairs corresponding to each sample in matched samples according to a transcriptome matrix of miRNA of the matched samples, transcriptome matrixes of a first target gene and a second target gene of the matched samples and priori miRNA and target gene regulation and control relation data; and fusing the plurality of ceRNA competition relationship pairs corresponding to each sample to obtain a ceRNA network corresponding to each sample. According to the method, the ceRNA network of the single sample is obtained based on the miRNA of the matched sample, the transcriptome matrix of the first target gene and the transcriptome matrix of the second target gene and the regulation and control relation data of the priori miRNA and the target genes, the gene regulation and control mechanism in the single sample can be disclosed, and the competitive relation between the miRNA target genes under the single sample can be reflected.
Owner:DALI UNIV
Using miRNA-gene co-expression networks to predict abnormal gap junctional communication in anthracycline-treated cardiomyocytes
The invention relates to a method for predicting anthracycline drug cardiomyocyte gap junction communication abnormality using miRNA-gene co-expression network, comprising the following steps: Step 1: Experimental grouping; Step 2: RNA extraction; Step 3: miRNA-gene chip data Differential expression analysis; step 4: miRNA target gene analysis and regulatory network construction. The present invention uses the miRNA-gene co-expression network to predict the abnormality of anthracycline drug cardiomyocyte gap junction communication, and predicts the abnormal cardiomyocyte gap junction communication through the miRNA-gene co-expression network, which is a new molecular marker for discovering anthracycline drug cardiotoxicity A new approach is provided, which provides a reference for predicting its prognosis.
Owner:JINZHOU MEDICAL UNIV
Nucleic Acids and Their Uses
The invention provides a separated nucleic acid with a miRNA target gene binding site sequence. By the separated nucleic acid in an embodiment, miRNA reduction can be realized. Especially, as for high-CG-content miRNA with CG content being up to 90%, reduction efficiency of the nucleic acid is remarkably improved as compared with that in the prior art.
Owner:FIELD OPERATION BLOOD TRANSFUSION INST OF PLA SCI ACAD OF MILITARY +1
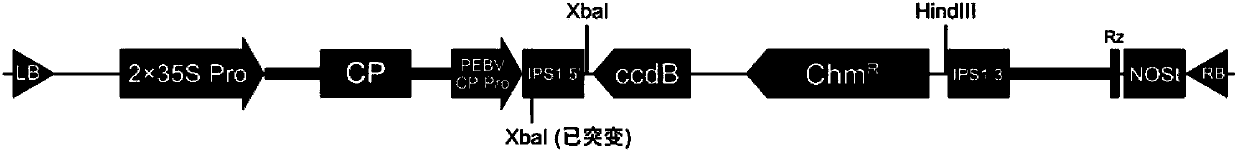
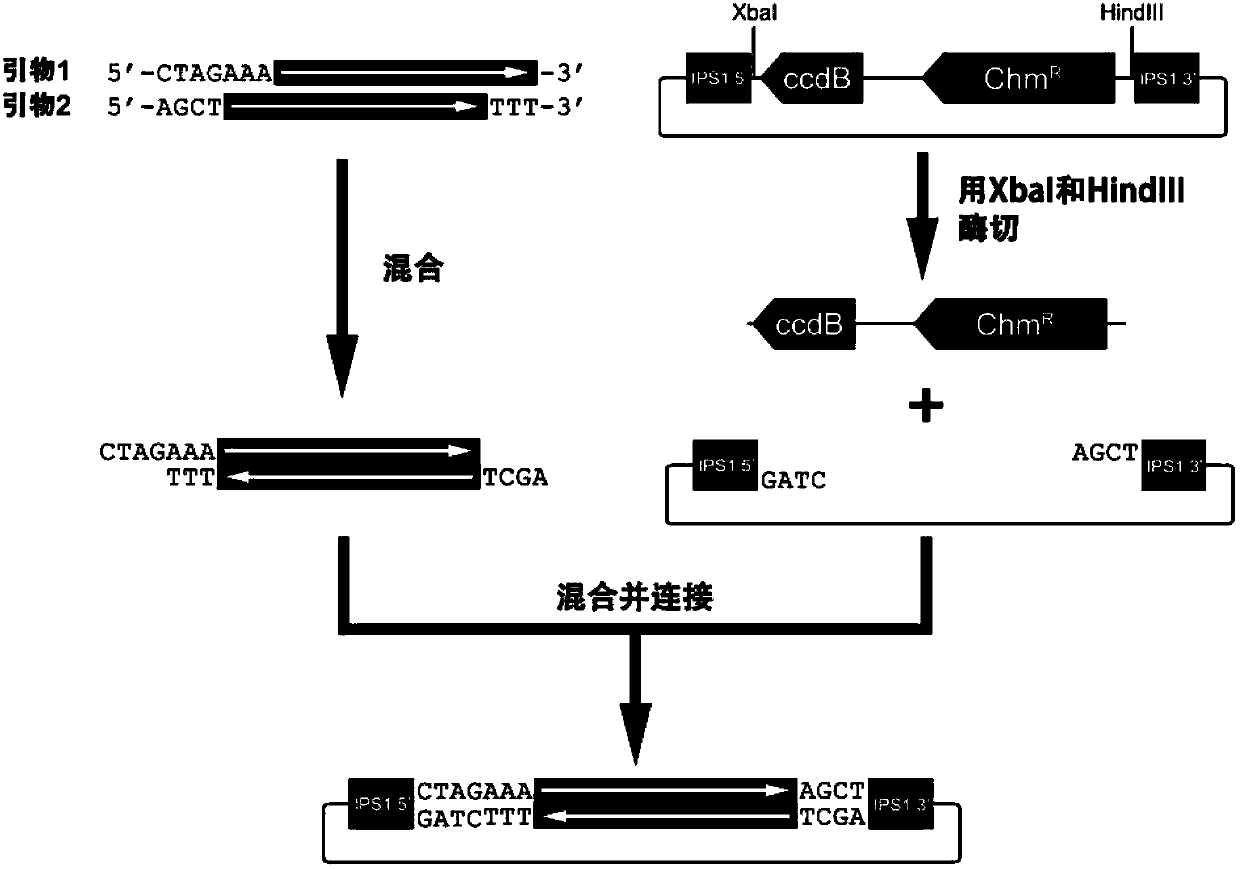
Method for constructing and expressing micro ribonucleic acid (miRNA) target simulation sequence by using plant virus vector
ActiveCN102703500BHigh expressionSimple methodFermentationVector-based foreign material introductionChemical synthesisNicotiana tabacum
The invention discloses a method for constructing and expressing a micro ribonucleic acid (miRNA) target simulation sequence by using a plant virus vector. Experiments prove that deoxyribonucleic acid (DNA) molecules which are chemically synthesized and encoded and have the miRNA158 or miRNA172 target simulation sequences are inserted into the plant virus expression vector pTY102 to obtain a recombinant expression vector, the recombinant expression vector is imported into tobaccos leaf, and after three weeks, the phenotypic modulation of tobacco and the raising of the expression of the miRNA target genes can be detected. According to the method, bridge polymerase chain reaction (PCR) is not needed; the method is simple; and by using the characteristics of high expression, wide host range and high speed of spreading in the whole plant of virus, the expression level of plant endogenous miRNA can be reduced quickly and conveniently, and a new way is provided for researching the plant endogenous miRNA function.
Owner:TSINGHUA UNIV
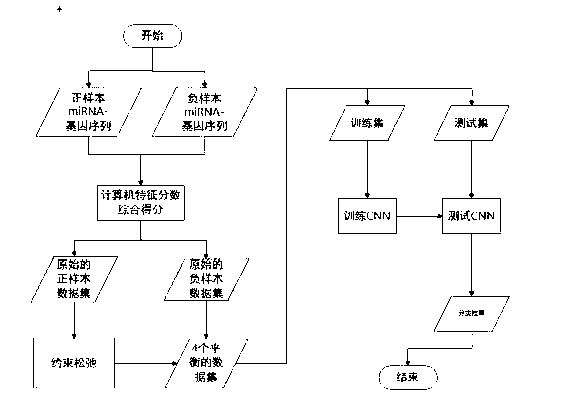
Prediction method of miRNA target genes based on convolutional neural network
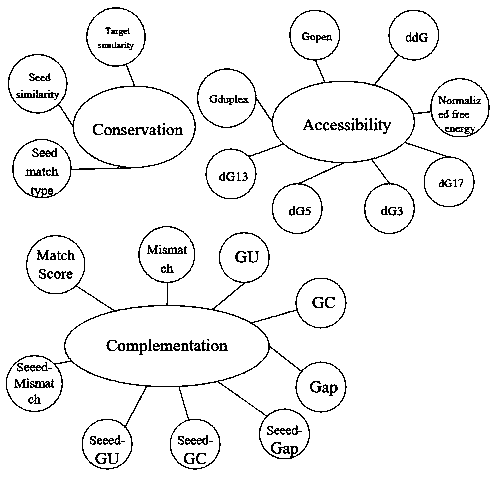
The invention provides a new algorithm (CNNmiRT) to predict miRNA target genes by utilizing the features of complementation, conservation and accessibility between miRNA-target genes. Because experiment support on negative interaction is not published commonly, and is not recorded in a database, therefore the number of loci of verified negative samples is far lower than that of loci of positive samples. A restraint release method is used to build four kinds of balanced experimentally-verified training data sets in order to compensate, and the four kinds of balanced experimentally-verified training data sets comprise one highly-conservative positive sample data set, one completely-complementary positive sample data set, one accessible positive sample data set and one negative sample data set. The method not only avoids wrong filtering of real targets which do not meet certain feature thresholds, but also solves the problem of disequilibrium of the experimentally-verified data sets. Then, the miRNA target genes are predicted by applying the convolutional neural network.
Owner:SUN YAT SEN UNIV
A method for releasing miRNA repressive function to promote target gene expression
ActiveCN110157730BEasy to operateLow running costVector-based foreign material introductionBase JGenome editing
The invention discloses a strategy for releasing the inhibitory function of miRNA to promote the expression of target genes, and provides a specific implementation method based on genome editing technology. The recognition sequence of the miRNA target gene is edited by gene editing technology, the base substitution mutations and amino acid deletion mutations with normal function of the target gene are screened, and the mutants whose recognition sequence cannot be suppressed by the upstream miRNA is destroyed. The invention is to promote the function of the target gene Research and functional applications provide an easy and efficient way.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com