A recommendation model-based method for predicting miRNA target genes
A target gene prediction, target gene technology, applied in the field of systems biology, can solve the problem of complex miRNA relationship
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
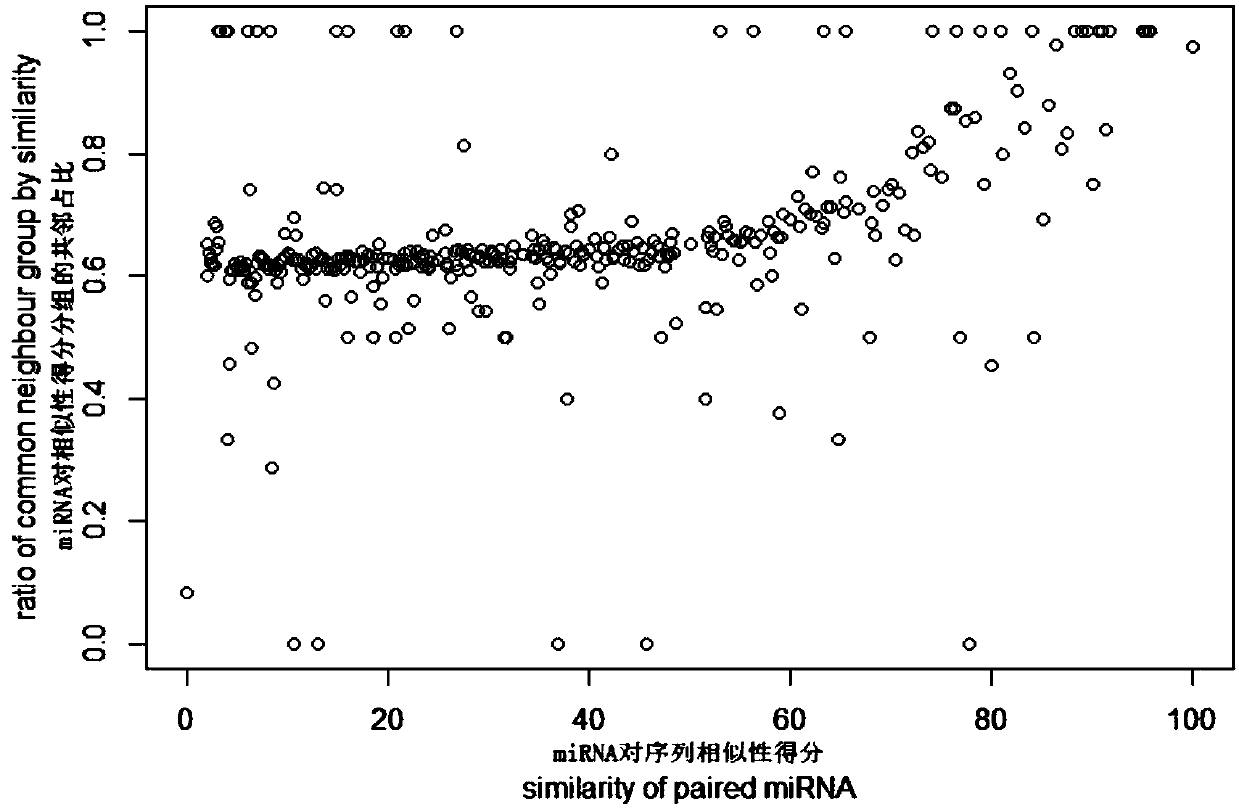
[0055] see Figure 1-8 , the miRNA target gene prediction method based on the recommended model is implemented in the following steps:
[0056] 1. Analysis of miRNA target gene data distribution characteristics (based on miRTarBase database)
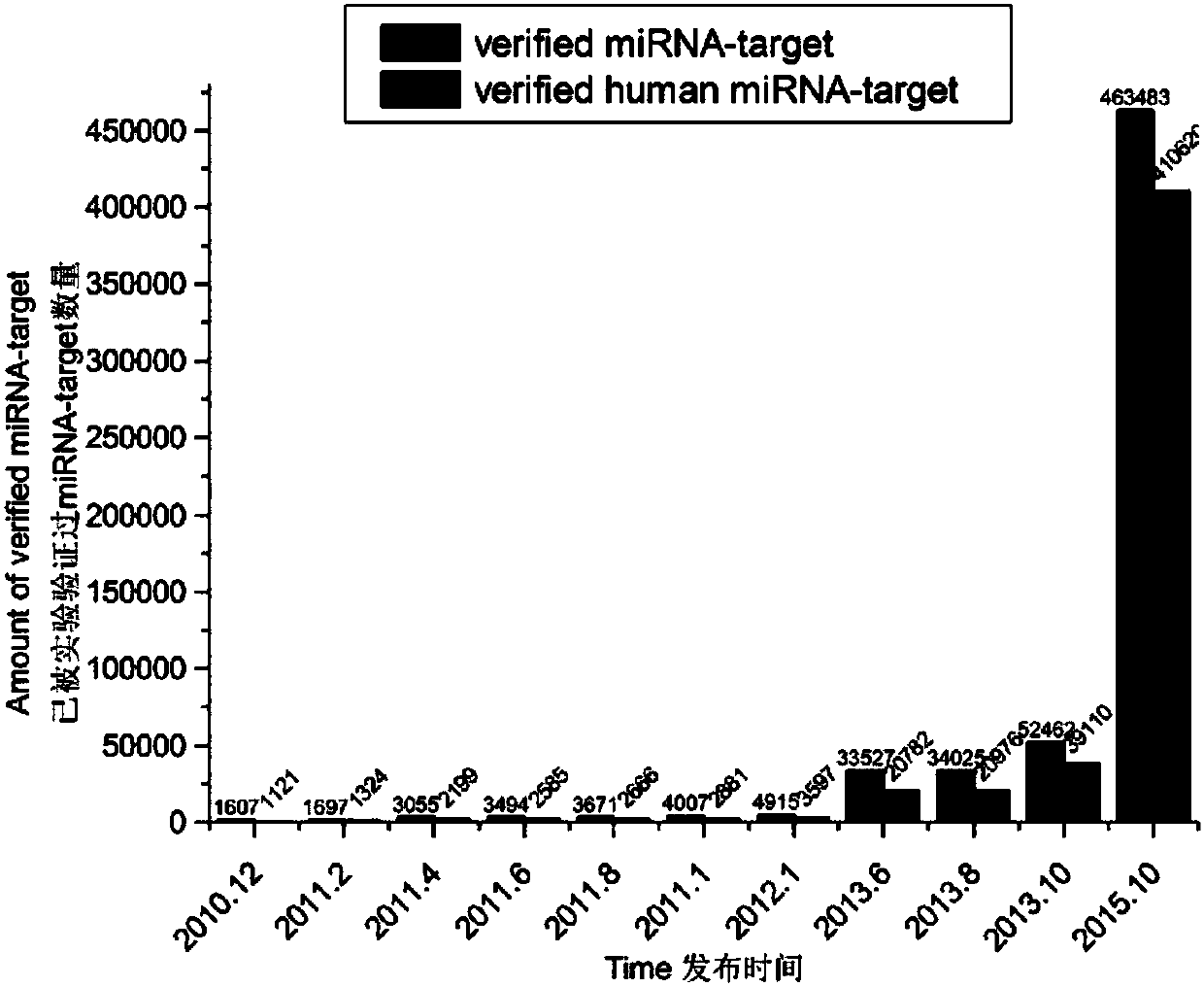
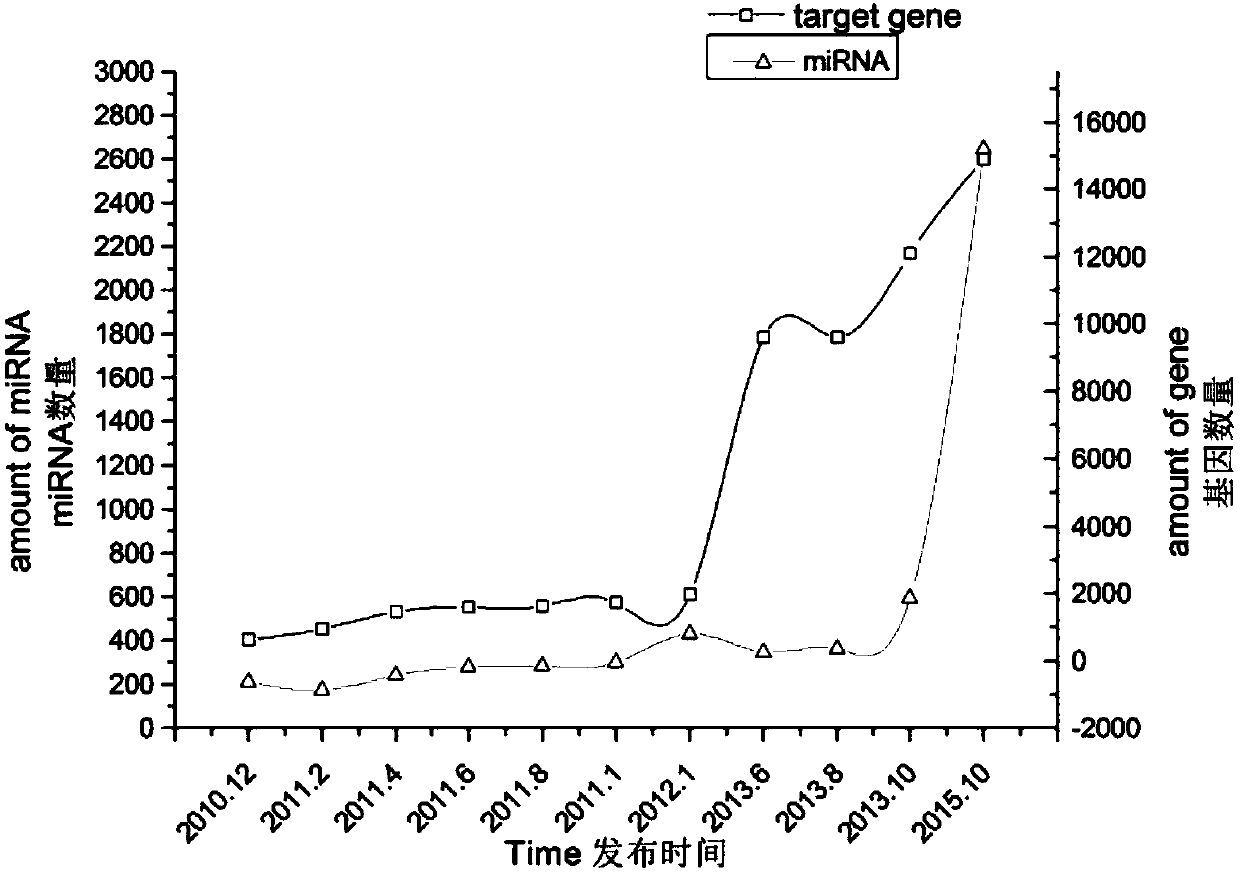
[0057] At present, the databases that record miRNA target genes that have been verified by biological experiments include miRTarBase, miRwalk, TarBase, etc. Among them, miRTarBase is the most commonly used, so statistical analysis is performed on it. Since the release of the first version 1.6 in December 2010, the miRTarBase database has released a total of 11 versions to version 6.1 in October 2015. The miRTarBase V6.1 database contains 463,483 miRNA target gene relationship data of 18 species including humans, among which the human miRNA target gene relationship data reaches 410,620. figure 1 Shown is the histogram of the number of miRNA target genes in these 11 versions. It can be seen from the figure that, especially in the past tw...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com