Analysis and identification method on miRNA key target gene of blood fluke in specific growth period
A developmental stage and key gene technology, applied in the field of life medical science, can solve problems such as difficult to accurately determine key target genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0050] A method for analyzing and identifying key target genes of Schistosoma japonicum miRNA at a specific developmental stage, comprising the following steps:
[0051] Step 1: Analysis of miRNA expression characteristic of Schistosoma japonicum at specific developmental stages:
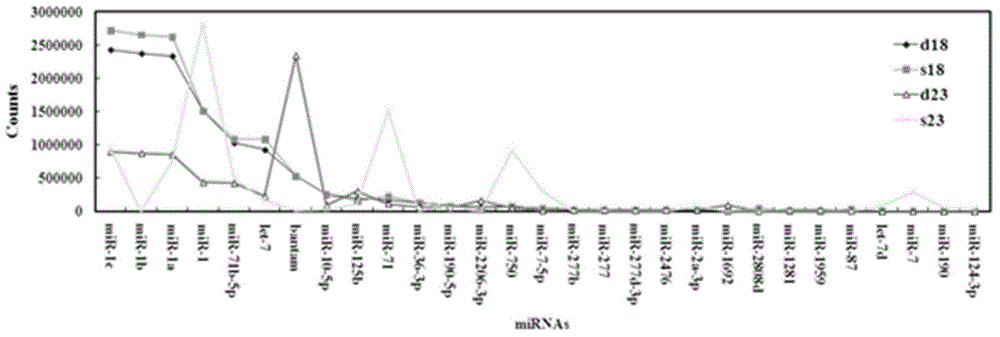
[0052]Through Solexa analysis, compare the miRNA expression characteristics of Schistosoma japonicum in different developmental stages, and determine the characteristic miRNA of Schistosoma japonicum in specific developmental stages; Body miRNA expression profiles. Characteristic miRNAs differentially expressed in different periods were analyzed. Such as figure 1 . from figure 1 It can be seen that the miRNA expression profiles are different in different developmental stages of worms, suggesting that each stage has its own characteristics. For example, at 18 days after infection, the expression of miRNA-1c, miRNA-1b and miRNA-1a increased in single-sex (s18) or double-sex (d18) infected females...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com