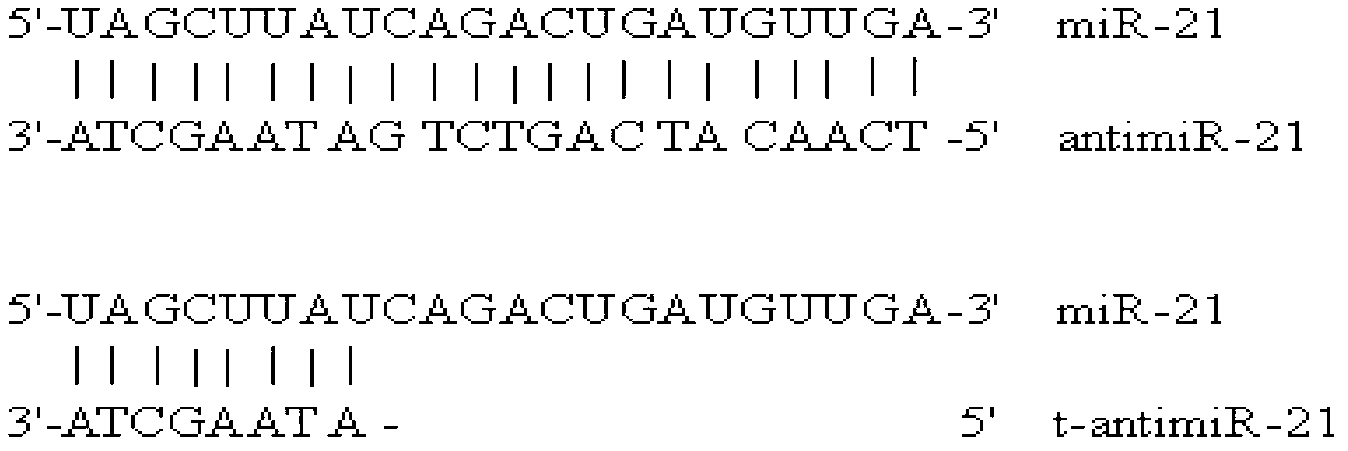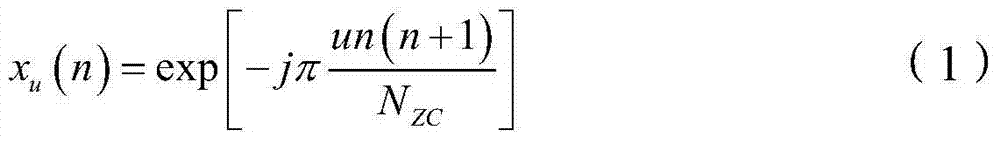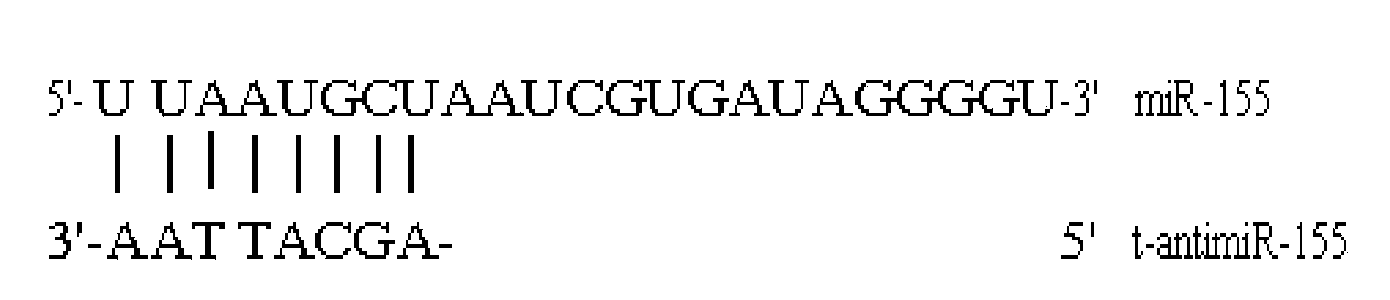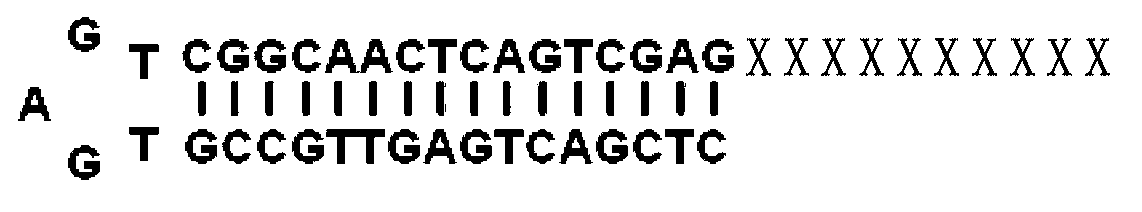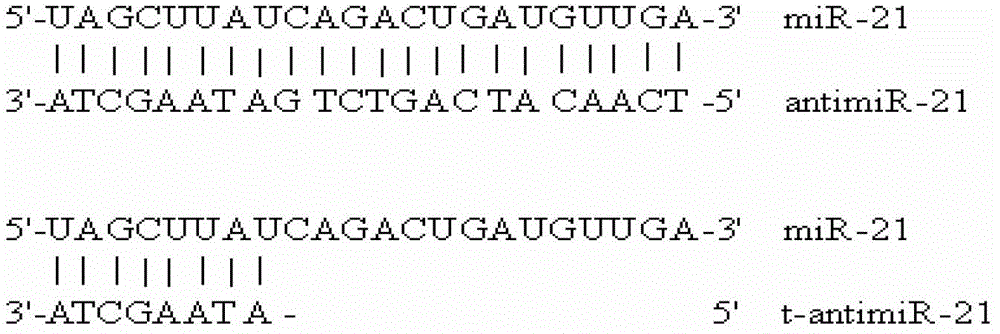Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
56 results about "Seed sequence" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Seed sequence. A seed sequence is an object that uses a sequence of integer values to produce a series of unsigned integer values with 32 significant bits that can be used to seed a pseudo-random generator engine.
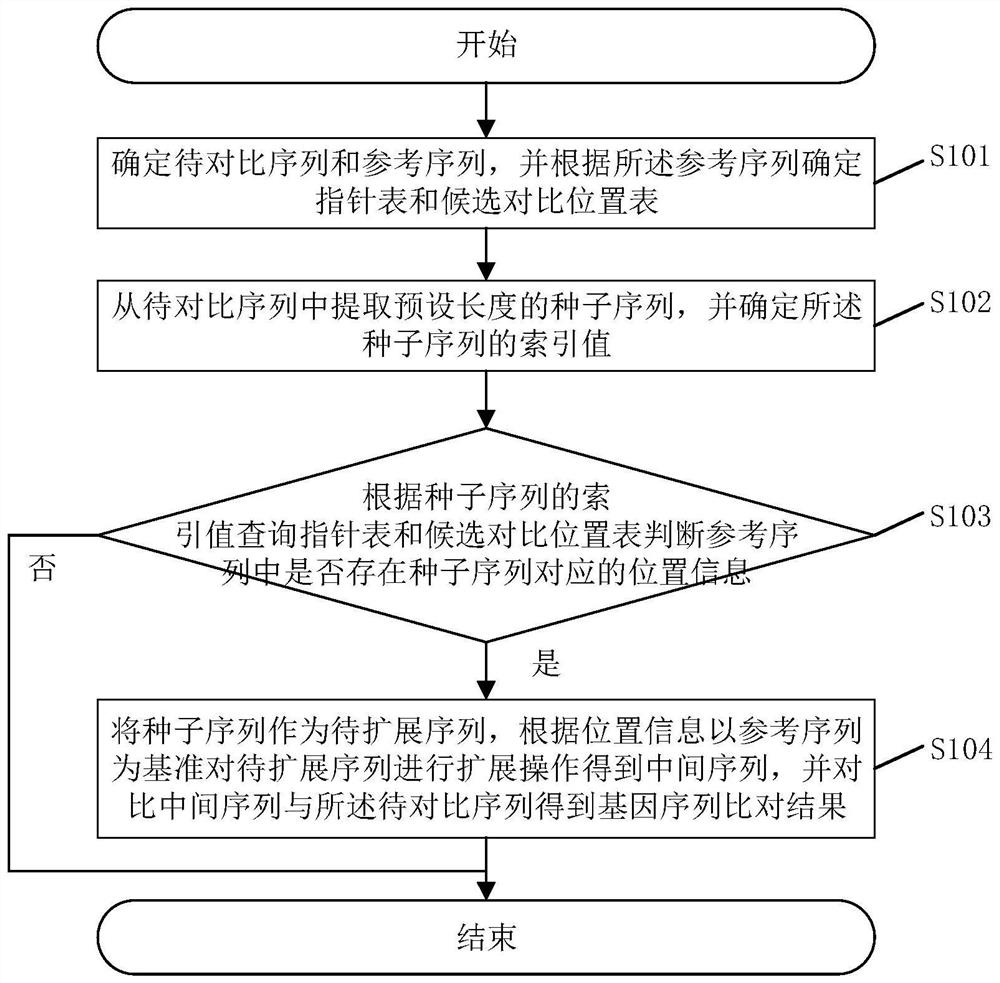
Gene sequence comparison method and system, and related assembly
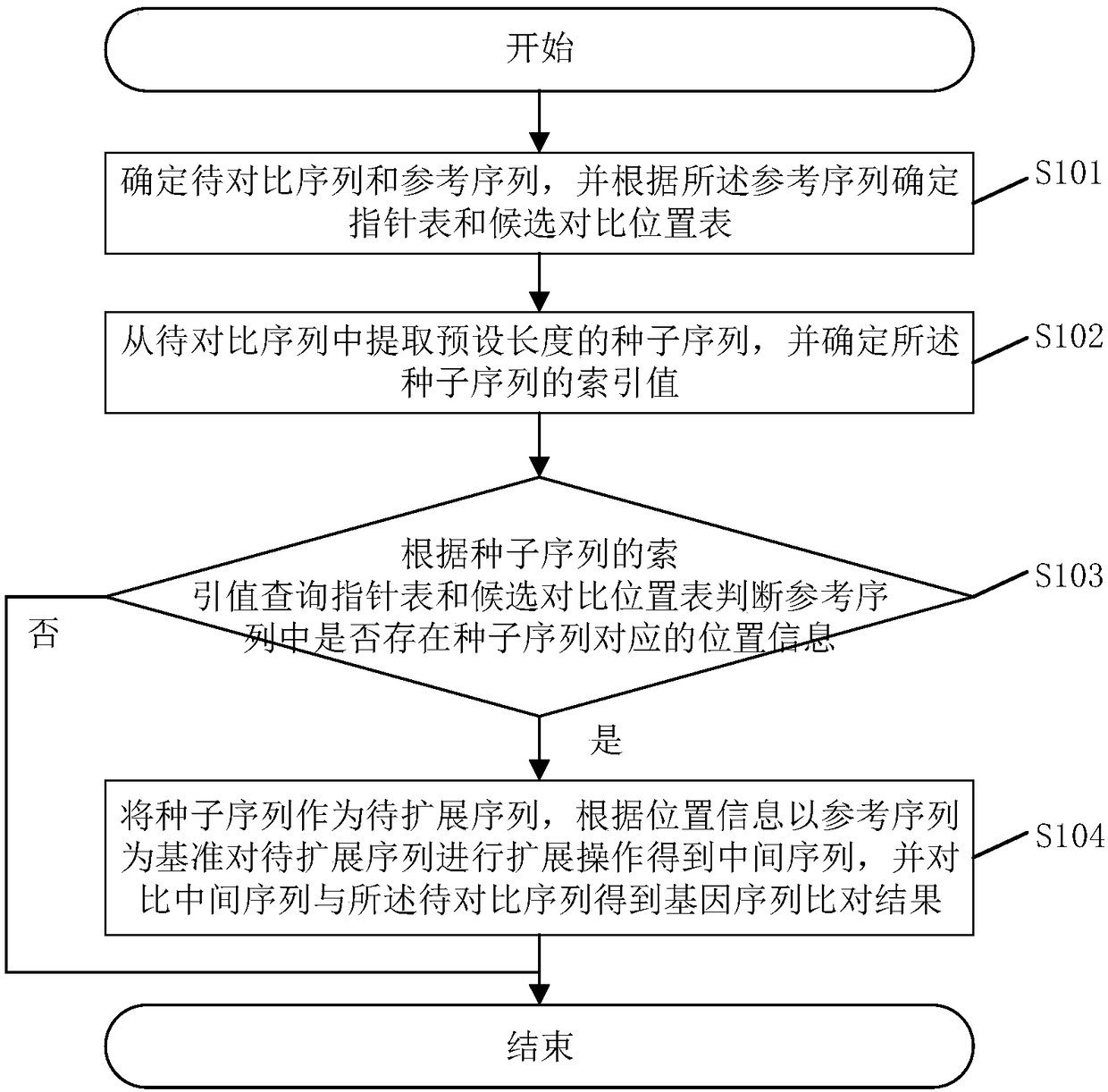
The invention discloses a gene sequence comparison method. The method comprises steps that a to-be-compared sequence and a reference sequence are determined, and a pointer table and a candidate comparison position table are determined according to the reference sequence; a seed sequence of a preset length is extracted from the to-be-compared sequence, and an index value of the seed sequence is determined; the pointer table and the candidate comparison position table are queried according to the index value of the seed sequence; whether the position information corresponding to the seed sequence exists in the reference sequence is determined; if yes, the seed sequence is used as a to-be-extended sequence, according to the position information, on the basis of the reference sequence, the to-be-extended sequence is extended to obtain an intermediate sequence, and the intermediate sequence and the to-be-compared sequence are compared to obtain the gene sequence comparison result. The method is advantaged in that the storage space of the table storing the reference sequence index value can be reduced, and the gene sequence comparison rate can be increased. The invention further discloses a gene sequence comparison system, a computer readable storage medium and an electronic device. The advantages are realized.
Owner:ZHENGZHOU YUNHAI INFORMATION TECH CO LTD
Automatic steppe burn scar detection method based on MODIS data
InactiveCN1763560AEasy to observeAccurate extractionElectromagnetic wave reradiationPoint sequenceGrassland
The invention discloses a MODIS data self-detection method of grassland fire hazard land, which comprises the following steps: MODIS data receiving step from satellite, fire hazard land index GEMI-B construction step, fire point detection step, point sequence step, comparison step. If the comparison result doesn't approximate the statistic feature value, the data removes from the seed point sequence; if approximates, the data spreads outsides at the center of land pixel and adds the pixel into the seed sequence, which is removed from the seed point sequence. If the seed point quantity is not zero, the operation returns to the comparison step; if zero, the operation precedes the land image output step.
Owner:INST OF AGRI RESOURCES & REGIONAL PLANNING CHINESE ACADEMY OF AGRI SCI
Alignment method, apparatus and system
ActiveCN107403075AEfficient and accurate processingIncreased sensitivityBiostatisticsHybridisationTheoretical computer scienceComputer science
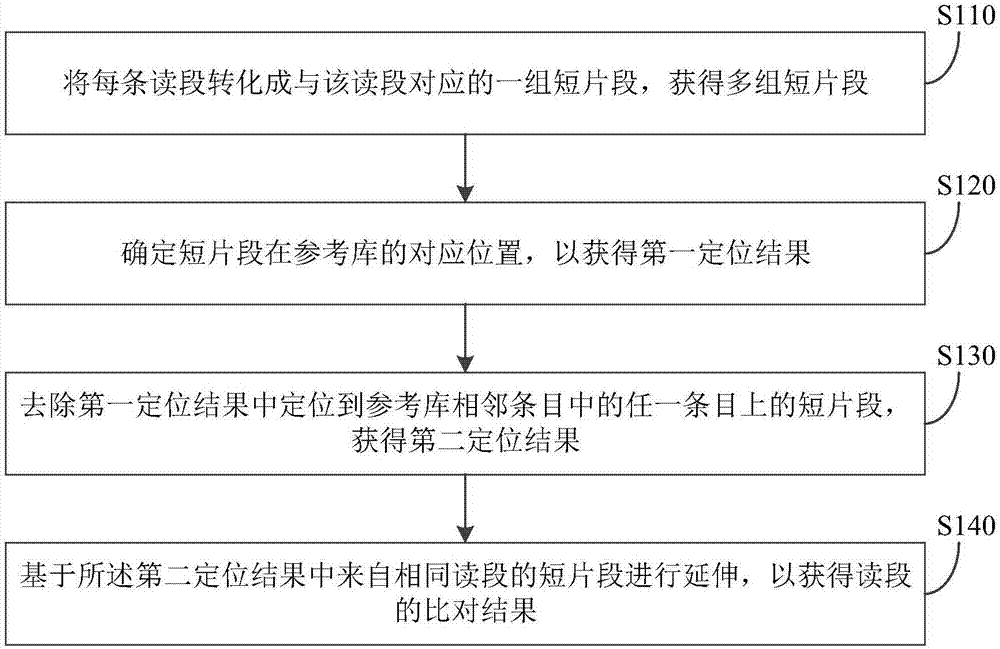
The invention discloses an alignment method, apparatus and system. The alignment method comprises converting each read into a set of short fragments corresponding to the read, and obtaining multiple sets of short fragments; determining corresponding positions of the short fragments in a reference library so as to obtain a first positioning result, wherein the reference library is a Hash table constructed on basis of a reference sequence, the reference library includes a plurality of entries, each entry of the reference library is corresponding to a seed sequence, each seed sequence can be matched with at least one sequence fragment on the reference sequence, and a distance between two seed sequences corresponding to two adjacent entries of the reference library on the reference sequence is less than the length of the short fragments; removing the short fragment positioned to any one of the adjacent entries of the reference library in the first positioning result, and obtaining a second positioning result; and performing extending based on common parts of the short fragments from the same read in the second positioning result so as to obtaining an alignment result of the read. Sequencing data can be processed and positioned efficiently and accurately by the alignment method.
Owner:GENEMIND BIOSCIENCES CO LTD
Methods, libraries and computer program products for determining whether siRNA induced phenotypes are due to off-target effects
InactiveUS20080009012A1Reduce the possibilitySugar derivativesActivity regulationP PHENOTYPEOff targets
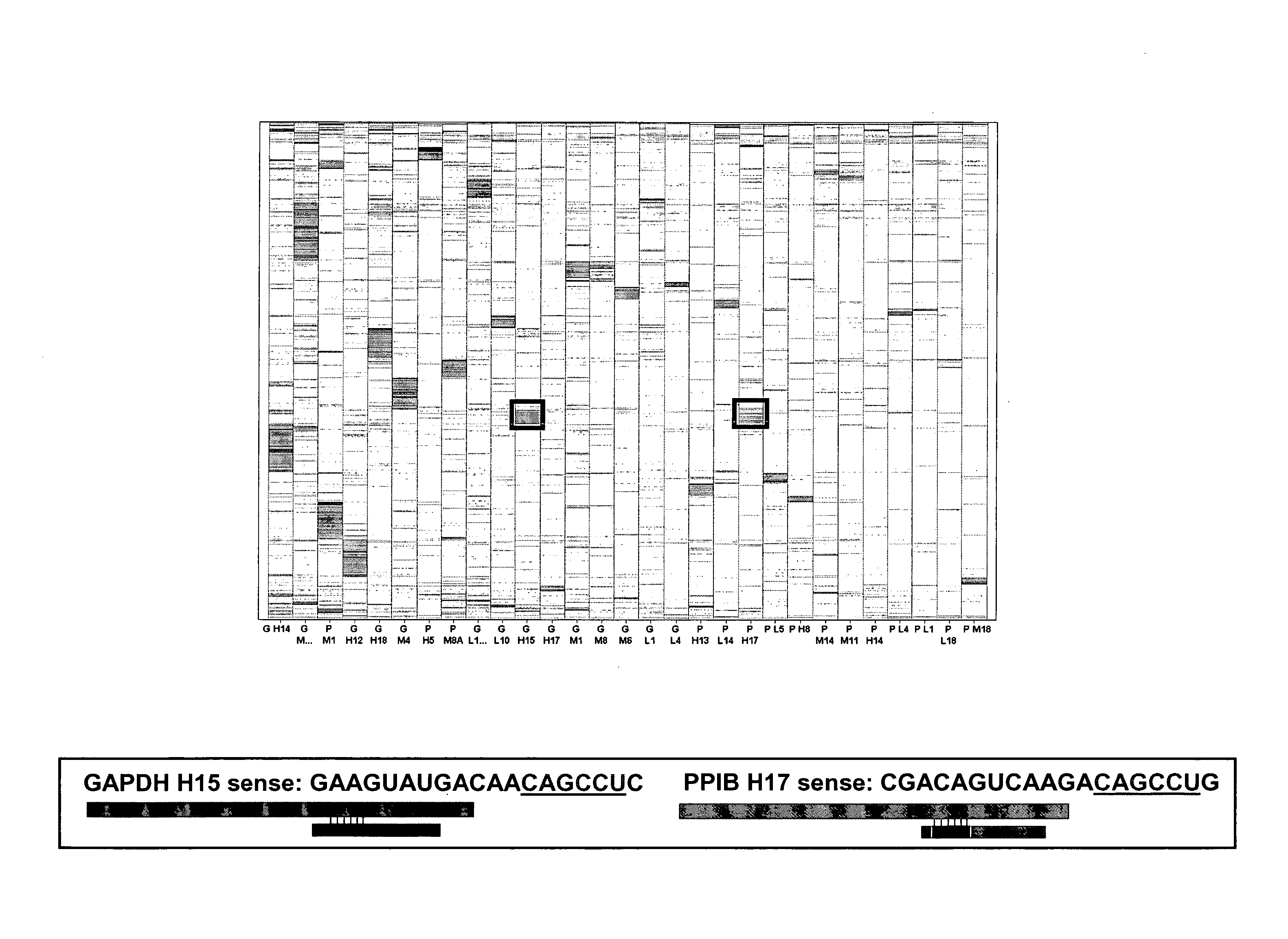
The present disclosure provides methods, libraries and computer program products for determining whether a phenotype induced by a candidate siRNA for a target gene in an RNAi experiment is target specific or a false positive. Through the use of a control siRNA that has one or two seed sequences of six or seven bases in combination with a neutral scaffolding sequence, a distinction can be made between false positive and true positive analyses of functionality.
Owner:DHARMACON INC
Compression method for next generation sequencing data
ActiveCN105760706ASave storage spaceProcessing speedSpecial data processing applicationsCompression methodParallel processing
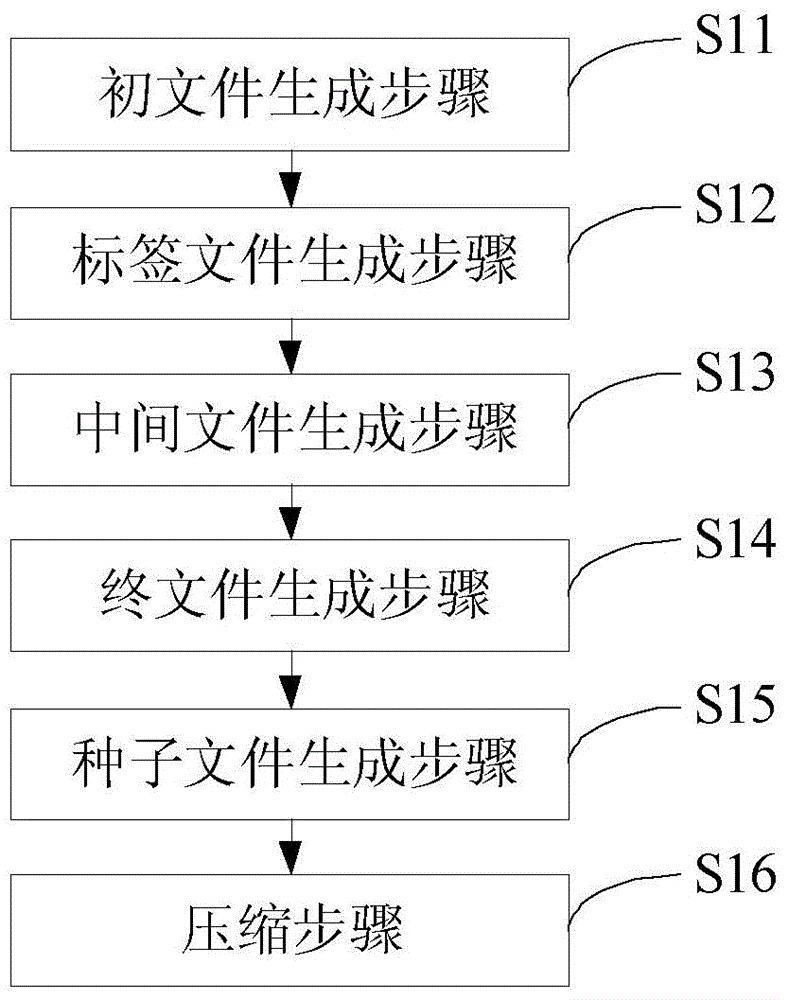
The invention discloses a compression method for next generation sequencing data. The method comprises: dividing the next generation sequencing data of each sample according to a first preset length, to generate a BSSL original file; according to a second preset length, establishing a cutting tag file; according to the cutting tag file, processing the BSSL original file, to obtain BSSL intermediate files; combining the BSSL intermediate files to obtain a BSSL final file; counting a frequency distribution result of a seed sequence in the BSSL final file, to obtain a seed file according to the result; combined with the format characteristics of the sequencing data, determining compression rules, and based on the seed file, compressing the next generation sequencing data of each sample. Through dividing the next generation sequencing data and performing parallel processing, processing speed is improved, and combined with seed sequence selection, the seed file is obtained, and the next generation sequencing data is compressed according to the format characteristics of the sequencing data and the seed file, so that storage space of the next generation sequencing data is greatly reduced.
Owner:SHENZHEN HUADA GENE INST
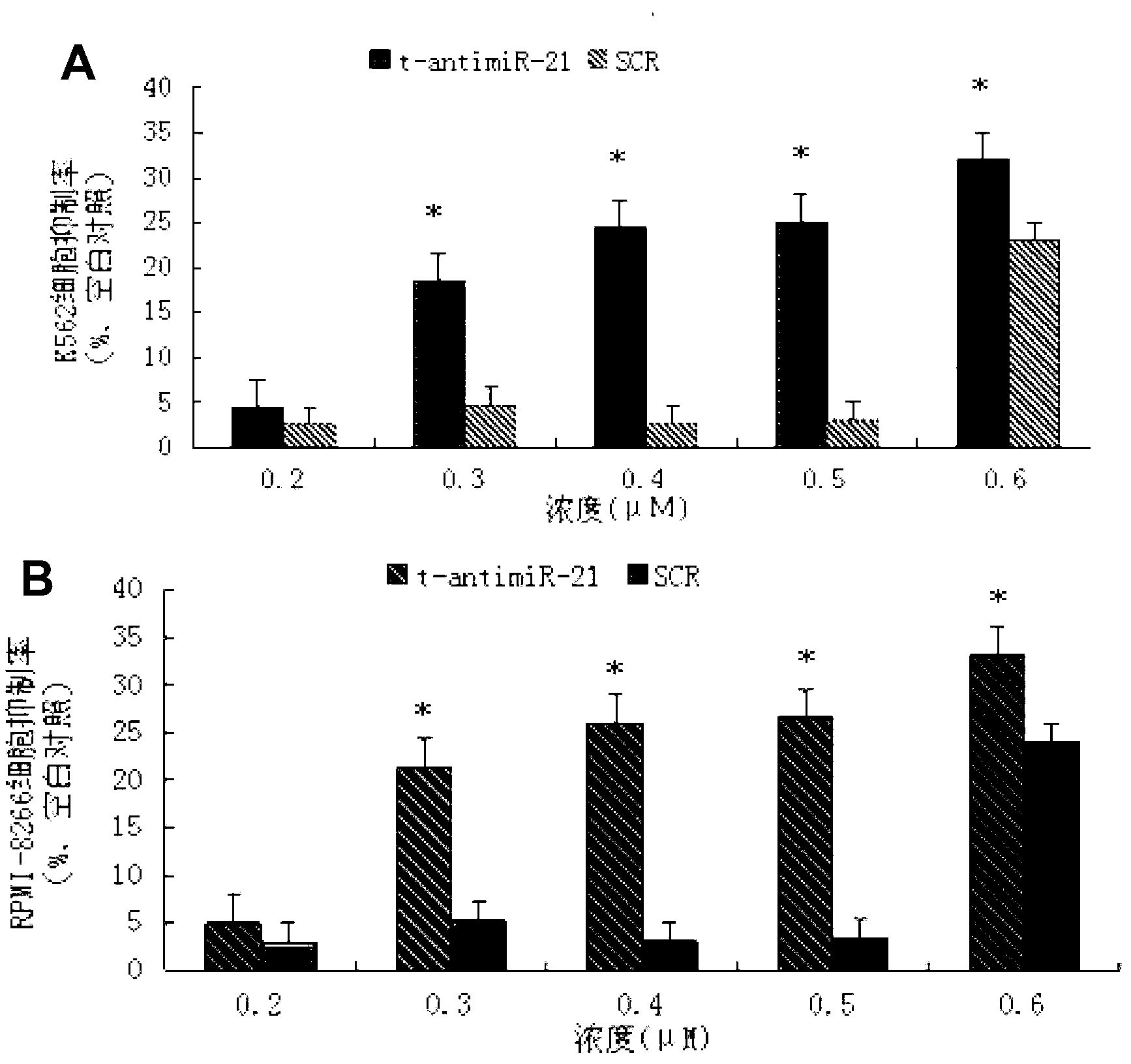
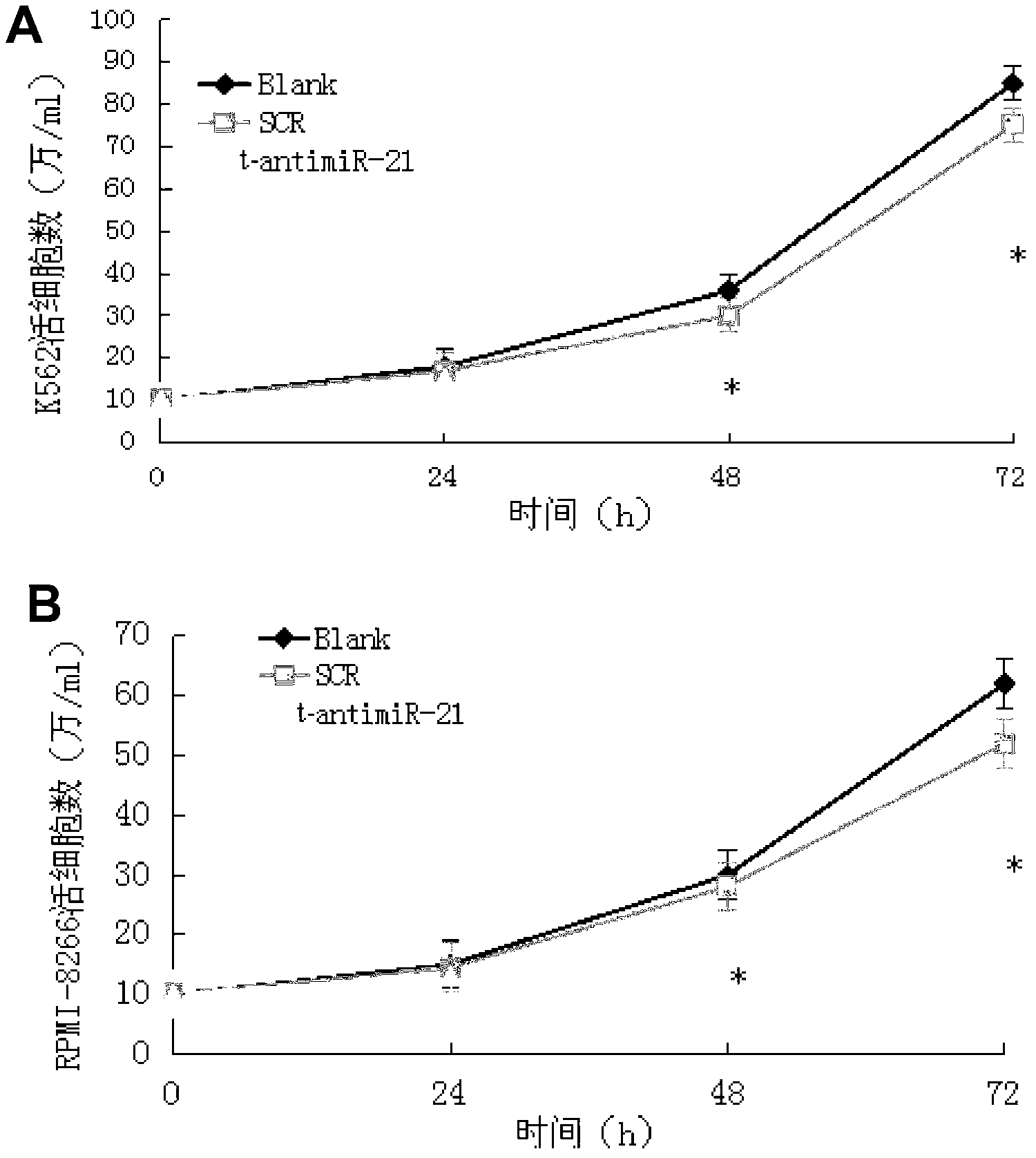
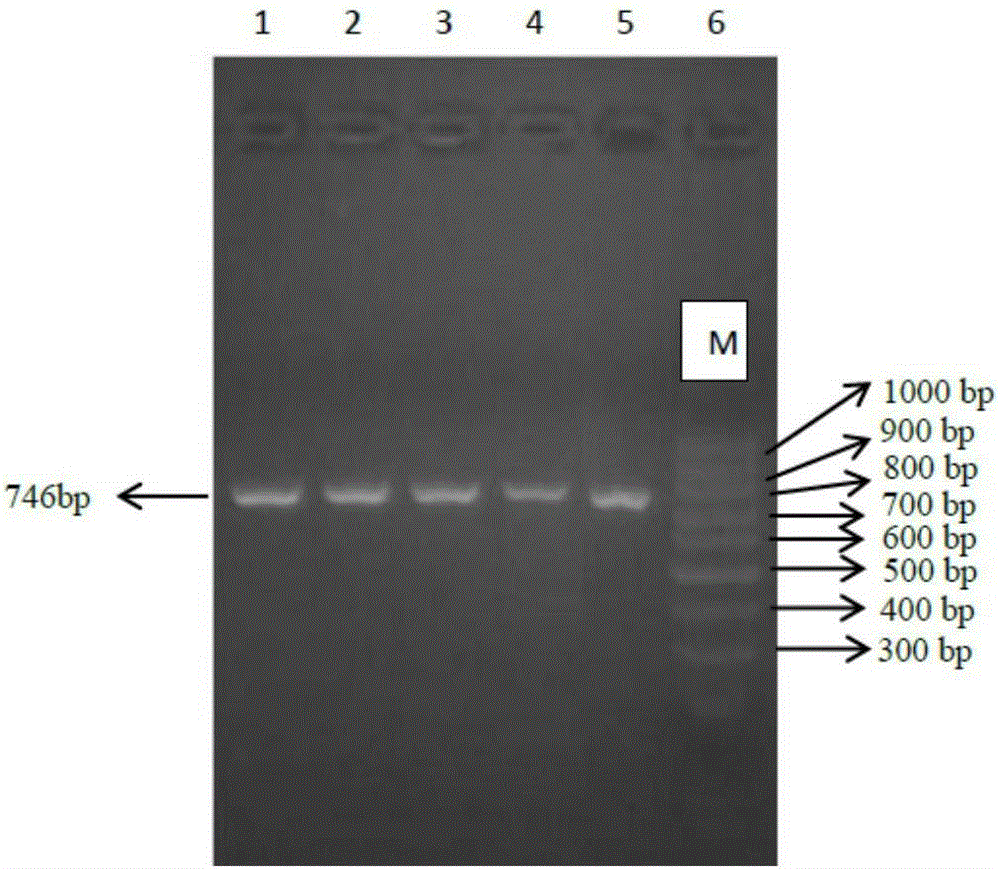
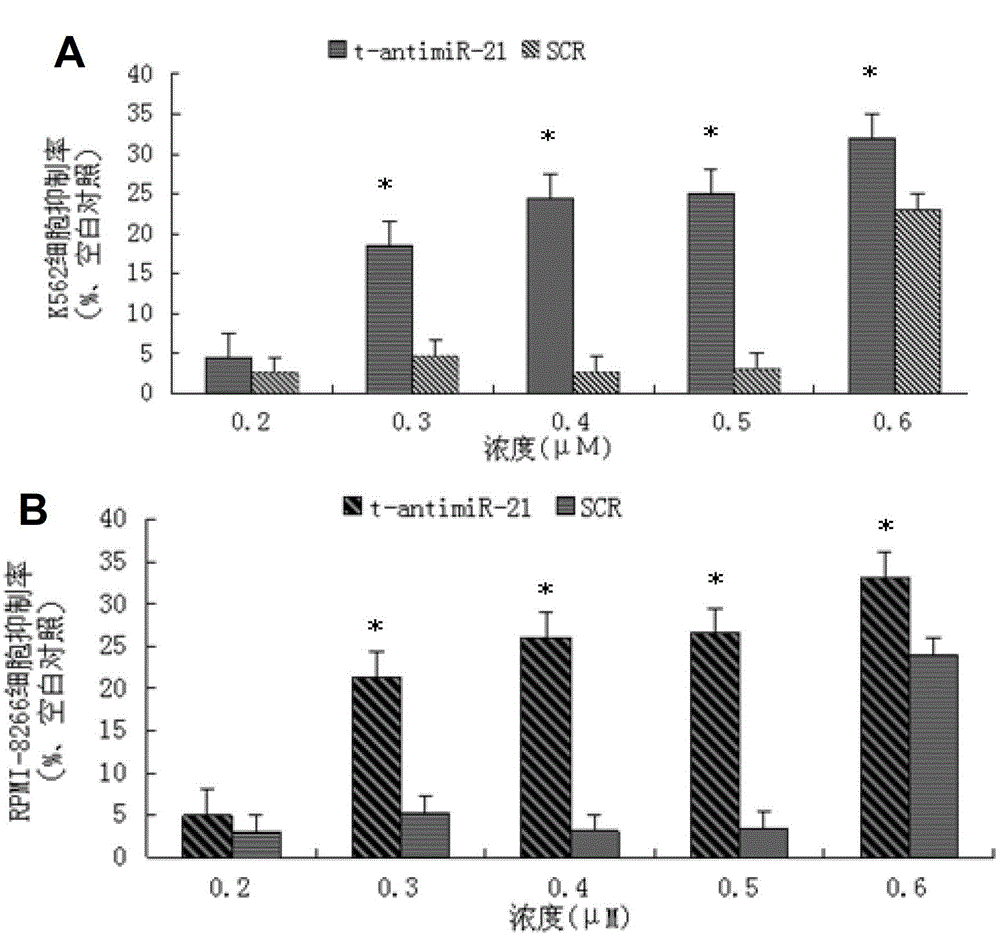
Antisense oligonucleotide for tiny RNA-21 seed sequence and application thereof
ActiveCN103320444AGrowth inhibitionInhibitionGenetic material ingredientsAntineoplastic agentsAntisense nucleic acidChronic granulocytic leukemia
The invention discloses an antisense oligonucleotide for tiny RNA-21 seed sequence and an application thereof and belongs to the field of medicine preparation. According to the research, tiny antisense nucleic acid t-antimiR-21 of 8 basic groups is designed and synthetized for miR-21 seed sequence. It shows through a t-antimiR-21 transfection cell experiment that t-antimiR-21 targets miR-21 of K562 cell and RPMI-8266 cell strain, cell growth is inhibited and cell apoptosis is obviously increased. It shows that lymphoma cell growth can be effectively inhibited through interfering with miRNA-21 expression by t-antimiR-21. Meanwhile, it shows that miRNA-21 might be used as a potential target for lymphoma gene therapy. t-antimiR-21 can be used in the preparation of an antineoplastic medicine, especially in the preparation of a medicine against chronic granulocytic leukemia and multiple myeloma.
Owner:广州安镝声生物医药科技有限公司
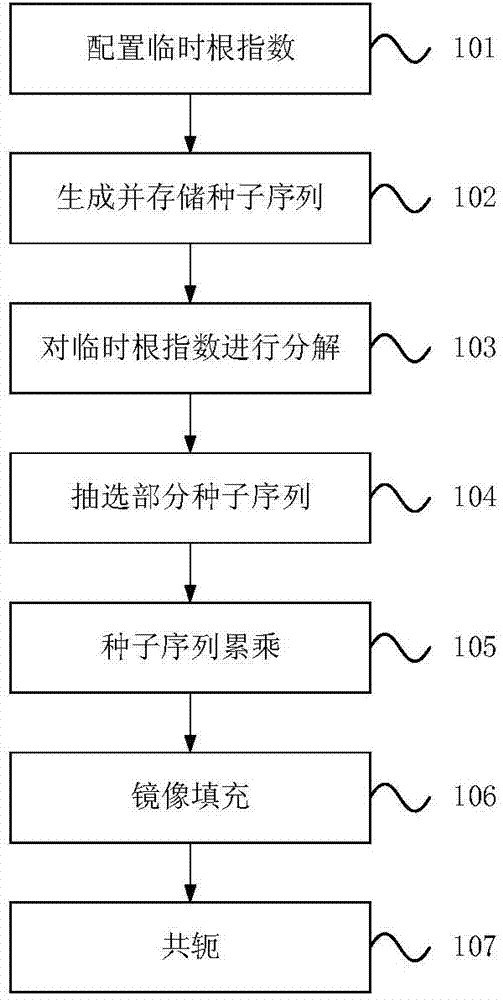
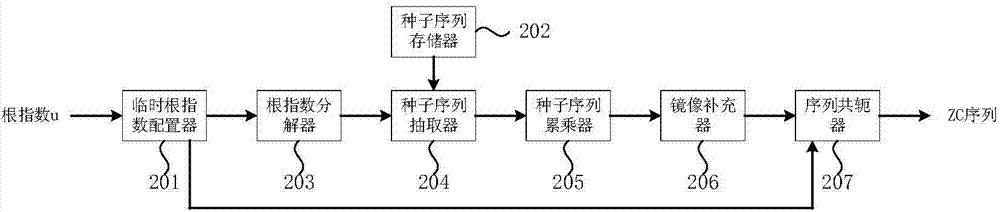
Method and device for generating ZC (Zadoff-Chu) sequence
ActiveCN104506271ASave storage spaceProduce quicklyMultiplex code generationWeight coefficientComputer science
The invention provides a method for generating a ZC (Zadoff-Chu) sequence. The method comprises the following steps: (1) determining a current radical exponent according to the radical exponent of the ZC sequence needing to be generated; (2) decomposing the current radial exponent into an expression of the weights and forms of seed sequence radical exponents in a seed sequence set, wherein weight coefficients in the expression are non-negative integers; the seed sequence set is a set of pre-stored ZC sequences having different radical exponents; and each ZC sequence stored in the seed sequence set is a seed sequence; and (3) multiplying corresponding seed sequences according to the weight coefficients obtained in the step (2) to obtain a ZC sequence corresponding to the current radical exponent in order to obtain the ZC sequence needing to be generated. The invention further provides a corresponding device for generating the ZC sequence. Through adoption of the method and the device, ZC sequences of any radical exponent can be generated rapidly and accurately; storage space required in the generating process of the ZC sequences can be reduced; and the method and the device are particularly suitable for rapidly generating the ZC sequences through a DSP (Digital Signal Processor).
Owner:北京中科晶上科技股份有限公司
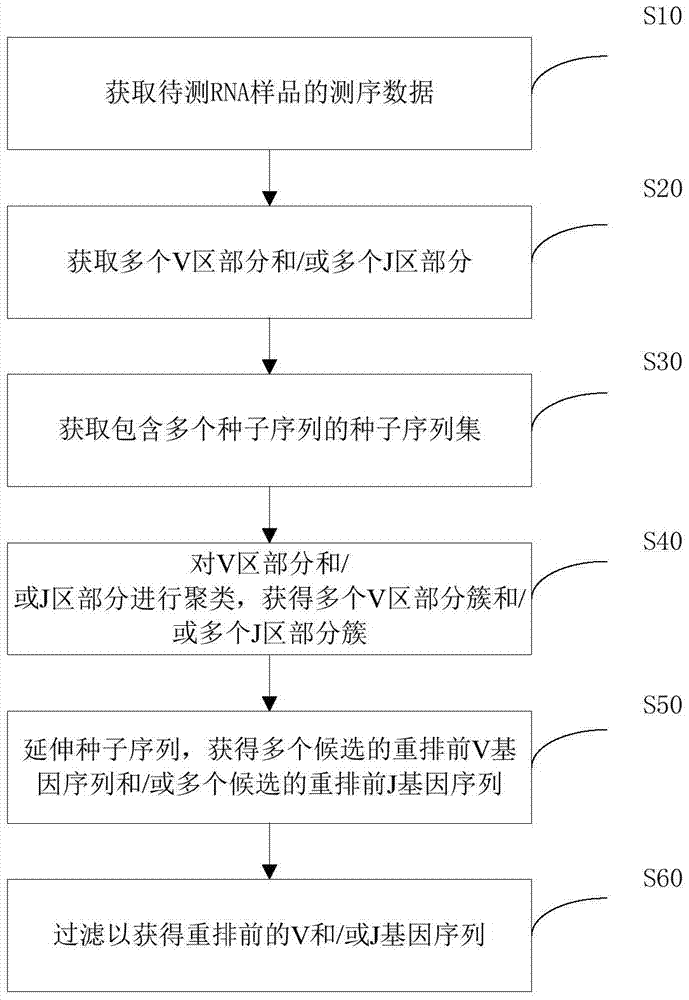
Method and device for determining pre-rearrangement V/J gene sequences
ActiveCN107038349AReduce difficultyShorten the timeMicrobiological testing/measurementProteomicsSupport conditionComputational biology
The invention discloses a method for determining pre-rearrangement V and / or J gene sequences. The method comprises the following steps of: (1) obtaining sequencing data of a to-be-tested RNA sample, wherein the sequencing data comprises a plurality of reads from variable regions of TCR, BCT and / or Ig; (2) determining parts, from V / J gene fragments, on the reads according to arrangement relationship of V, J and C gene fragments in the variable regions on the basis of the sequencing data, so as to obtain a plurality of V / J region parts; (3) taking out at least one section of sequence from each V / J region part as a seed sequence, so as to obtain a seed sequence set comprising a plurality of seed sequences; (4) clustering the V / J region parts according to the difference of support number of the V / J region part of each seed sequence, so as to obtain a plurality of V / J region part clusters; (5) extending seed sequence supported by each V / J region part cluster by utilizing each V / J region part cluster, so as to obtain a plurality of candidate pre-rearrangement V / J gene sequences; and (6) filtering support conditions of the candidate pre-rearrangement V / J gene sequences by utilizing the reads, so as to obtain a pre-rearrangement V / J gee sequence.
Owner:SHENZHEN HUADA GENE INST
INHIBITORS OF MICRORNAs THAT REGULATE PRODUCTION OF ATRIAL NATRIURETIC PEPTIDE (ANP) AS THERAPEUTICS AND USES THEREOF
InactiveUS20150133521A1Increase probabilityImprove high blood pressureOrganic active ingredientsSugar derivativesHypertension riskBlood plasma
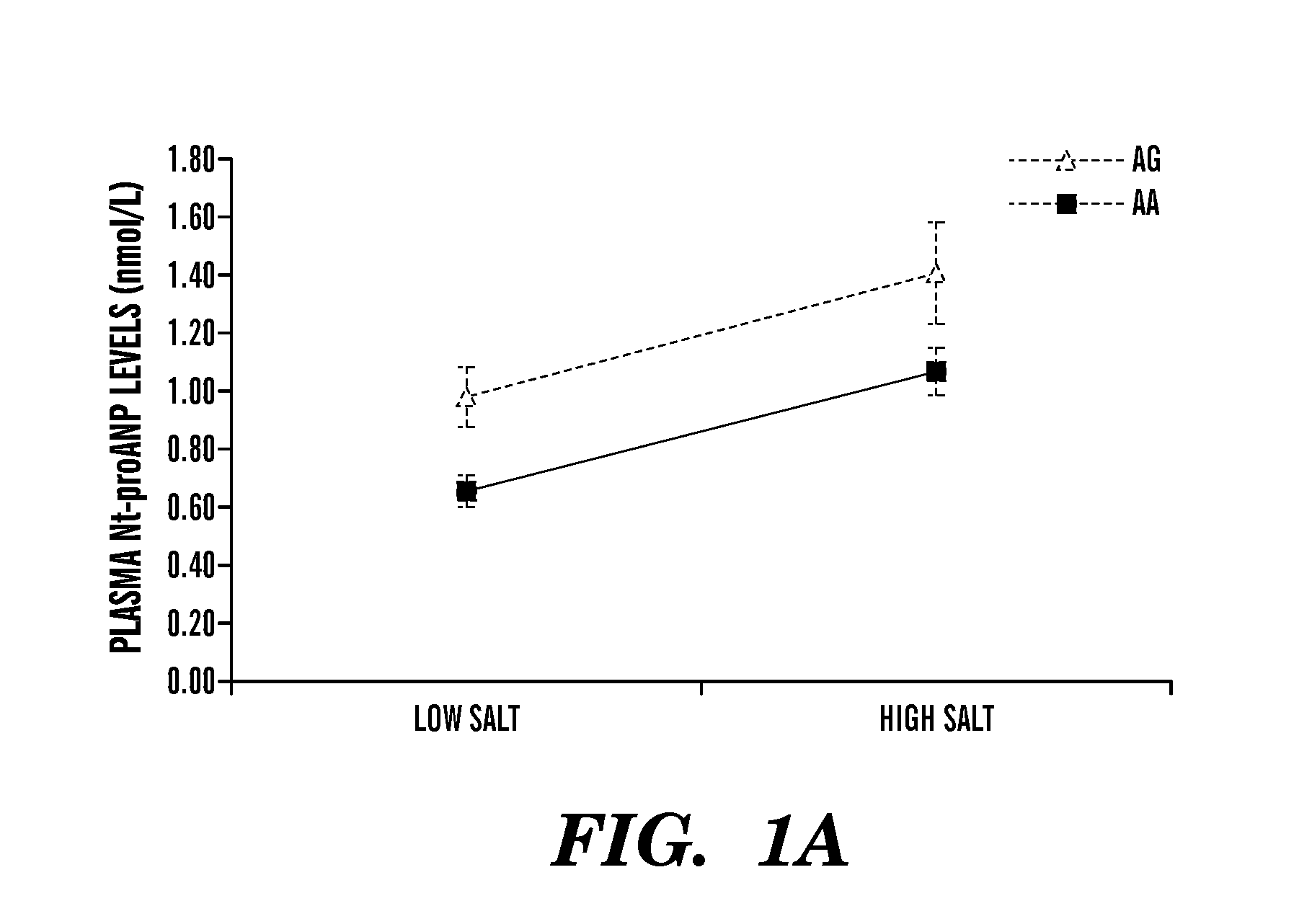
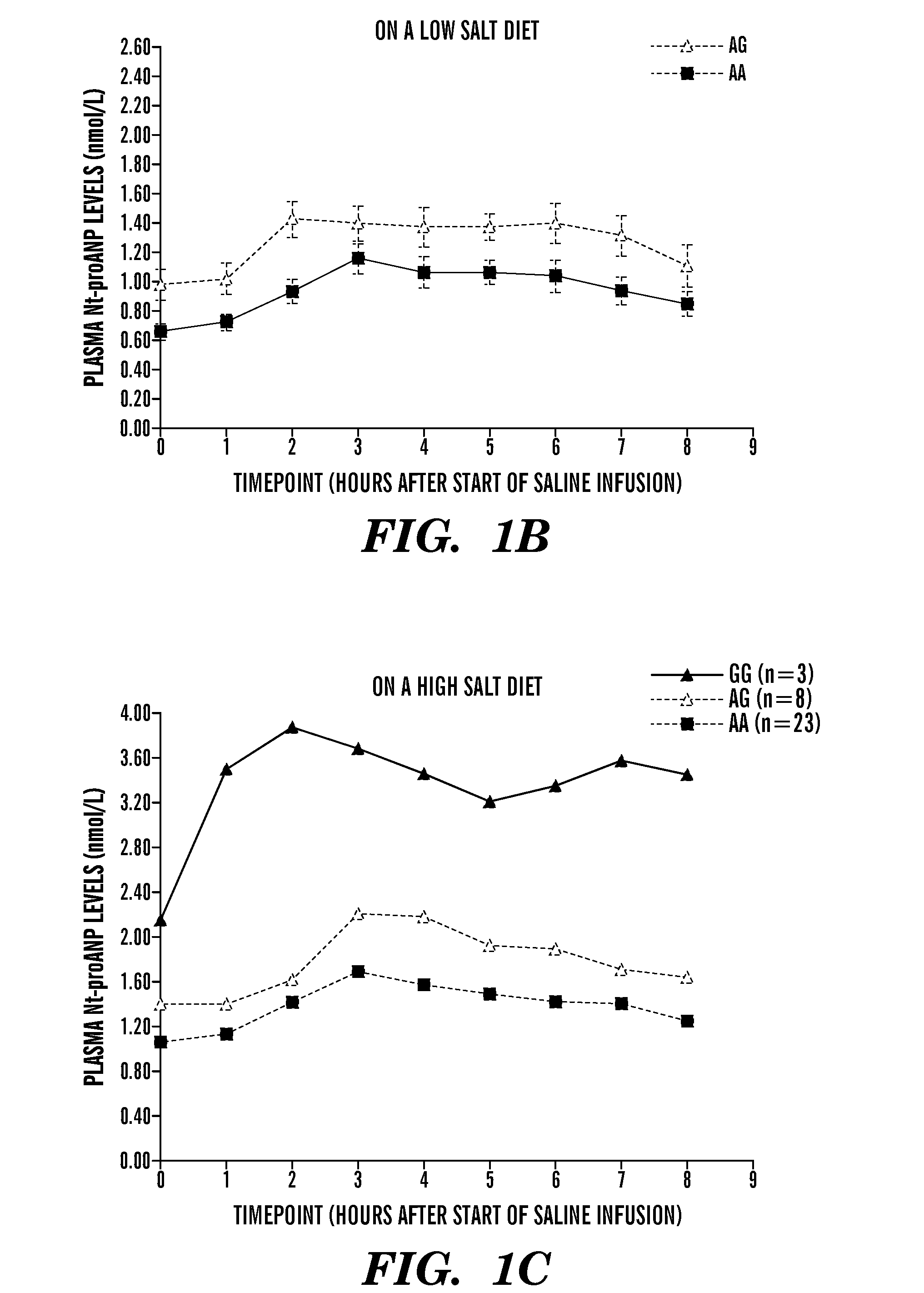
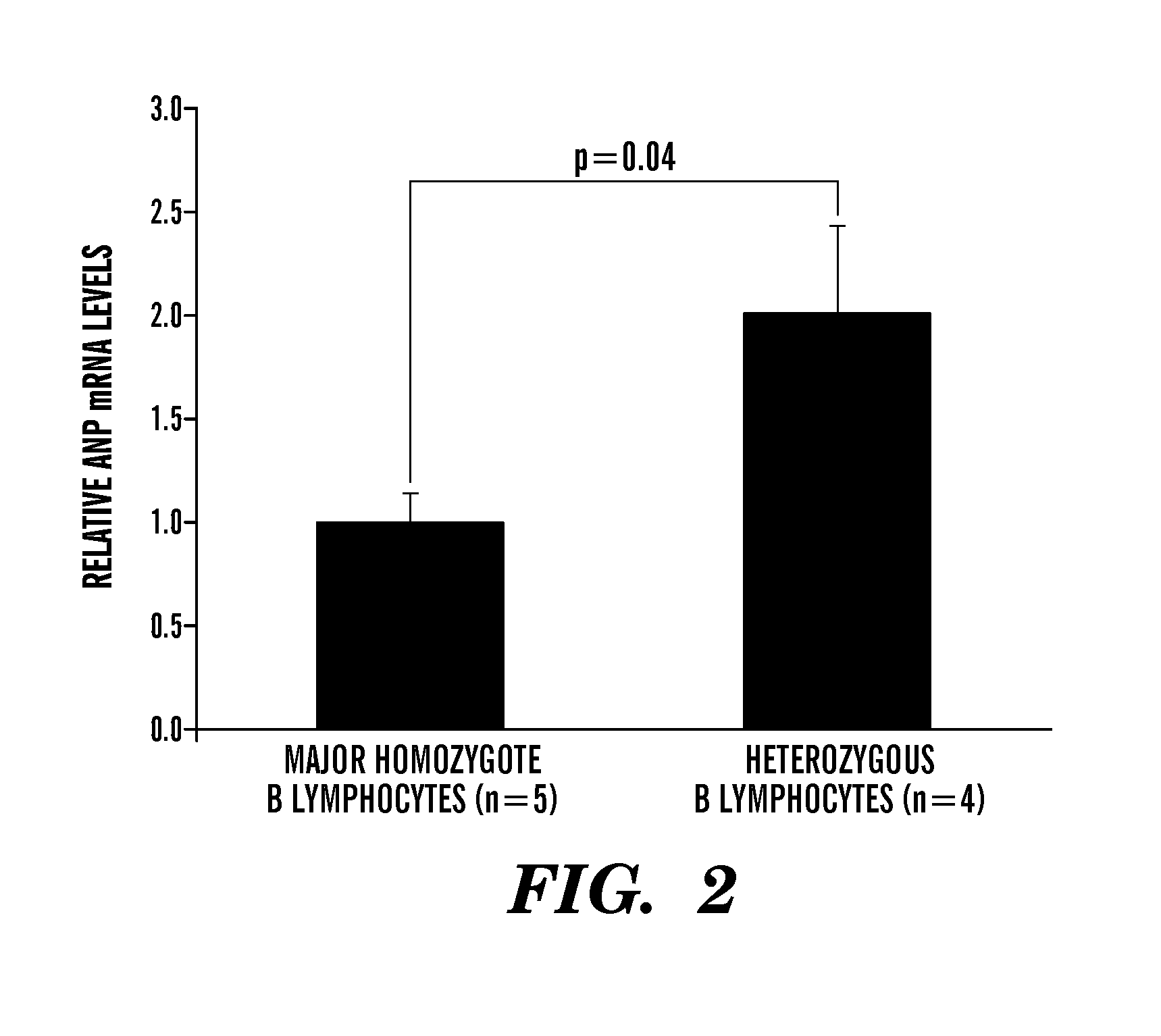
The present invention relates to methods, kits and compositions to treat hypertension and other cardiovascular diseases in a subject, in particular, a method of treating or preventing a cardiovascular disease in a subject comprising administering to a subject at least one anti-miR agent to miRNA-425. In some embodiments, an anti-miR agent is a small molecule or an oligonucleotide complementary to at least part of the miR-425 of SEQ ID NO: 1, or an anti-miR complementary to at least part of the miRNA seed sequence AUGACA (SEQ ID NO: 2). Another aspect of the present invention relates to methods, kits and compositions to treat low blood pressure in a subject comprising administering a composition comprising a miR-425 agent to decrease ANP levels in the subject. Other aspects of the present invention relates to assays, methods and systems to identify a subject at risk of hypertension, or identifying a subject suitable to administration of an anti-miR-425 agent for treatment of hypertension, the assay comprising assessing if a subject is homozygous or heterozygous for the major (A) allele of rs5068 SNP, and / or assaying for levels of miR-425 and / or assaying for levels of NT-proANP and / or levels of ANP in the plasma of a subject.
Owner:THE GENERAL HOSPITAL CORP
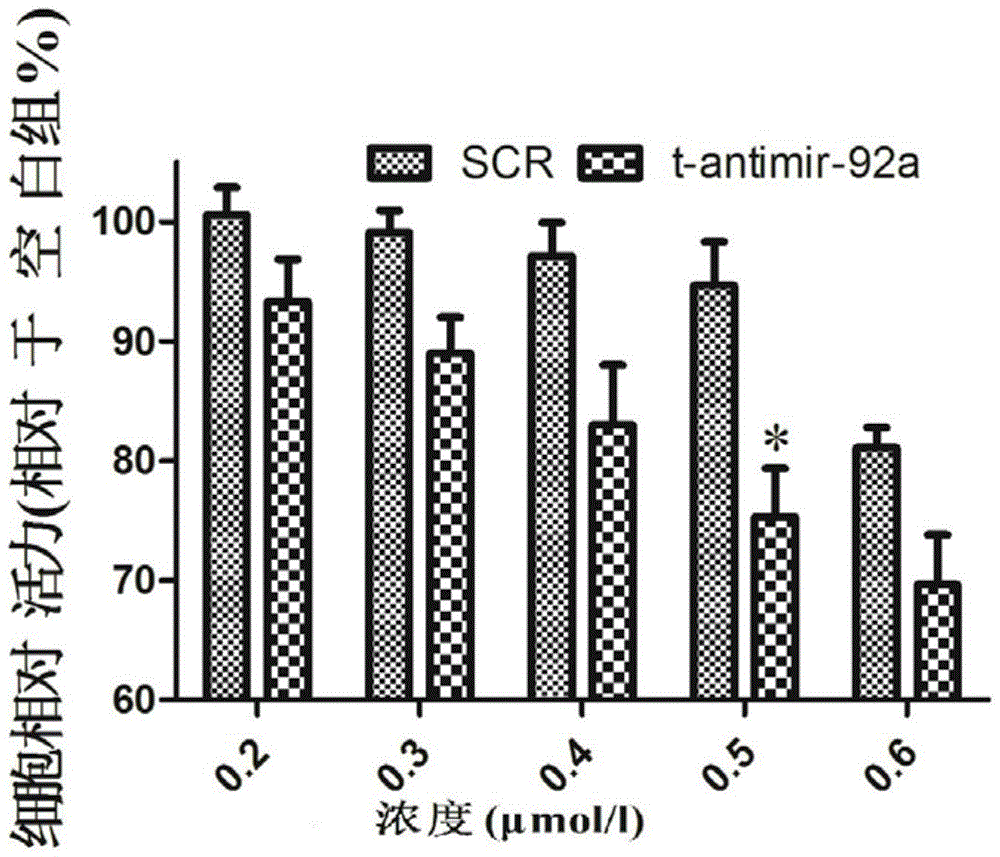
Small antisense oligonucleotide aiming at miR-92a seed sequence and application
InactiveCN104531706ASmall molecular weightLow toxicityGenetic material ingredientsAntineoplastic agentsNucleotideNucleotide sequencing
The invention belongs to the field of medicine preparation, and particularly relates to a small antisense oligonucleotide aiming at a miR-92a seed sequence and an application. A nucleotide sequence of the small antisense oligonucleotide is 5'-AGTGCAAT-3'; the small antisense oligonucleotide is subjected to perthio modification, targets on miR-92a in multiple myeloma cells RPMI-8266, and has an inhibiting effect on the multiple myeloma cells, so that the small antisense oligonucleotide can be applied to preparation of medicines for preventing or treating tumors, and has a wide prospect in clinical treatment.
Owner:JINAN UNIVERSITY
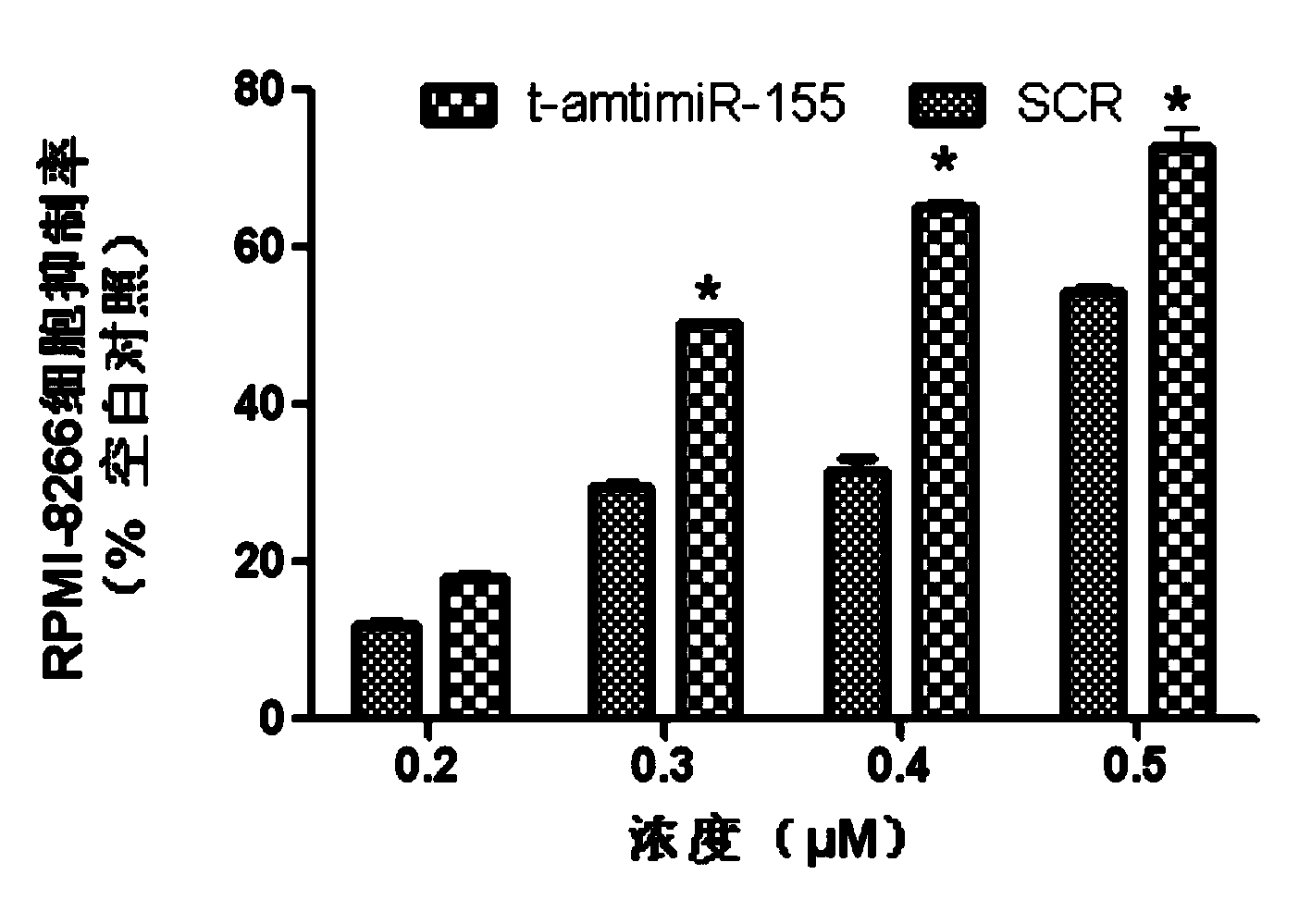
Antisense oligodeoxynucleotide for micro RNA-155 seed sequence and application
InactiveCN103882020AInhibit progressPrevent proliferationGenetic material ingredientsAntineoplastic agentsAntisense nucleic acidApoptosis
The invention discloses an antisense oligodeoxynucleotide for a micro RNA-155 seed sequence and an application. Micro antisense nucleic acids t-antimiR-155 of eight basic groups are designed and synthesized for the miR-155 seed sequence. T-antimiR-155 transfection cell test shows that t-antimiR-155 is targeted to miRNA-155 of a RPMI-8266 cell strain, so that cell growth is remarkably inhibited and apoptosis is remarkably increased. The test shows that t-antimiR-155 can effectively inhibit growth of lymphoma cells by interfering expression of the miRNA-155, and meanwhile, the test shows that the miRNA-155 can probably be taken as a potential target spot for gene therapy of lymphoma. The t-antimiR-155 can be used for preparing anti-tumor medicines, and particularly medicines for treating multiple myeloma.
Owner:JINAN UNIVERSITY
Method of deriving mature hepatocytes from human embryonic stem cells
ActiveUS20130217752A1Liver inflammation is reducedReduce incidenceHepatocytesPeptide/protein ingredientsDrug metabolismVirus type
A method for producing mature hepatocytes having functional hepatic enzyme activity from human pluripotent cells is disclosed. The method includes the step of transferring an external vector comprising the DNA sequence coding for a microRNA having the seed sequence of the microRNA miR-122, the DNA sequence coding for a microRNA having the seed sequence of the microRNA miR-let-7c, a microRNA having the seed sequence of the microRNA miR-122, a microRNA having the seed sequence of the microRNA miR-let-7c, or a combination thereof into one or more fetal hepatocytes. The resulting cells differentiate into mature hepatocytes that exhibit functional hepatic enzyme activity, and can be used in drug metabolism and toxicity testing, in the study of viruses that target hepatic tissue, and as therapeutics.A related method of maintaining the functional hepatic enzyme activity of primary hepatocytes over time is also disclosed. The method includes the step of transferring an external vector comprising the DNA sequence coding for a microRNA having the seed sequence of the microRNA miR-122 into one or more cultured primary hepatocytes.
Owner:WISCONSIN ALUMNI RES FOUND
Novel method for gene assembly using improved CRISPR-Cpf1
ActiveCN109825520ASticky end longSuitable for high-throughput cloningStable introduction of DNAVector-based foreign material introductionTularemiaNucleotide
The invention discloses a novel method for gene assembly using improved CRISPR-Cpf1. The method includes utilizing a DNA gene editing enzyme FnCpf1 from a Francisella tularemia noticed U112 strain (gene sequence number: MF193599) for gene recombinant cloning. The seed sequence of crRNA (an RNA sequence generated by transcription of regular clustering spaced CRISPR) from 23 nucleotides reported inthe literature to 18 without affecting the FnCpf1 cleavage efficiency. At the same time, oligo(dN)6 (oligonucleotide hexamer) is used as the viscous end of the cleavage target site, the problem of terminal heterogeneity after FnCpf1 digestion is solved, the assembly efficiency of exogenous DNA fragments is remarkably improved, and the DNA fragment assembly efficiency reaches about 85%. The gene editing enzyme FnCpf1 can be replaced with other nucleic acid editing enzymes Cpf1 such as AsCpf1 and LbCpf1.
Owner:HUBEI UNIV
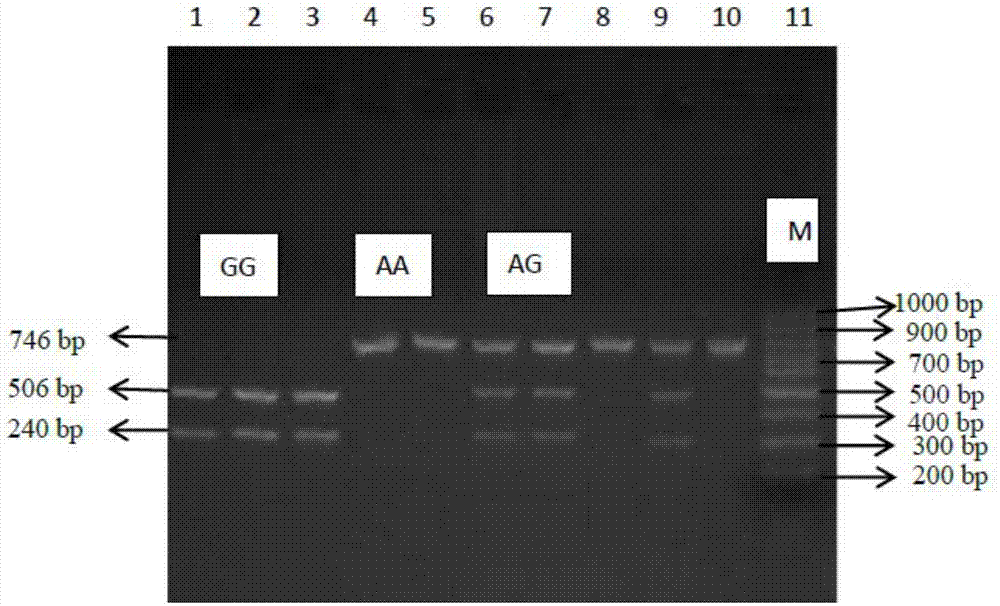
Molecular cloning of GLP2R gene fragments associated with pork quality traits and application of GLP2R gene fragments
The invention belongs to the technical field of livestock molecular biology and particularly relates to a molecular cloning of GLP2R gene fragments associated with pork quality traits and application of GLP2R gene fragments. The human GLP2R gene sequence is taken as a seed sequence, specific primers are designed by BLASTN in GenBank and referring to swine EST of which the homology is above 80% and swine GLP2R gene cDNA full-length is cloned by RACE. DNA sequence of the swine GLP2R gene fragments is shown in SEQ ID NO:1. GLP2R gene polymorphism is determined by designing a primer based on human GLP2R gene via comparative genomics method, amplifying by swine genomic DNA as template, screening SNP from amplification fragment via sequencing, and genotyping by PCR-RFLP. A / G base mutation resulting in PCR-RFLP-BstEII polymorphism is found at 244bp, a new molecular marker is provided for swine marker-assisted selection and the situation of pig breeds at home and abroad can be detected by the marker.
Owner:HUNAN AGRICULTURAL UNIV
Method for obtaining miRNA (Ribose Nucleic Acid) candidate target gene and special reverse transcription primer for method
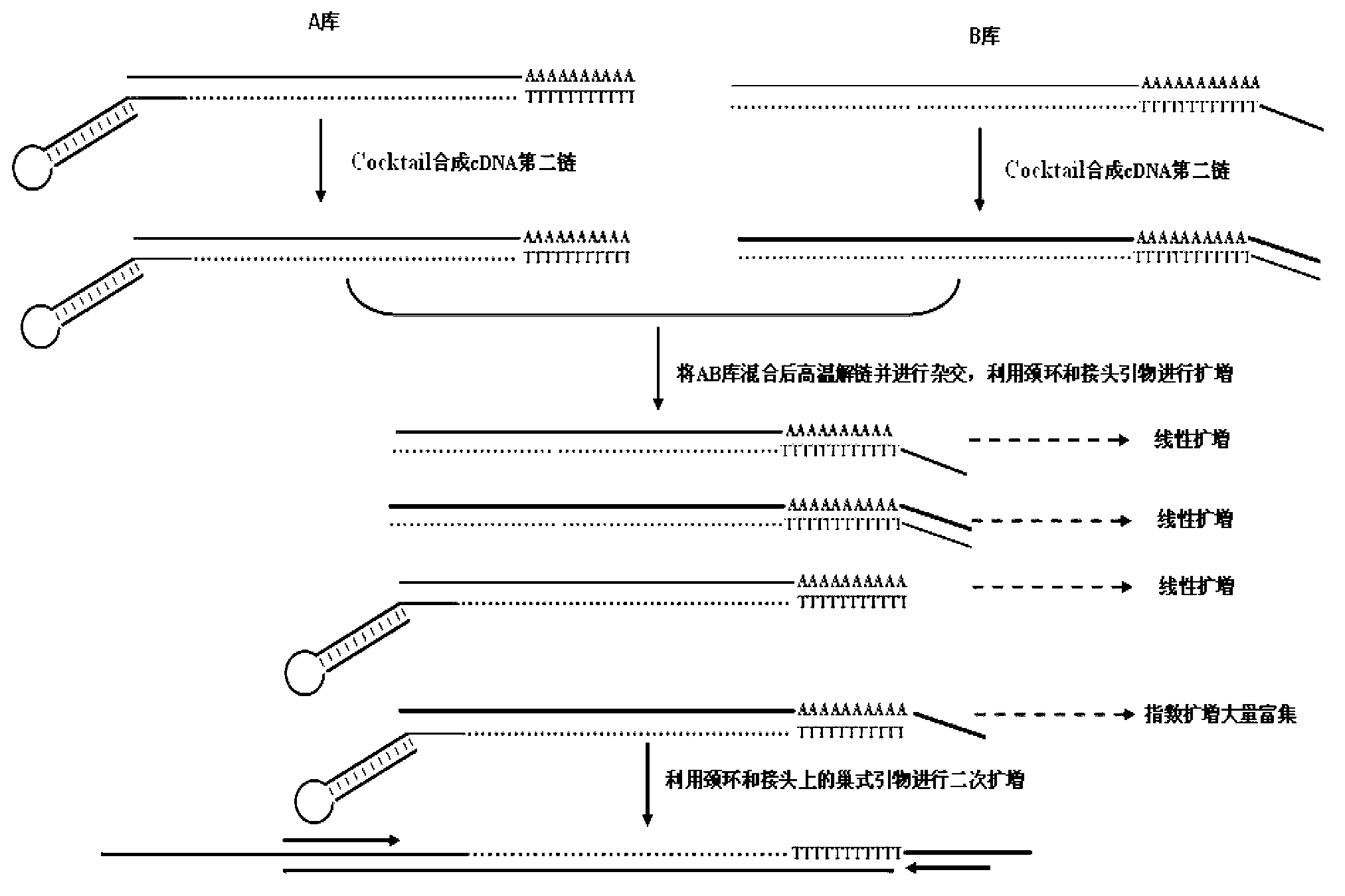
The invention discloses a method for obtaining a miRNA (Ribose Nucleic Acid) candidate target gene and a special reverse transcription primer for the method. The method comprises the following steps of: (1), acquiring cDNA (Deoxyribonucleic Acid) first chains corresponding to mRNA which is complementary to a target miRNA seed sequence to obtain a storeroom A by using the DNA with a stem-loop structure and the target miRNA seed sequence as a reverse transcription primer; (2), acquiring cNDA first chains corresponding to mRNA of the eukaryote target gene tissue to obtain a storeroom B by using the DNA with a special joint and 14-20 deoxidized thymine nucleotides as a reverse transcription primer, wherein the structure of the reverse transcription primer B is the special joint-T14-20; and (3), synthesizing the storeroom A and the storeroom B to obtain a double-chain cDNA, then hybridizing to obtain a hybrid DNA molecule, amplifying the DNA segment between the target miRNA seed sequence and the special joint on the hybrid DNA module and obtaining the target miRNA candidate target gene according to the amplification products.
Owner:XINJIANG ACADEMY OF AGRI & RECLAMATION SCI
DNA methylation data detection method and device thereof based on seed sequence information
ActiveCN108182348AQuick comparisonIncrease profitHybridisationSpecial data processing applicationsA-DNAConducting system
The invention provides a DNA methylation data detection method and a device thereof based on seed sequence information. The method comprises the steps of building an index database; obtaining sequencing data of a target sample, and according to the preset seed sequence length, dividing the sequencing data to obtain divided seed sequence information; based on the index database, determining comparison candidate position information of each divided seed sequence information; conducting system evaluation on each comparison candidate position information to obtain a system evaluation result, and according to the system evaluation result, obtaining a DNA methylation locus of the target sample. Time of comparison operation which consumes most time in data analysis is greatly shortened, on the basis of guaranteeing the integrity of a methylation detection area locus, the utilization rate of data is increased by a large margin, the operation efficiency and accuracy of the data are improved bya large margin, and great convenience is brought to further research on DNA nucleobase modification information for scientific research staff in the field of life science.
Owner:THE THIRD AFFILIATED HOSPITAL OF GUANGZHOU MEDICAL UNIVERSITY
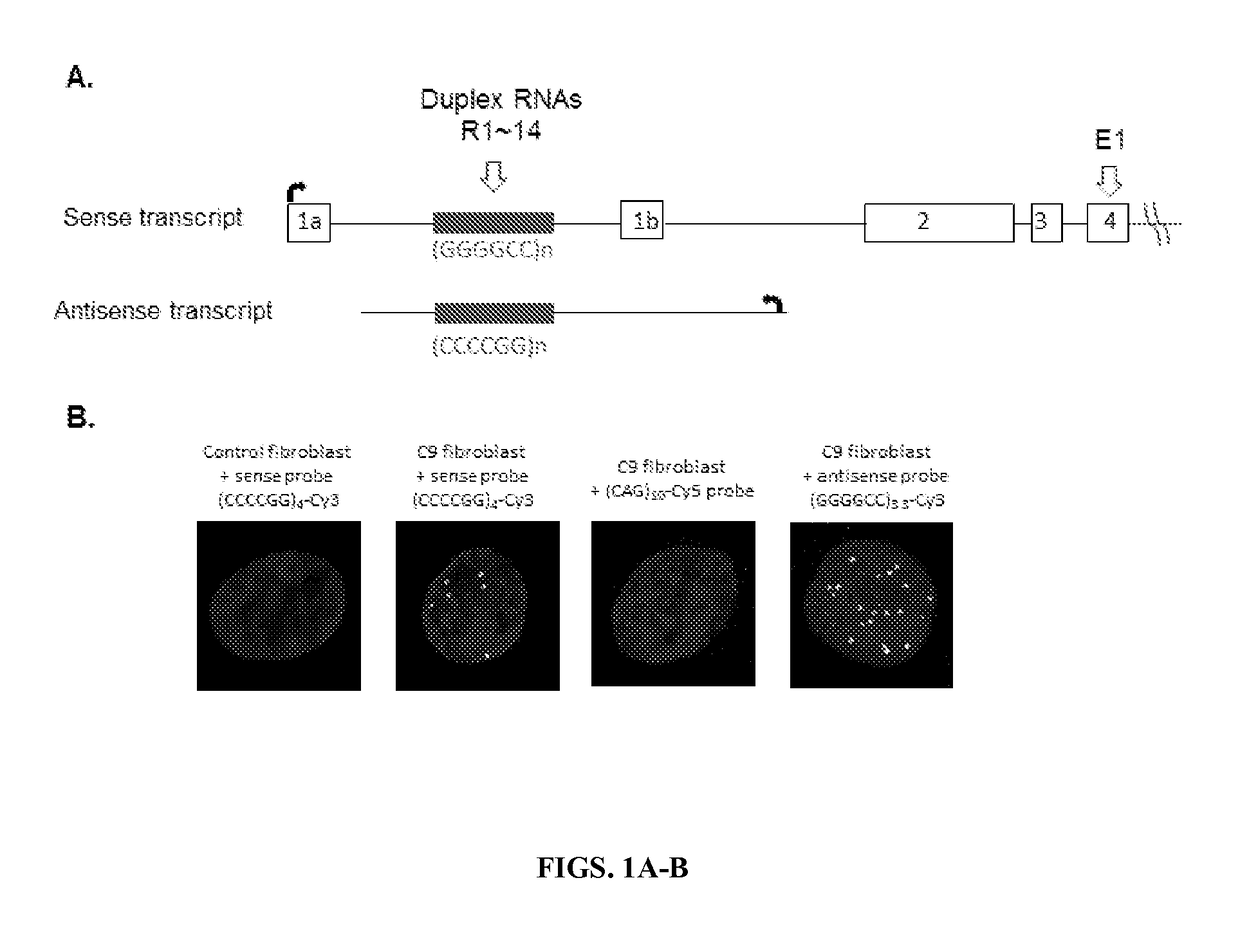
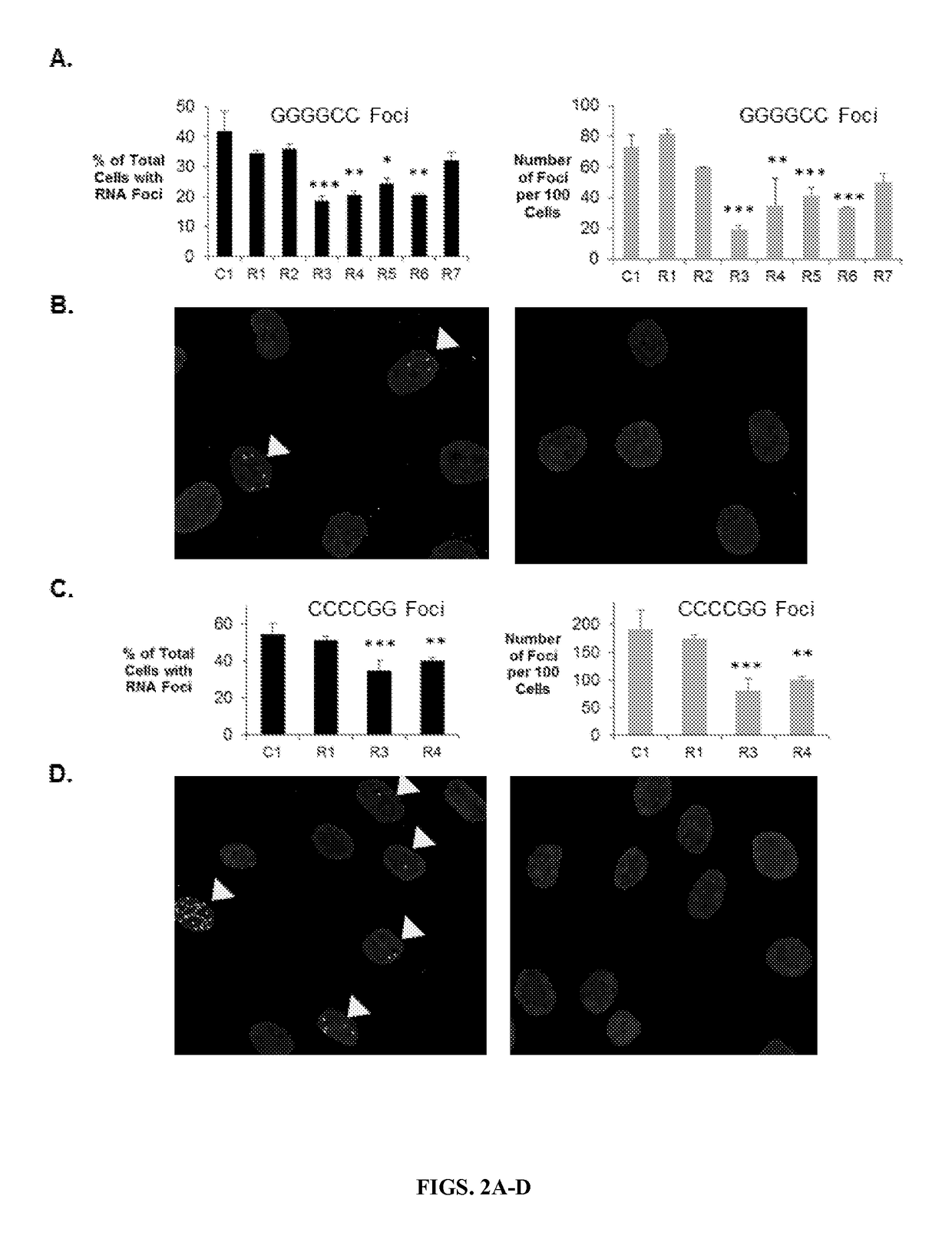
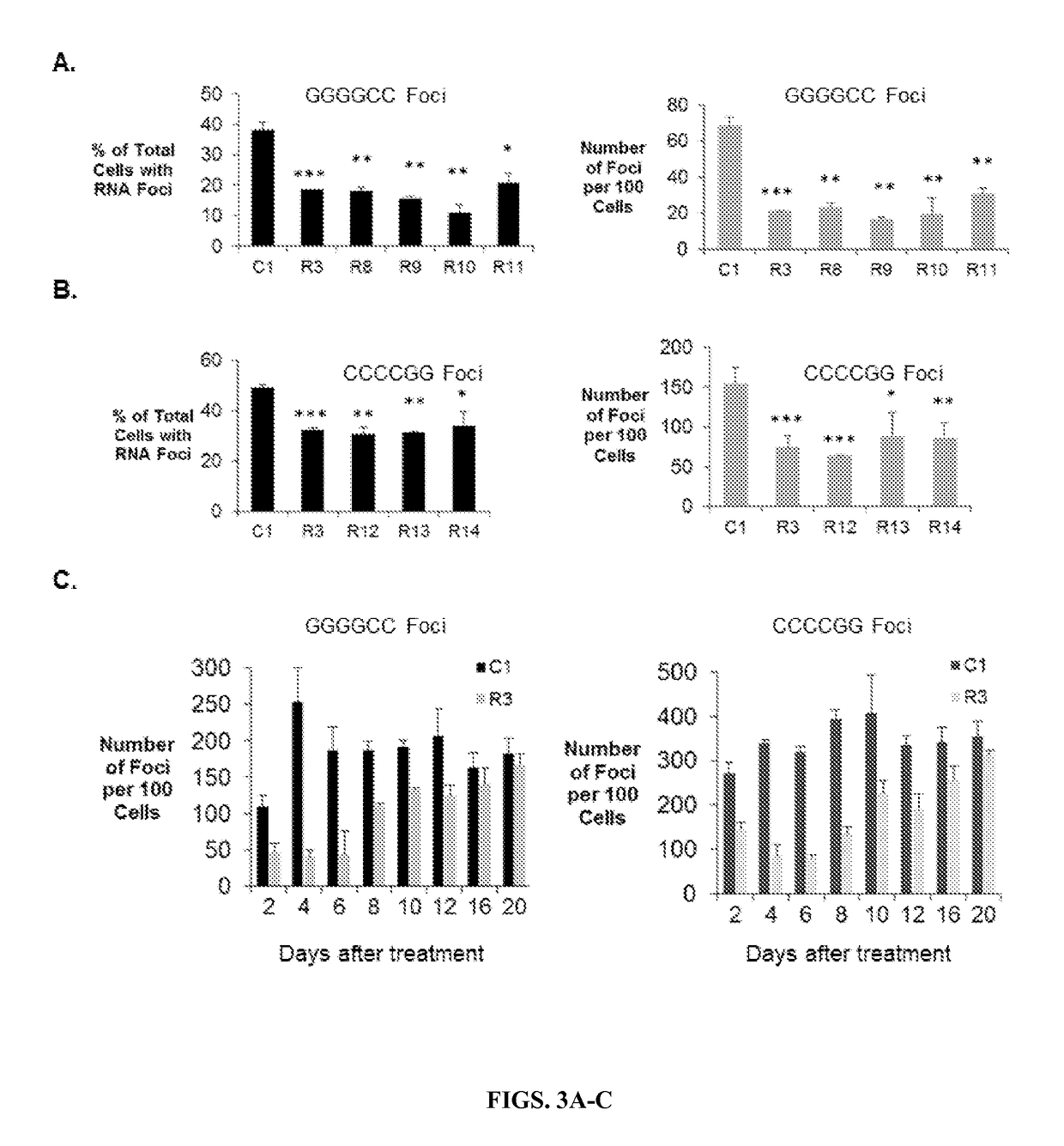
Allele selective inhibition of mutant c9orf72 foci expression by duplex rnas targeting the expanded hexanucleotide repeat
ActiveUS20170233735A1Reduce expressionSmall sizeOrganic active ingredientsPolymorphism usesGeneticsNucleobase
Provided herein are compositions and methods for reducing expression of C9orf72 transcripts in cells containing expanded intronic GGGGCC regions, including those in subjects having or at risk of developing amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Provided herein are a double-stranded oligonucleotides of 13 to 22 nucleobases in length targeting a GGGGCC expanded repeat region in an intron of C9orf72, comprises (a) 3-5 central mismatches (within bases 9-14) within a target sequence comprising the expanded repeat sequence, or (h) 3-5 mismatches outside of the seed sequence (bases 2-8 within the guide strand complementary to the expanded repeat sequence).
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
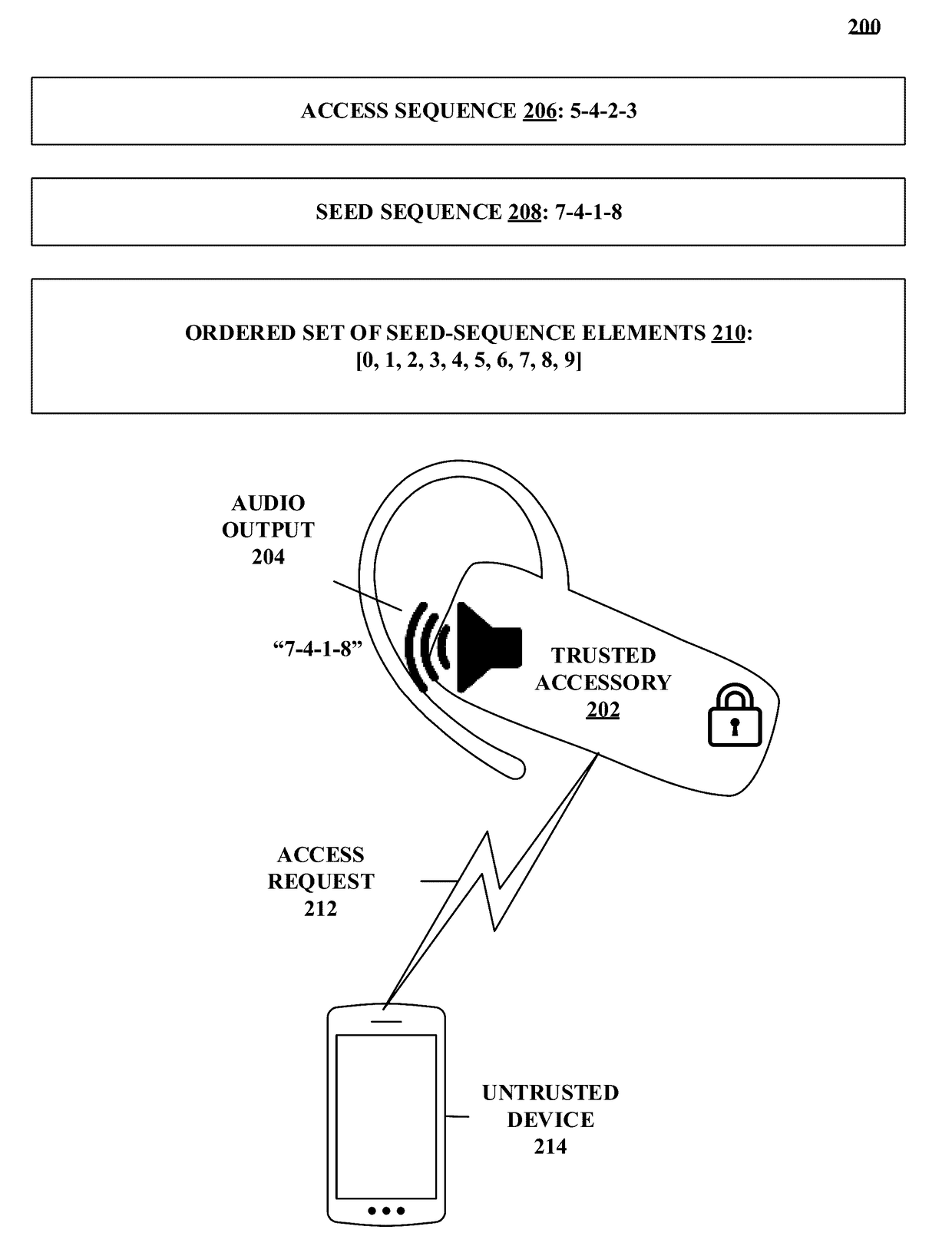
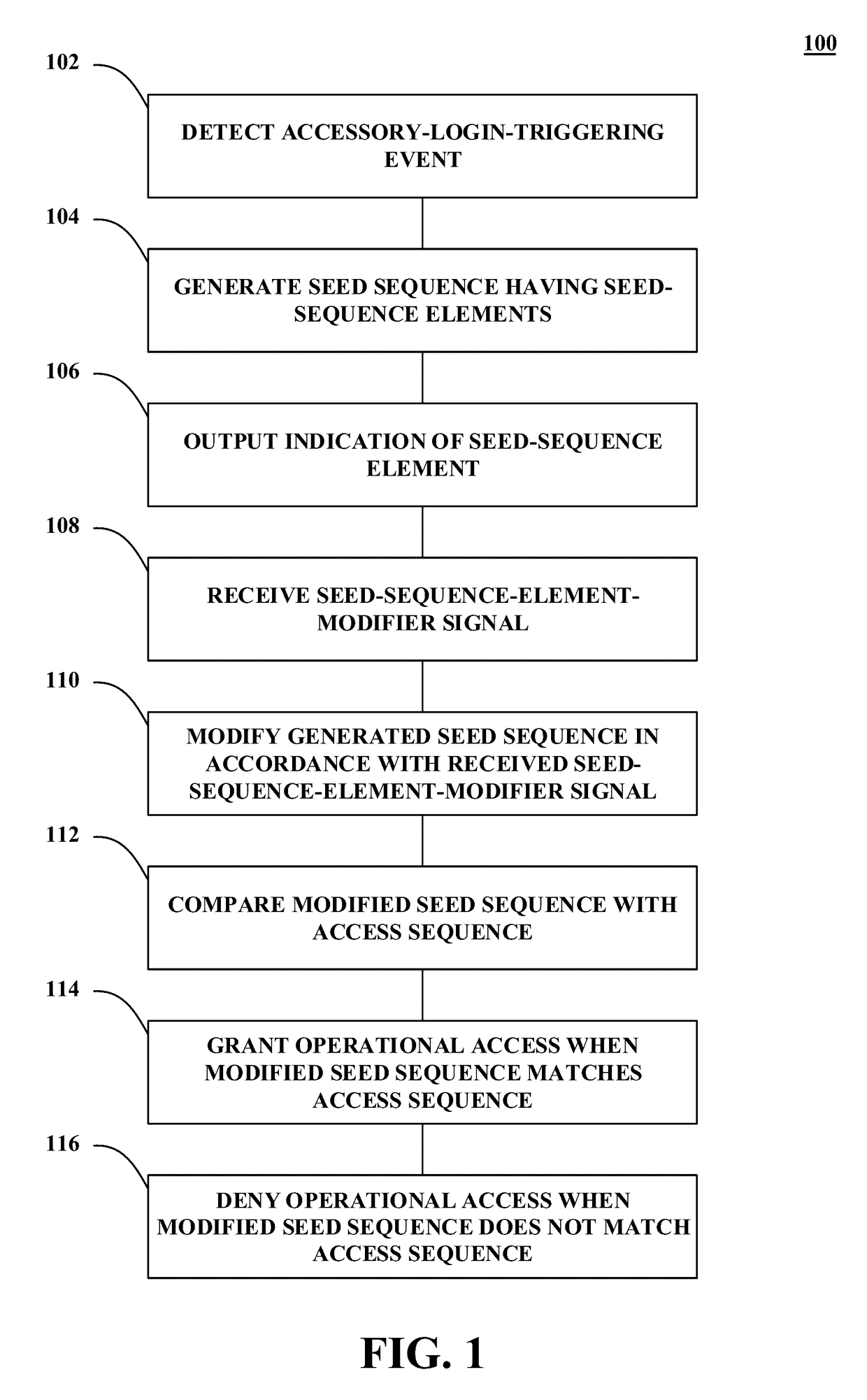
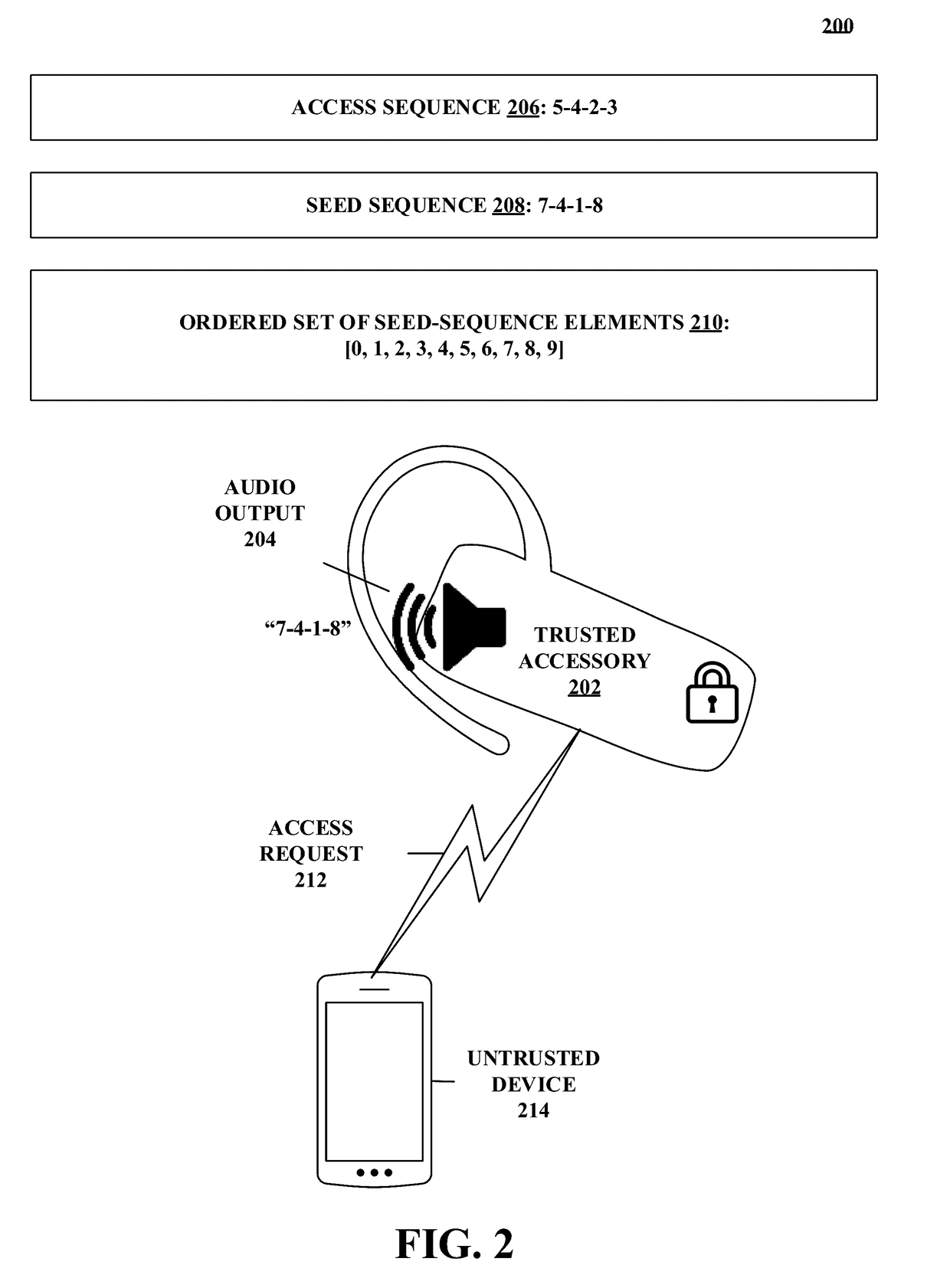
Methods and systems for authentication using zero-knowledge code
Disclosed herein are methods and systems for authentication using zero-knowledge code. One embodiment takes the form of a process that includes detecting an accessory-access-request event associated with a trusted accessory. The process includes generating a seed sequence having a first number of seed-sequence elements. The process includes outputting an indication of at least one seed-sequence element. The process includes receiving at least one seed-sequence-element-modifier signal for at least one of the seed-sequence elements. The process includes modifying the generated seed sequence in accordance with the at least one received seed-sequence-element-modifier signal. The process includes comparing the modified seed sequence with a stored access sequence. The process includes granting operational access to the trusted accessory when the modified seed sequence matches the stored access sequence. The process includes denying operational access to the trusted accessory when the modified seed sequence does not match the stored access sequence.
Owner:NAGRAVISION SA
Micro non-coding RNA antagonist and use thereof
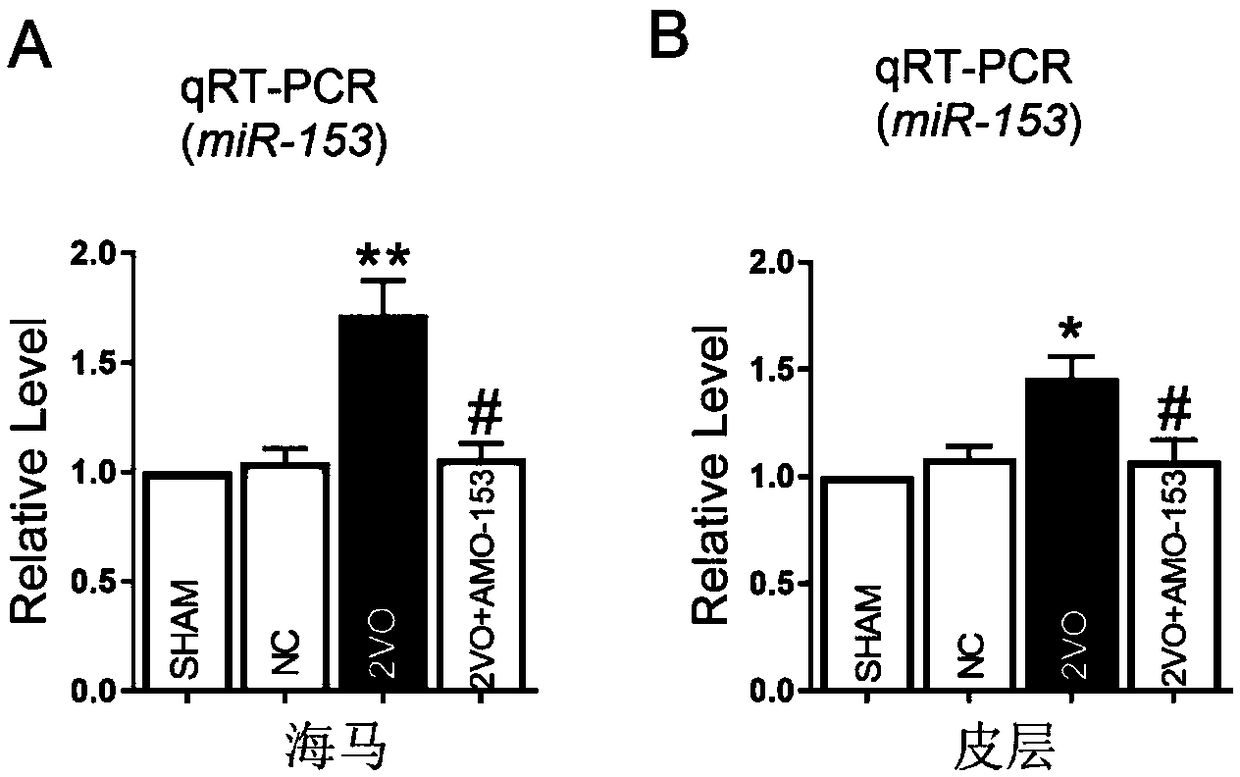
ActiveCN109223817AImprove cognitive impairmentInhibition releaseOrganic active ingredientsNervous disorderNervous systemRat model
The invention discloses a micro non-coding RNA antagonist and the use thereof. The micro non-coding RNA antagonist is miR-153-The antisense oligonucleotide (AMO 153 3p) of the 3p seed sequence is shown in SEQ ID NO. 1. Compared with the prior art, the invention has the advantage that a large number of experiments prove that in the long-term cerebral ischemia-hypoxia rat model induced by long-termcerebral hypoperfusion, miR-153-3p is overexpressed in the hippocampal cortex, which can regulate the expression of several neurotransmitter-releasing related proteins through multiple targets, and inhibit the release of neurotransmitters, thus promoting the occurrence of cognitive impairment in the brain. Targeted inhibition miR-153-The expression of 3p specifically blocked miR-153-Inhibitory effect of 3p on neurotransmitter release. Therefore, AMO-153-3p can be used for the prevention and treatment of various neurotransmitter release disorders induced by long-term cerebral ischemia and hypoxia.
Owner:HARBIN MEDICAL UNIVERSITY
Method for breeding sorgo seeds
The invention relates to a method for breeding sorgo seeds. The method comprises the following steps of (1) determining a breeding base; (2) determining water quality standards for farmland irrigation; (3) determining crops for rotation; (4) mixing diammonium phosphate, urea, potassium sulfate, zinc sulfate, manganese sulfate and ammonium molybdate in combination with tillage, applying 0 to 20 cm of soil layer, and ploughing levelly 5 to 7 days after winter water irrigation in every autumn; (5) covering mulching film after ploughing and soil preparation in the period of April 15th to April 25th of the next year; (6) when earth temperature of 5 cm deep is continuously stabilized to 10 DEG C, performing rectilinear seeding in the manners with a male parent plant spacing of 18 cm, a female parent plant spacing of 15 cm, a row spacing of 45 cm, a row ratio of 3:6 and a seeding depth of 2 to 3 cm, wherein a seeding sequence is according to a sequence of sterile line, first-phase maintainer line and second-phase maintainer line; (7) performing field management; (8) preventing diseases and pests; (9) harvesting, airing, and turning over once every five days; threshing, selecting, classifying, packaging and storing after the water content of fruit clusters is smaller than 13 percent. The method disclosed by the invention has the advantage that the problems of low yield and unstable yield in the process of breeding the sorgo seeds are effectively solved.
Owner:RES INST OF GANSU DUNHUANG SEED CO LTD
System and method for detecting non-reproducible pseudo-random test cases
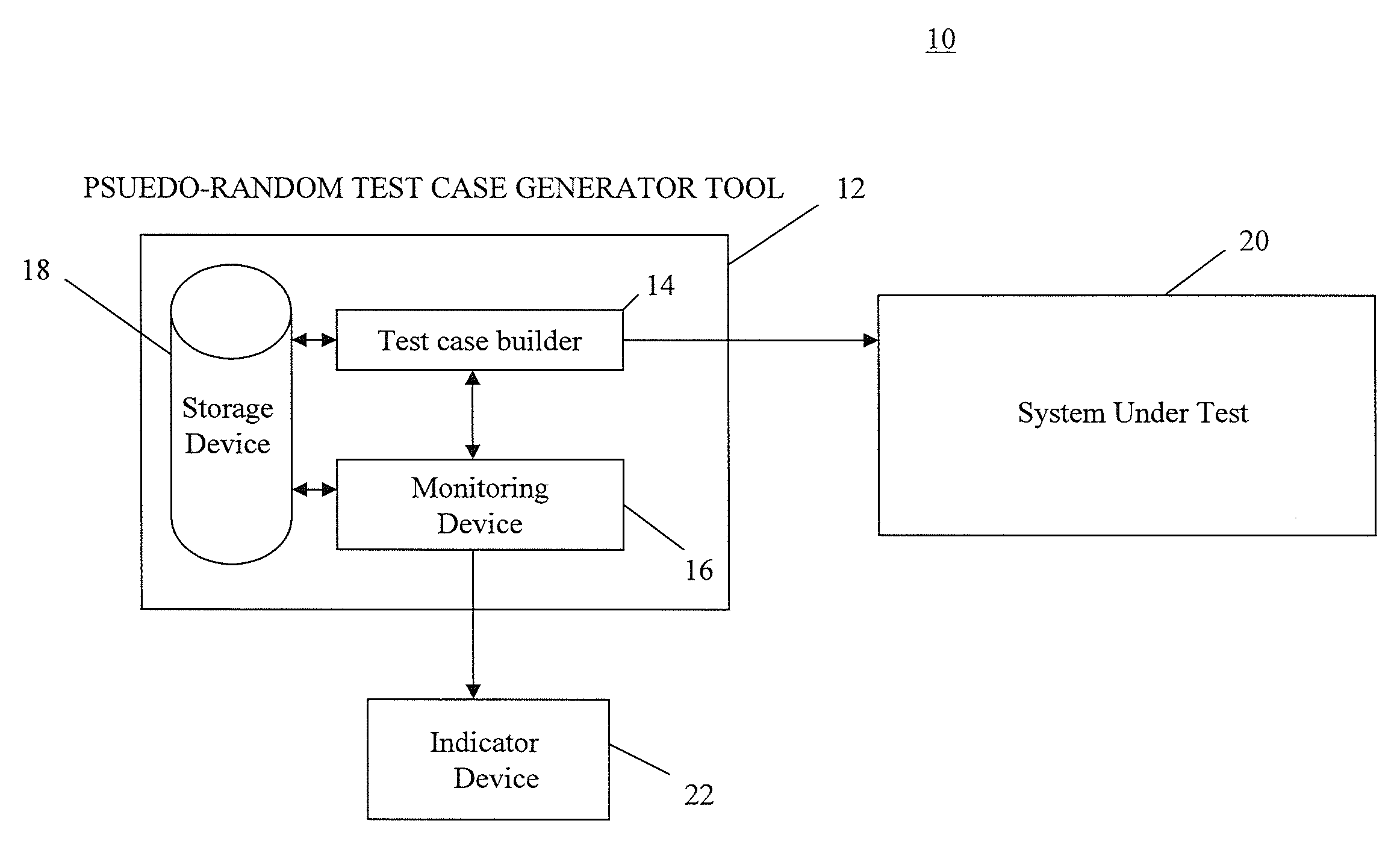
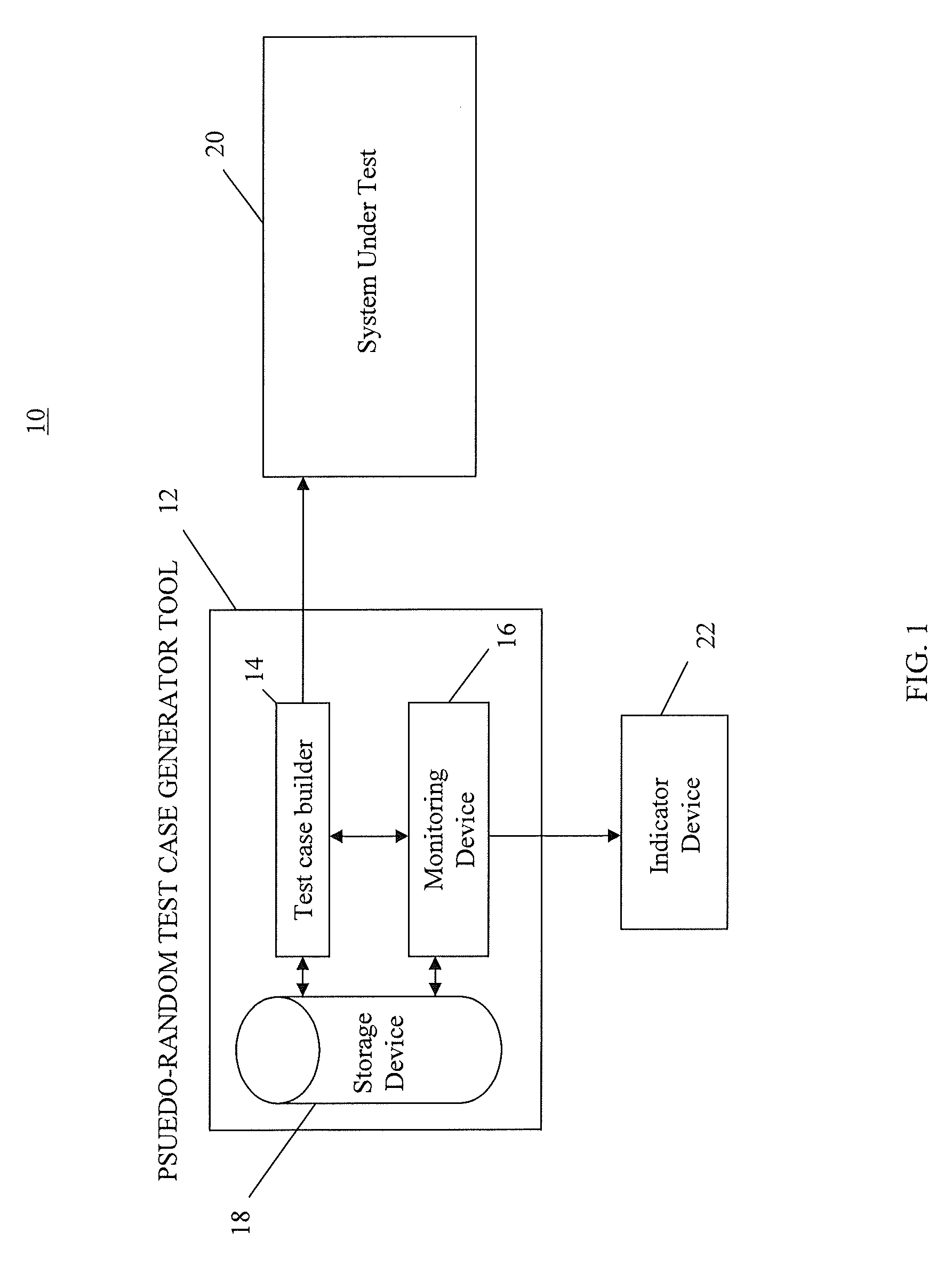
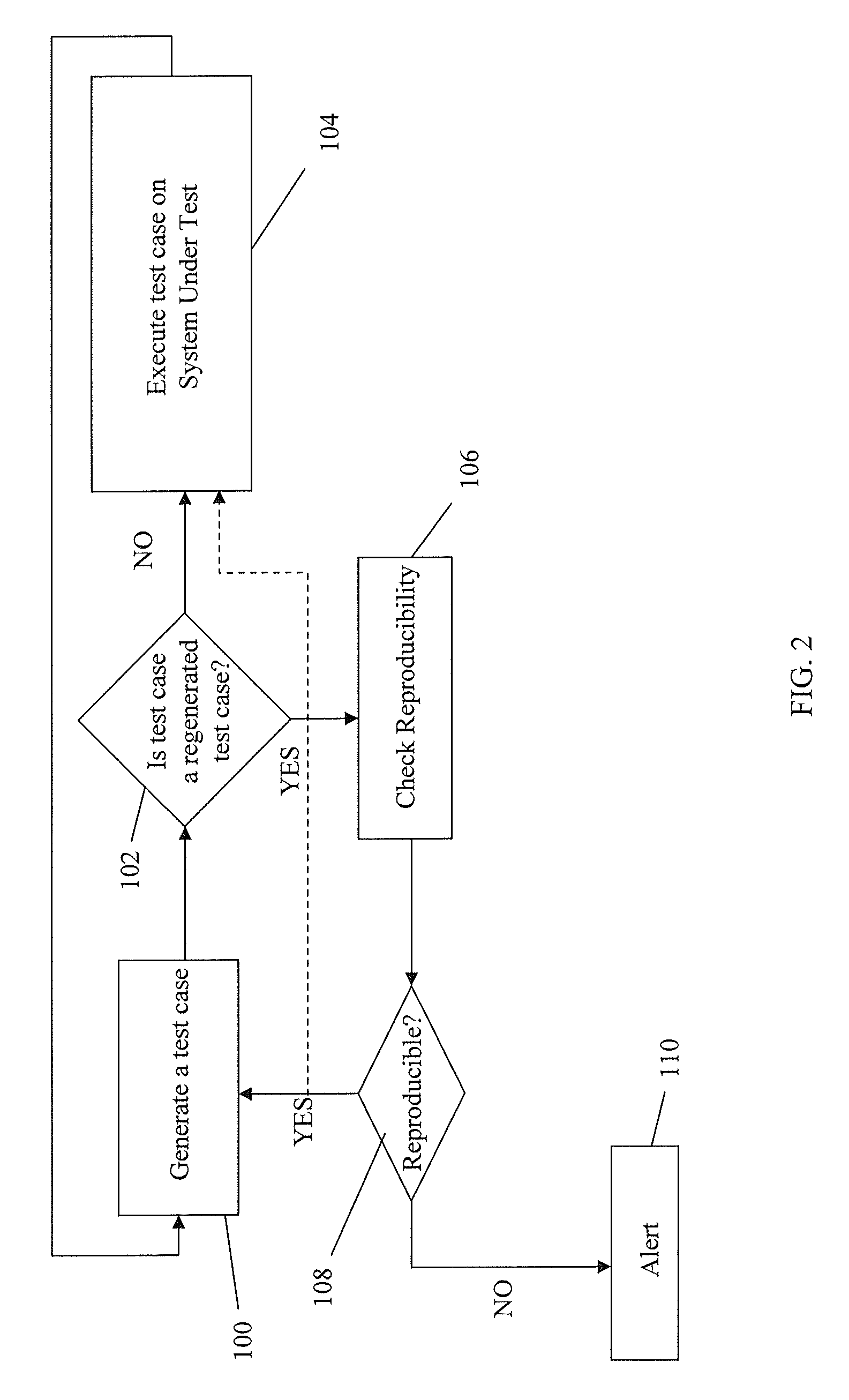
InactiveUS7774668B2Electronic circuit testingError detection/correctionTest case generatorComputer science
A method for monitoring a test case generator system by detecting non-reproducible pseudo-random test cases, comprising: building a first pseudo-random test case having a first sequence of seeds comprising a first starting seed and a first ending seed through the test case generator system; reproducing the first sequence of seeds of the first pseudo-random test case by building a second pseudo-random test case having a second sequence of seeds comprising a second starting seed and a second ending seed through the test case generator system when the test case generator system is operating in a reproduction mode, the first starting seed being used as the second starting seed of the second sequence of seeds; and comparing the first ending seed in the first sequence of seeds to the second ending seed in the second sequence of seeds.
Owner:IBM CORP
Compound side-filling precision seed-metering device
PendingCN111869377AImprove seed filling performanceAvoid missing broadcastSeed depositing seeder partsEngineeringMechanical engineering
The invention discloses a compound side-filling precision seed-metering device which comprises a fixed sequencing plate cover, a movable sequence-adjusting seed-filling plate, a saddle-shaped single (double)-side seed posture-adjusting queuing roller and a seed separating plate. A fixed sequencing plate is mounted on a main fixed shaft, and the movable sequence-adjusting seed-filling plate is assembled on the main fixed shaft by being in interference fit with a sliding bearing to form a seed cavity with the fixed sequencing plate cover; and after seeds enter the seed cavity from a seed inlet hole, the seeds enter a seed sequence-adjusting cavity under the action of the seed separating plate, a unique seed discharging opening is formed in the portion, corresponding to the seed throwing cavity, of the fixed sequencing plate cover, and under the combined action of the saddle-shaped single (double)-side seed posture-adjusting queuing roller and a sequencing V-shaped groove formed in the fixed sequencing plate cover, the seeds sequentially enter an axial primary seed filling U-shaped groove and a radial secondary seed filling hole in sequence, the movable sequence-adjusting seed-fillingplate rotates in the operation process, and when the movable sequence-adjusting seed-filling plate rotates to the unique seed discharging opening formed in the fixed sequencing plate cover, the seedsfall into a duckbilled part to complete seed filling. The seed-metering device is simple in structure, continuous and reliable in seed filling, and capable of realizing orderly seed filling and improving the seed filling performance of the seed metering device.
Owner:SHIHEZI UNIVERSITY
Method for suppressing tumors by mir-200 family inhibition
ActiveUS20180271895A1Reducing tumorigenicitySuppression of in vivo tumor growthOrganic active ingredientsGenetic material ingredientsAbnormal tissue growthIn vivo
Tumor growth was found to be significantly suppressed in vivo by inhibiting both miRNA containing 5′-AACACUG-3′ as a seed sequence and miRNA containing 5′-AAUACUG-3′ as a seed sequence. The inhibition significantly altered the proportion of subpopulations of tumor cells and reduced the tumorigenicity in all subpopulations. The inhibition also exerted a remarkable tumor-shrinking effect on already-formed tumors. The present invention provides novel therapeutic potential against tumor.
Owner:CHIBA UNIVERSITY
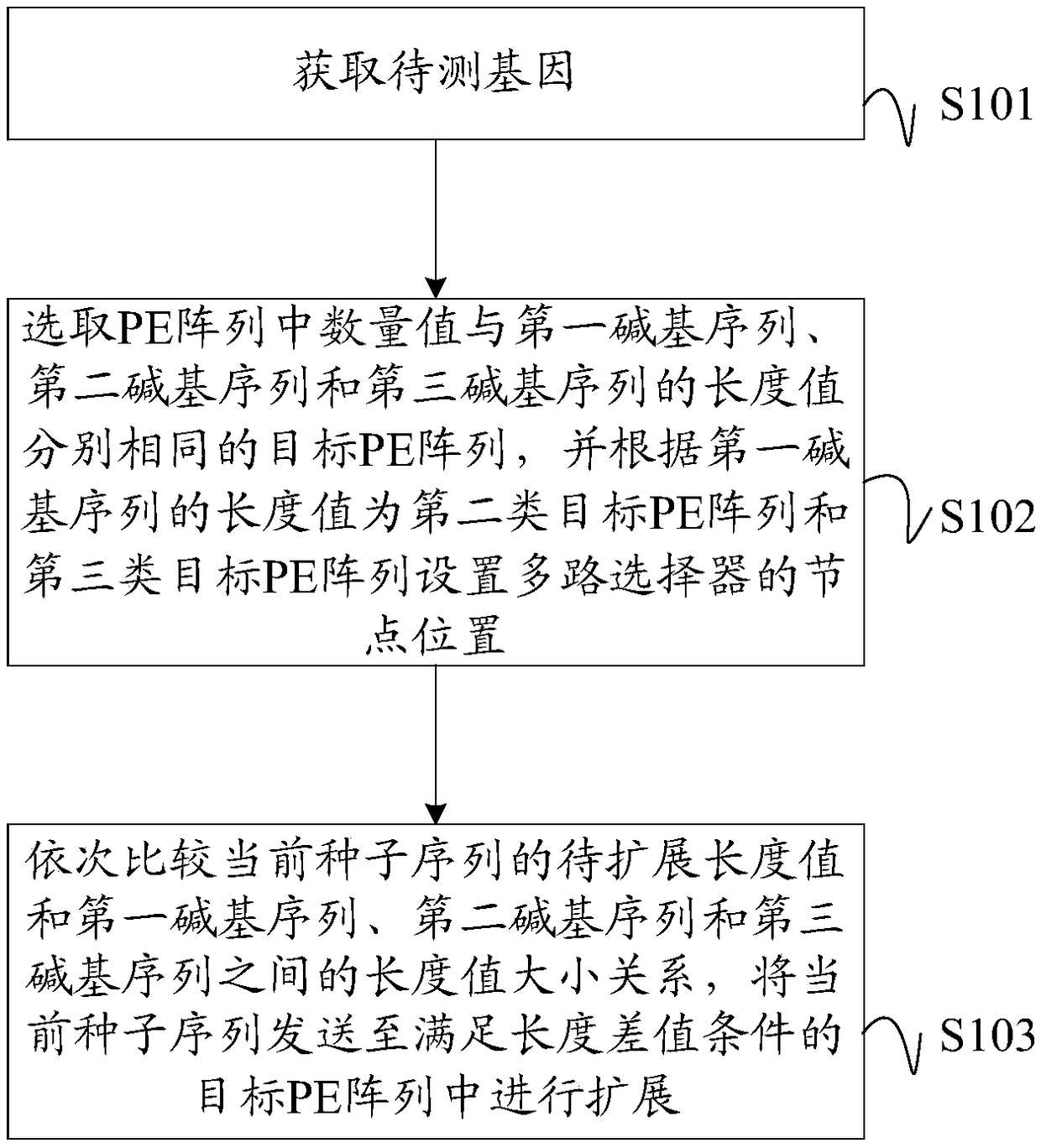
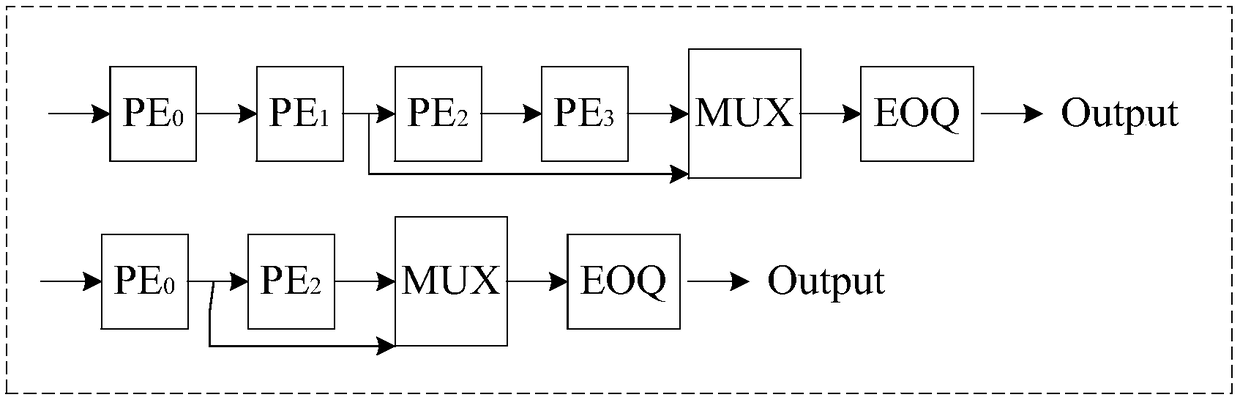
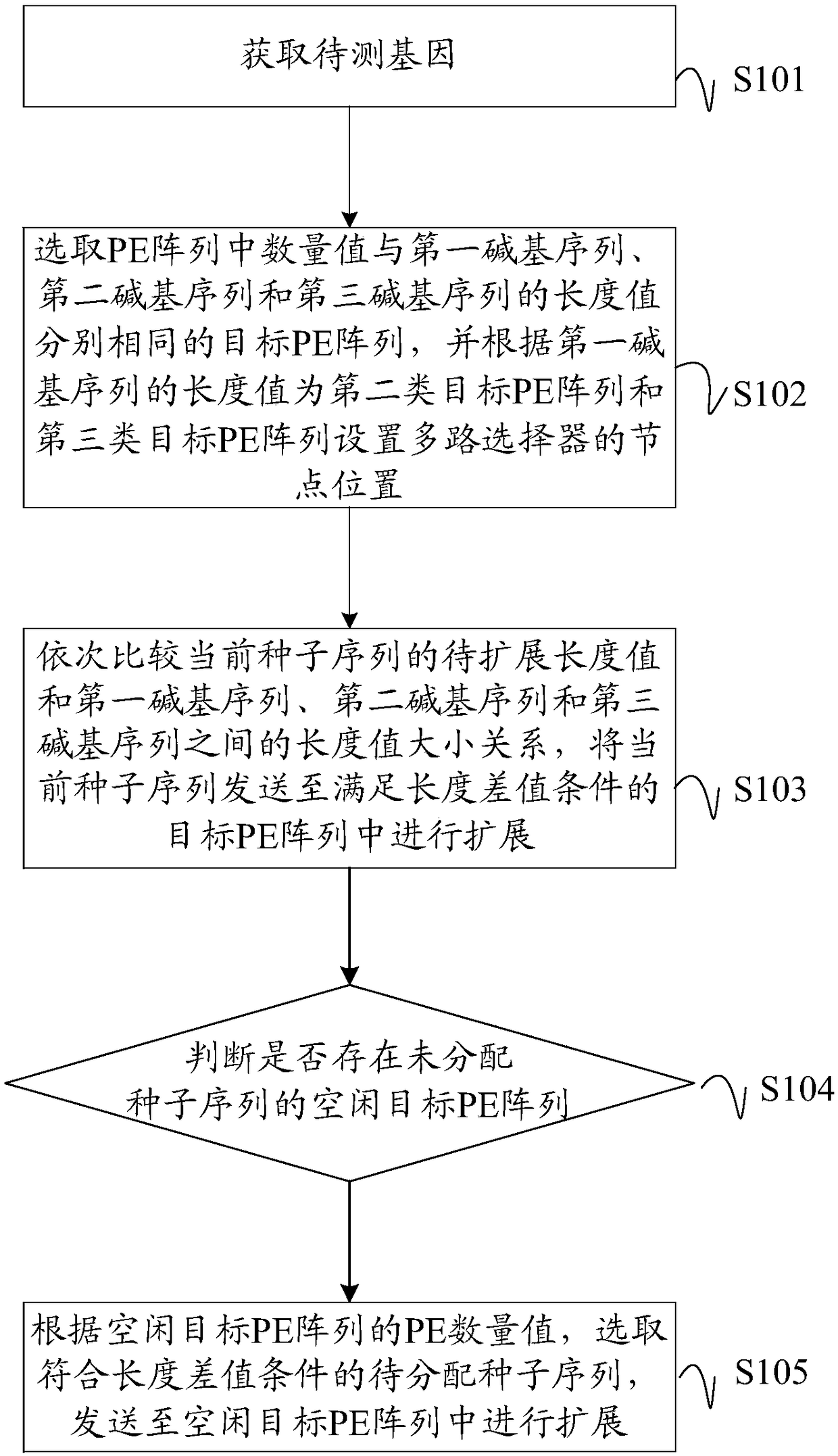
Gene sequence comparison method, PE configuration controller and computer readable storage medium
ActiveCN108875301AIncrease profitMinimize Latency WaitingSpecial data processing applicationsResource utilizationMultiplexer
Embodiments of the invention disclose a gene sequence comparison method, a PE configuration controller and a computer readable storage medium. The method comprises the steps of selecting target PE arrays with the quantity values as same as the length values of a first base sequence, a second base sequence and a third base sequence in PE arrays, and setting node positions of a multiplexer for the second target PE array and the third target PE array according to the length value of the first base sequence; and sequentially comparing relationships between the to-be-expanded length value of a current seed sequence and the length values of the first base sequence, the second base sequence and the third base sequence, and sending the current seed sequence to the target PE array meeting a lengthdifference value condition to perform expansion. According to the method, a pulse array with the variable logic length and physical length is set, and the target PE array with the appropriate PE quantity is selected according to the to-be-expanded length value of the seed sequence to perform expansion, so that the resource utilization rate of an FPGA is improved, and the processing performance ofa Smith-Waterman sequence comparison algorithm in a scoring stage is improved.
Owner:ZHENGZHOU YUNHAI INFORMATION TECH CO LTD
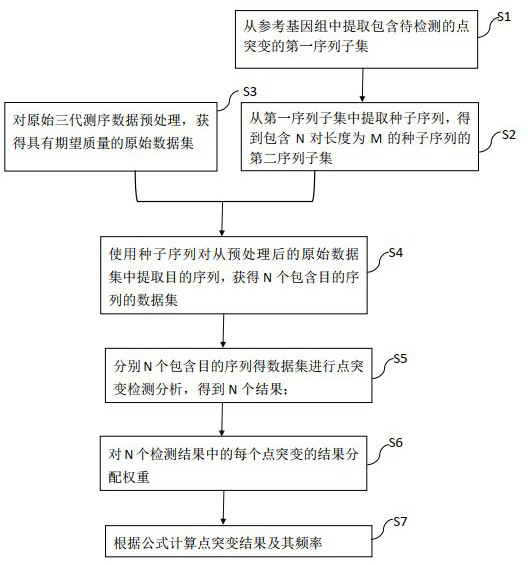
Analysis method and device for detecting point mutation based on third-generation sequencing data
ActiveCN114005489ARich technical meansSolve the current situation of insufficient mutation accuracyProteomicsGenomicsData setAlgorithm
The invention provides an analysis method and device for detecting point mutation based on third-generation sequencing data. The analysis method comprises the following steps: 1) extracting a first sequence subset containing to-be-detected point mutation; 2) extracting a seed sequence from the first sequence subset to obtain a second sequence subset; 3) obtaining an original data set with expected quality; 4) using the seed sequence pair of the second sequence subset to obtain N data sets containing target sequences; 5) carrying out point mutation detection analysis on the N data sets containing the target sequence; 6) allocating a weight W to the result of each point mutation in the N detection results; and 7) calculating a point mutation result and frequency thereof according to a formula. The invention further provides a device for detecting point mutation based on third-generation sequencing data. By using the method provided by the invention, the problem of false negative caused by low comparison rate due to random indel or relatively high sequencing error is effectively avoided from the aspect of data features, and meanwhile, the false positive result can be more effectively controlled.
Owner:QITAN TECH LTD CHENGDU
A method, system and related components for gene sequence comparison
Owner:ZHENGZHOU YUNHAI INFORMATION TECH CO LTD
A miRNA prediction method, a miRNA system and applications
InactiveCN109280698AConvenient researchImprove efficiencyMicrobiological testing/measurementProtein secondary structureGenome
The invention discloses a miRNA prediction method, a miRNA system and applications. The miRNA prediction method of the invention includes the following steps: (1) setting a length of a seed sequence;(2) fully arranging four bases A, T, C and G according to the set length to form a seed sequence set; (3) removing redundant sequences in the seed sequence set; (4) searching positions of the seed sequences on a genome to obtain a position set; (5) arranging the seed sequence position set from small to large; (6) traversing the positions of the position set, and if the sequences of the front and back positions complement each other and the distance is not greater than the length of the set precursor sequence, outputting the sequence; and (7) using secondary structure prediction software to perform secondary structure prediction on the output sequence to obtain miRNA, and generating a miRNA set of the species. The miRNA prediction method of the invention directly searches for the miRNA in the genome sequences, and so the obtained miRNA set is more complete. Furthermore, the miRNA prediction method is high in efficiency and more convenient, and lays the foundation for subsequent miRNA research.
Owner:BGI TECH SOLUTIONS
Molecular cloning and application of glp2r gene fragment related to pork quality traits
Owner:HUNAN AGRICULTURAL UNIV
A method for compressing next-generation sequencing data
ActiveCN105760706BSave storage spaceProcessing speedSpecial data processing applicationsParallel processingCompression method
The invention discloses a compression method for next generation sequencing data. The method comprises: dividing the next generation sequencing data of each sample according to a first preset length, to generate a BSSL original file; according to a second preset length, establishing a cutting tag file; according to the cutting tag file, processing the BSSL original file, to obtain BSSL intermediate files; combining the BSSL intermediate files to obtain a BSSL final file; counting a frequency distribution result of a seed sequence in the BSSL final file, to obtain a seed file according to the result; combined with the format characteristics of the sequencing data, determining compression rules, and based on the seed file, compressing the next generation sequencing data of each sample. Through dividing the next generation sequencing data and performing parallel processing, processing speed is improved, and combined with seed sequence selection, the seed file is obtained, and the next generation sequencing data is compressed according to the format characteristics of the sequencing data and the seed file, so that storage space of the next generation sequencing data is greatly reduced.
Owner:SHENZHEN HUADA GENE INST
Antisense oligonucleotides and applications directed at microrna-21 seed sequences
ActiveCN103320444BGrowth inhibitionInhibitionGenetic material ingredientsAntineoplastic agentsAntisense nucleic acidChronic granulocytic leukemia
The invention discloses an antisense oligonucleotide for tiny RNA-21 seed sequence and an application thereof and belongs to the field of medicine preparation. According to the research, tiny antisense nucleic acid t-antimiR-21 of 8 basic groups is designed and synthetized for miR-21 seed sequence. It shows through a t-antimiR-21 transfection cell experiment that t-antimiR-21 targets miR-21 of K562 cell and RPMI-8266 cell strain, cell growth is inhibited and cell apoptosis is obviously increased. It shows that lymphoma cell growth can be effectively inhibited through interfering with miRNA-21 expression by t-antimiR-21. Meanwhile, it shows that miRNA-21 might be used as a potential target for lymphoma gene therapy. t-antimiR-21 can be used in the preparation of an antineoplastic medicine, especially in the preparation of a medicine against chronic granulocytic leukemia and multiple myeloma.
Owner:广州安镝声生物医药科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com