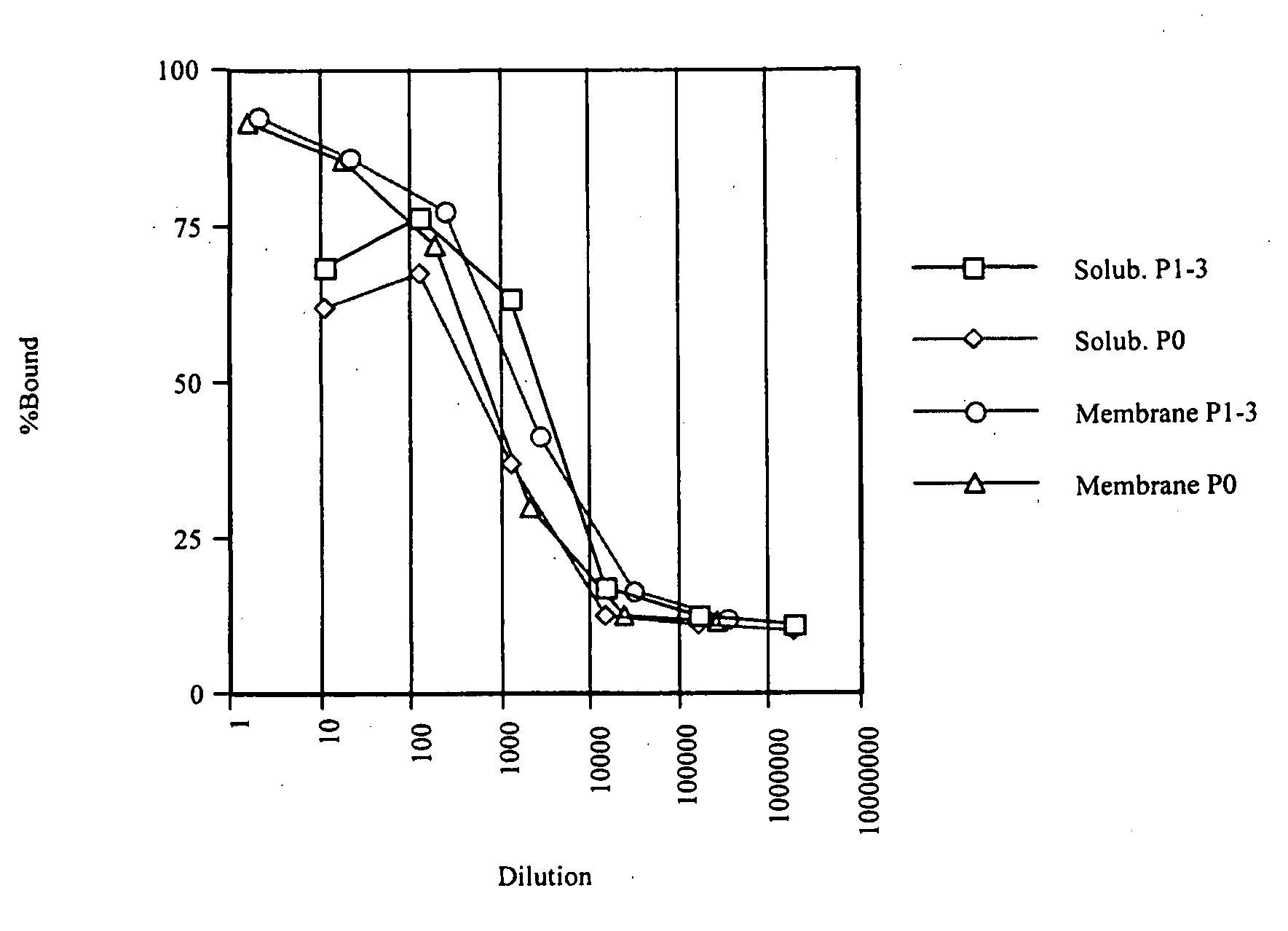
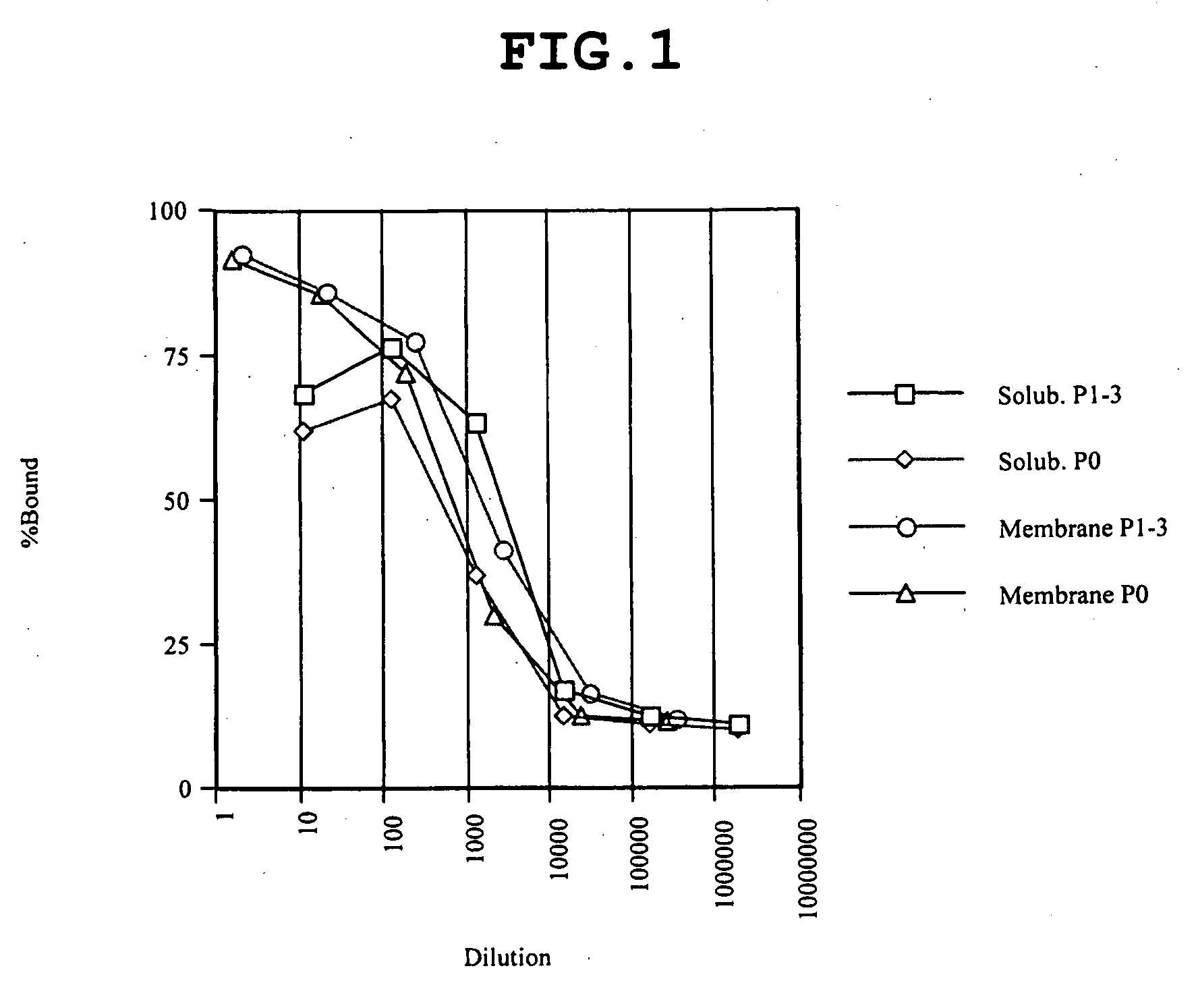
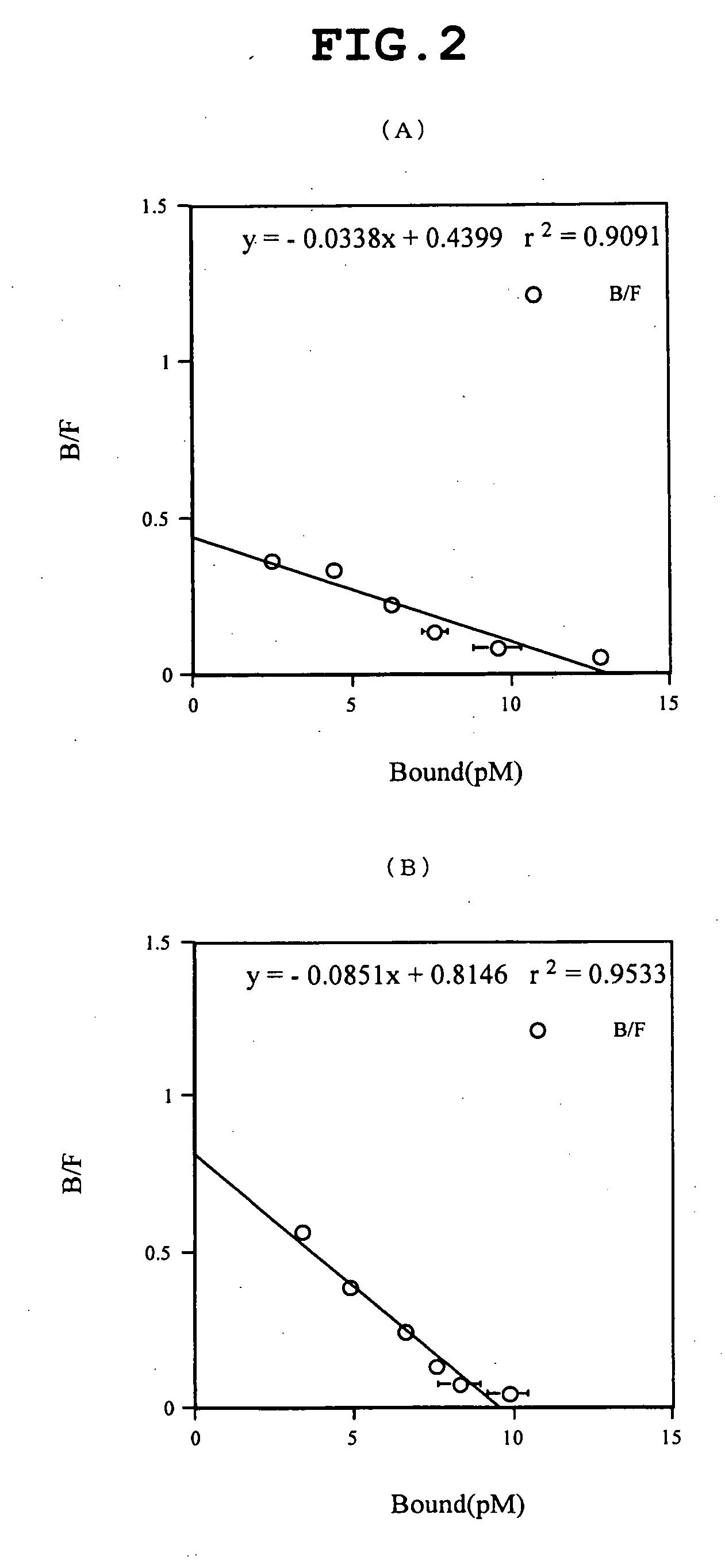
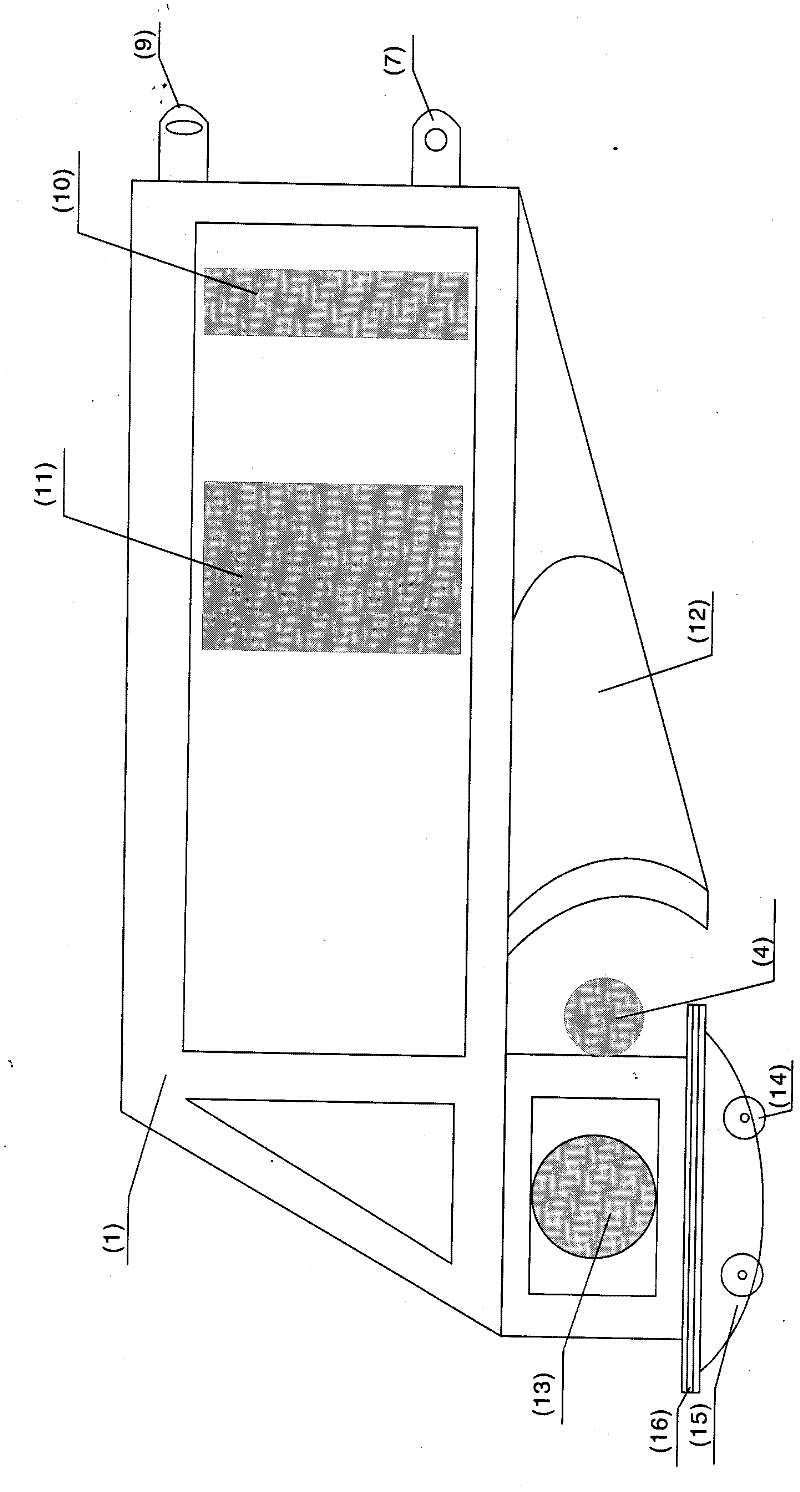
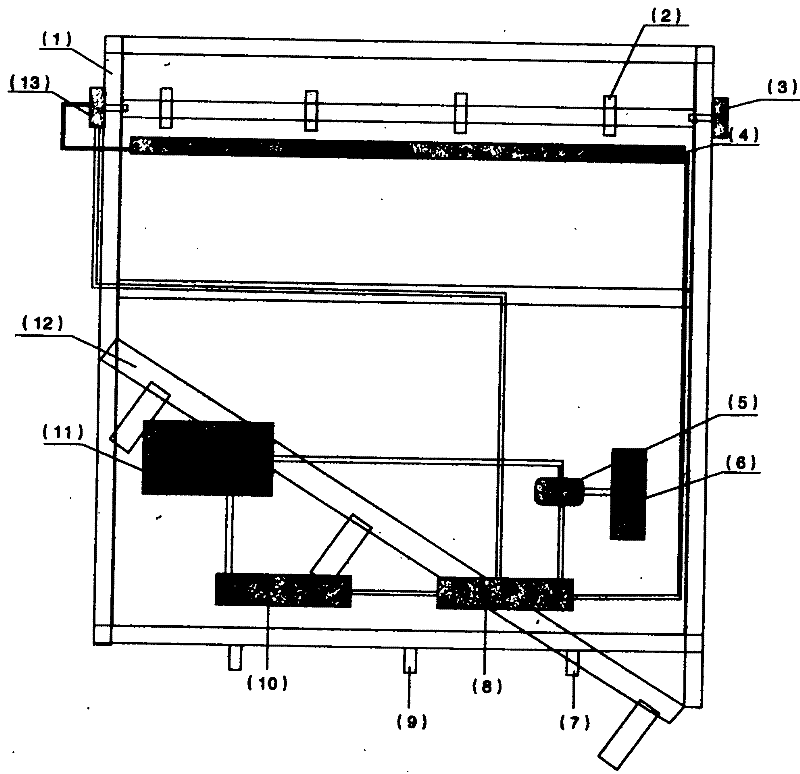
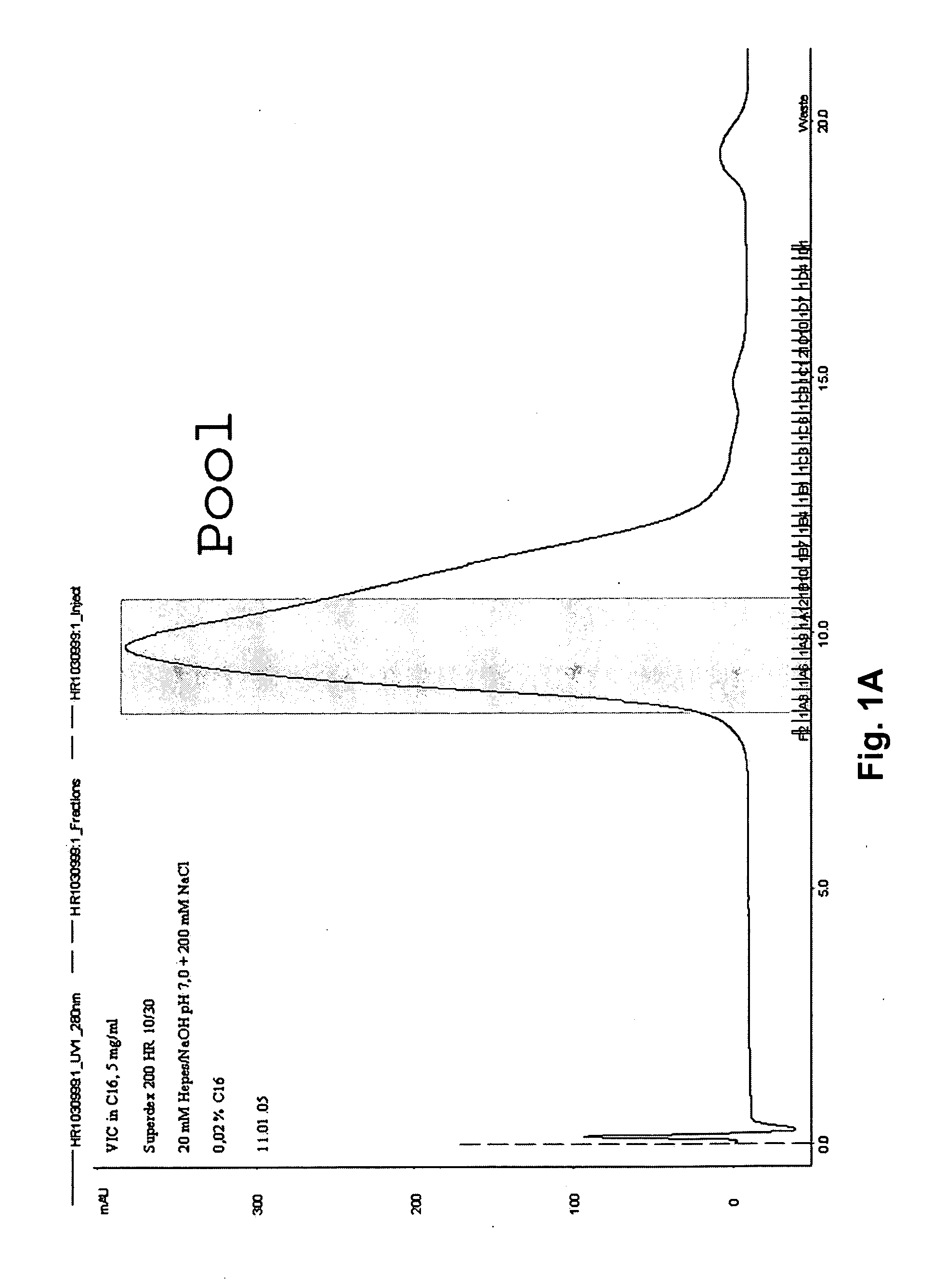
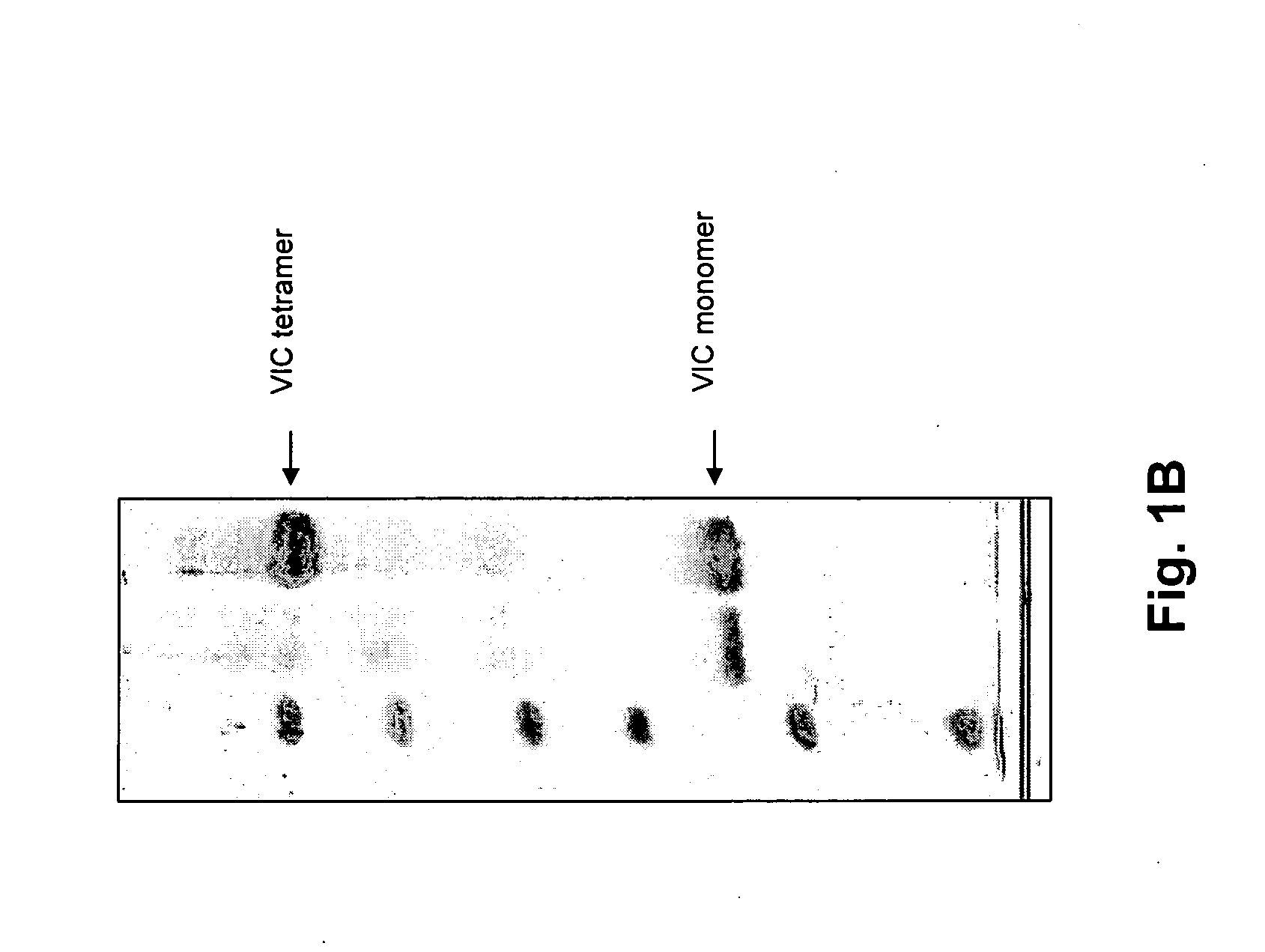
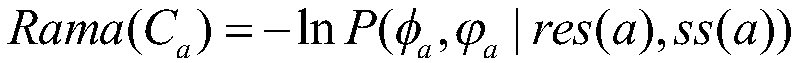Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
35 results about "Native structure" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
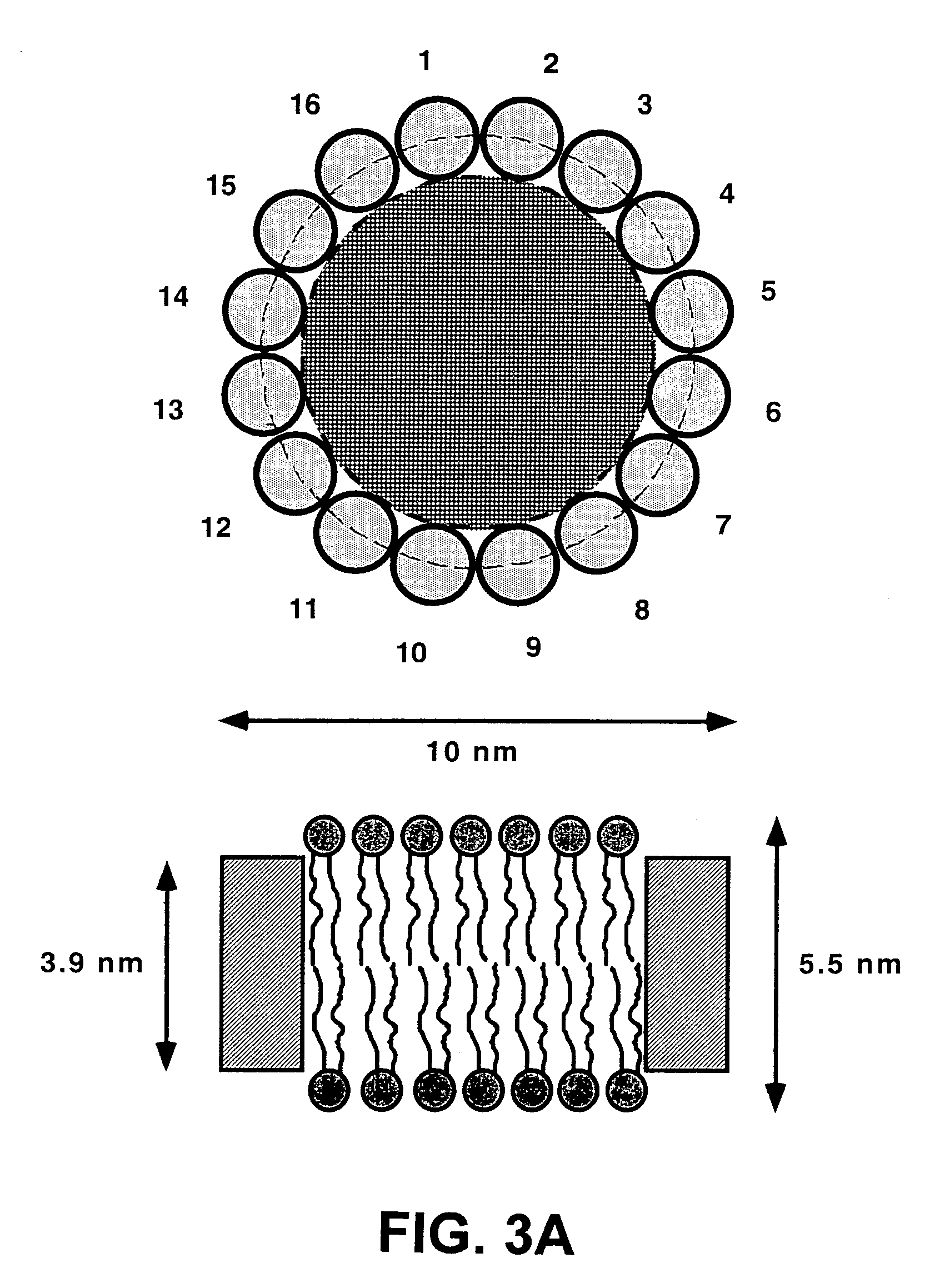
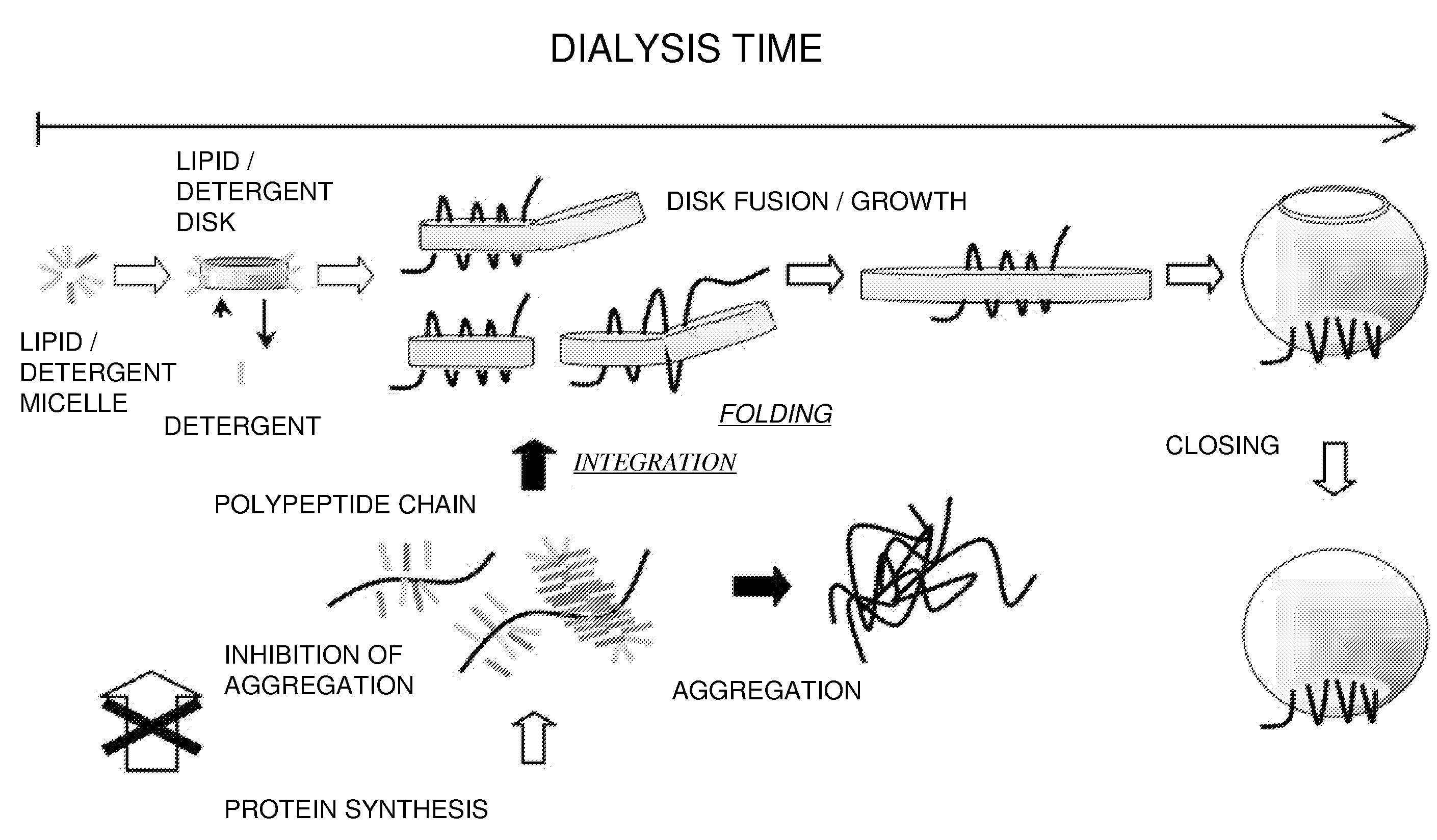
Membrane scaffold proteins
InactiveUS7083958B2Improve stabilityGood monodispersityDepsipeptidesPeptide preparation methodsNative structurePhospholipid
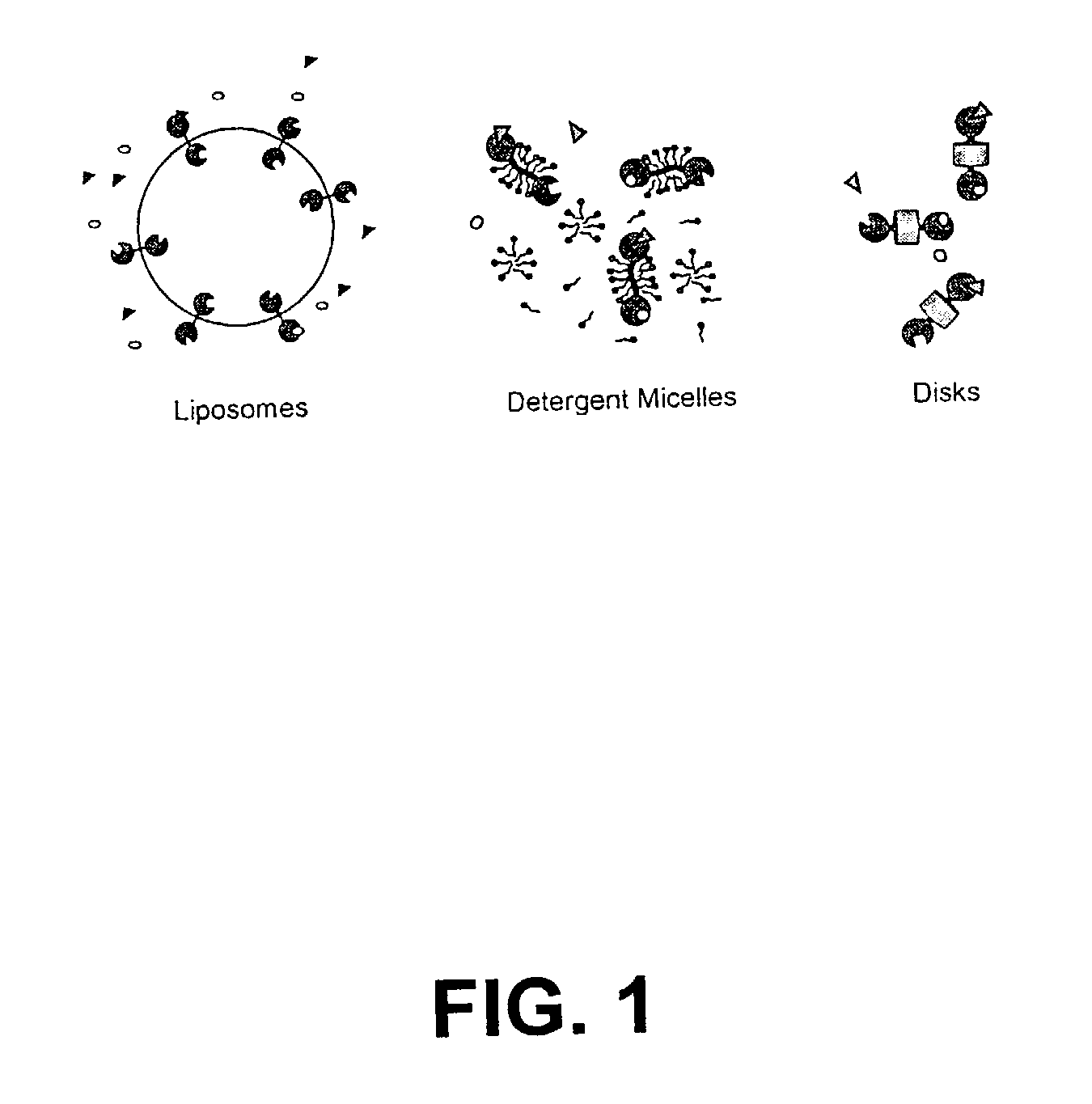
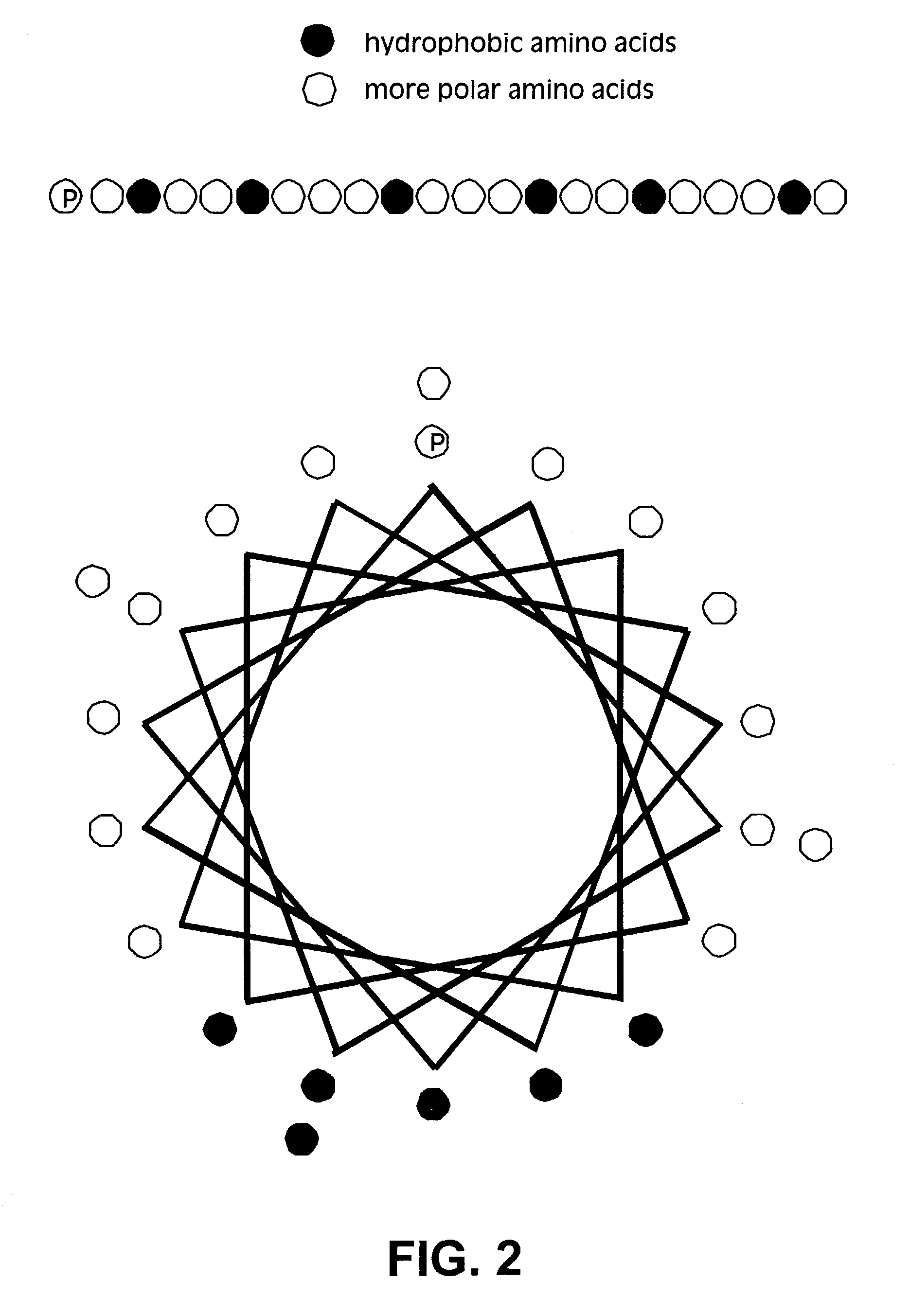
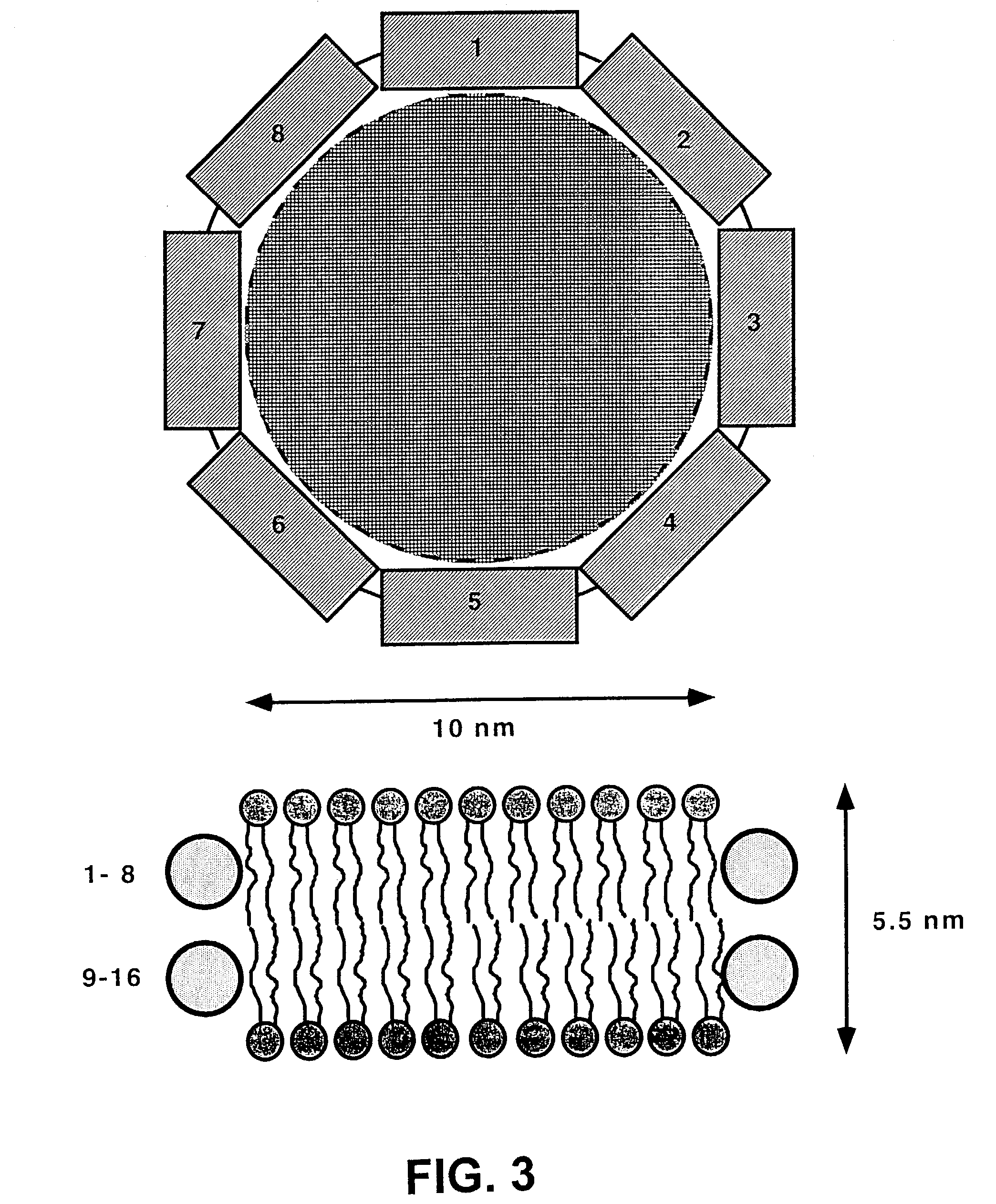
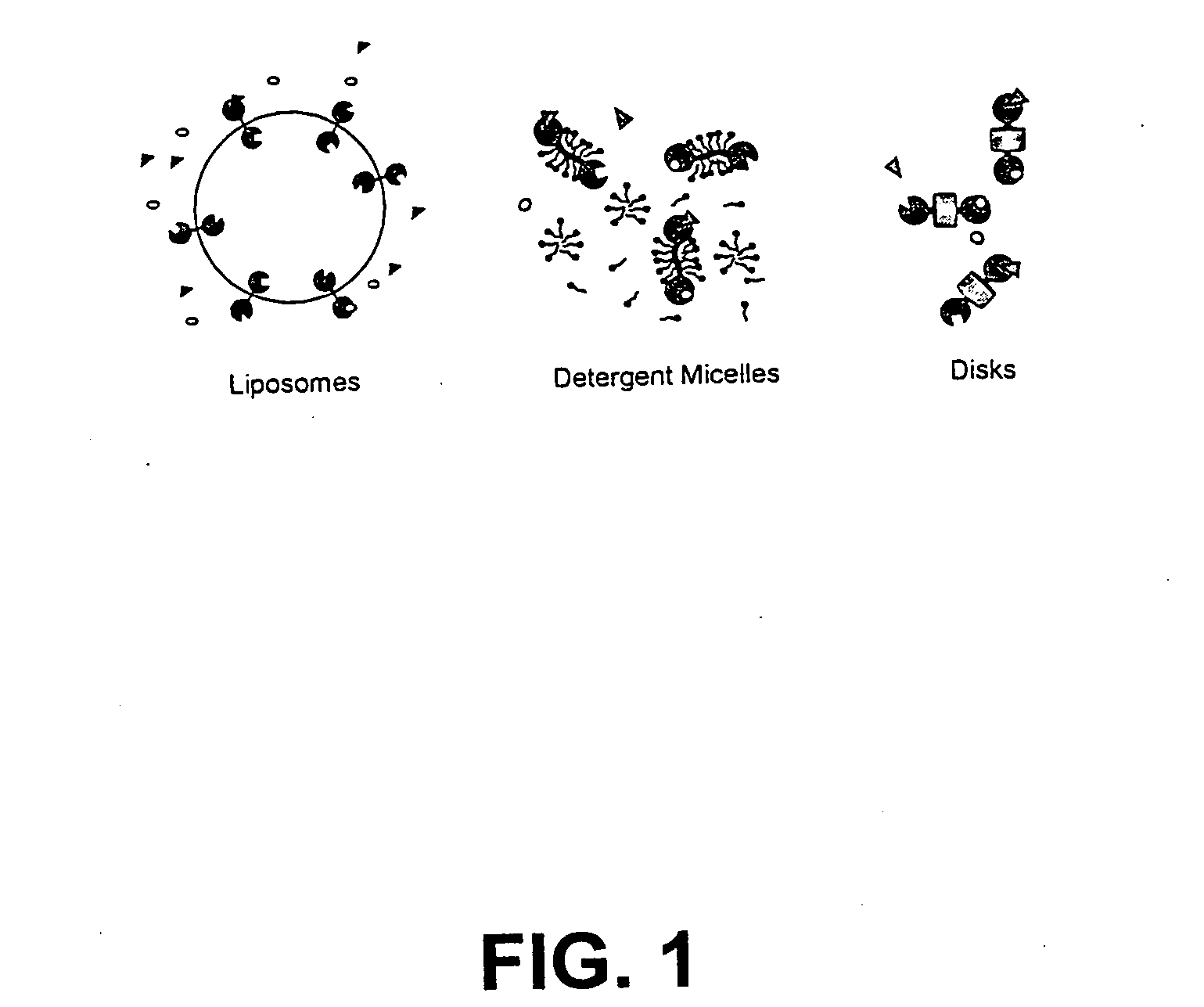
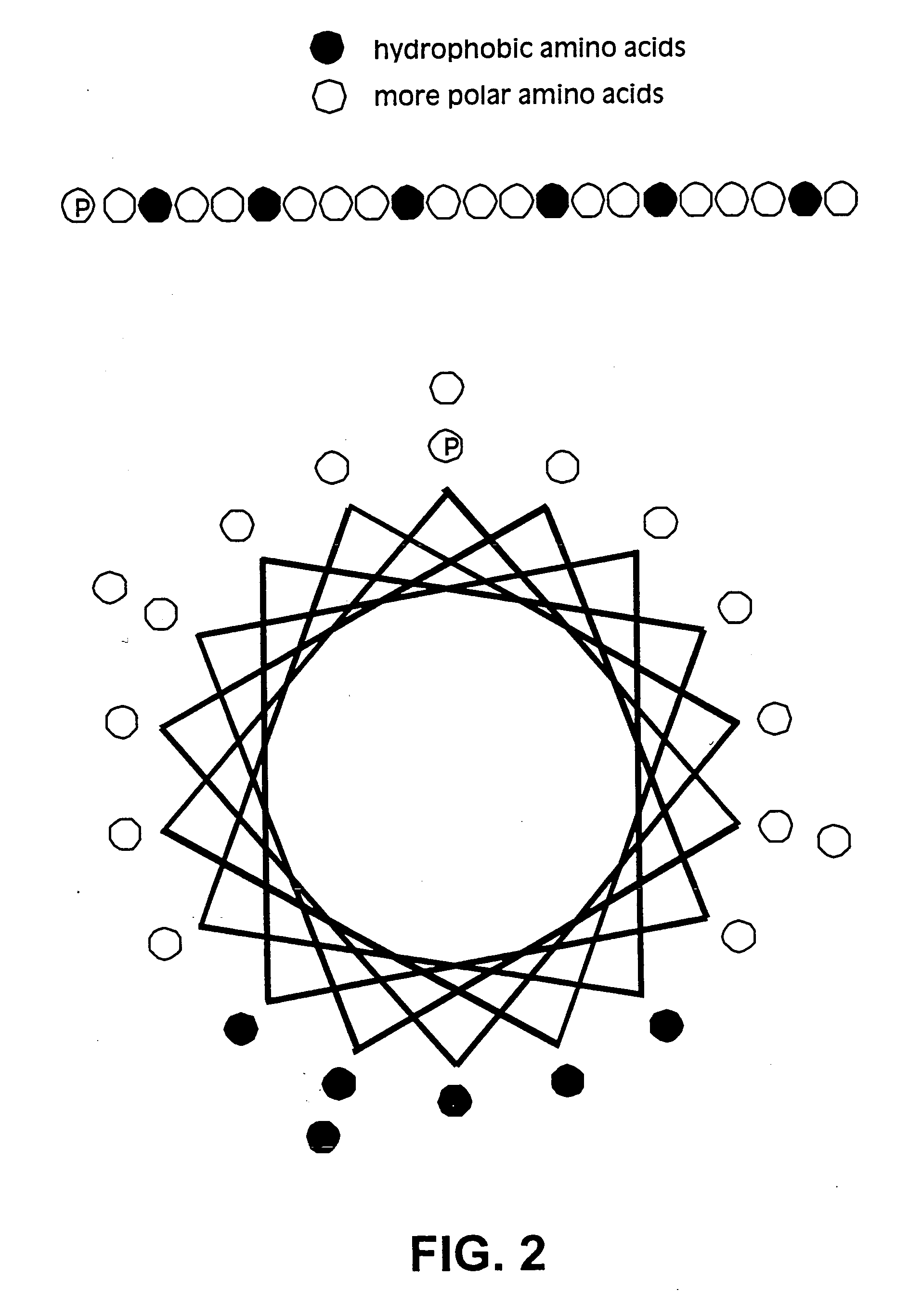
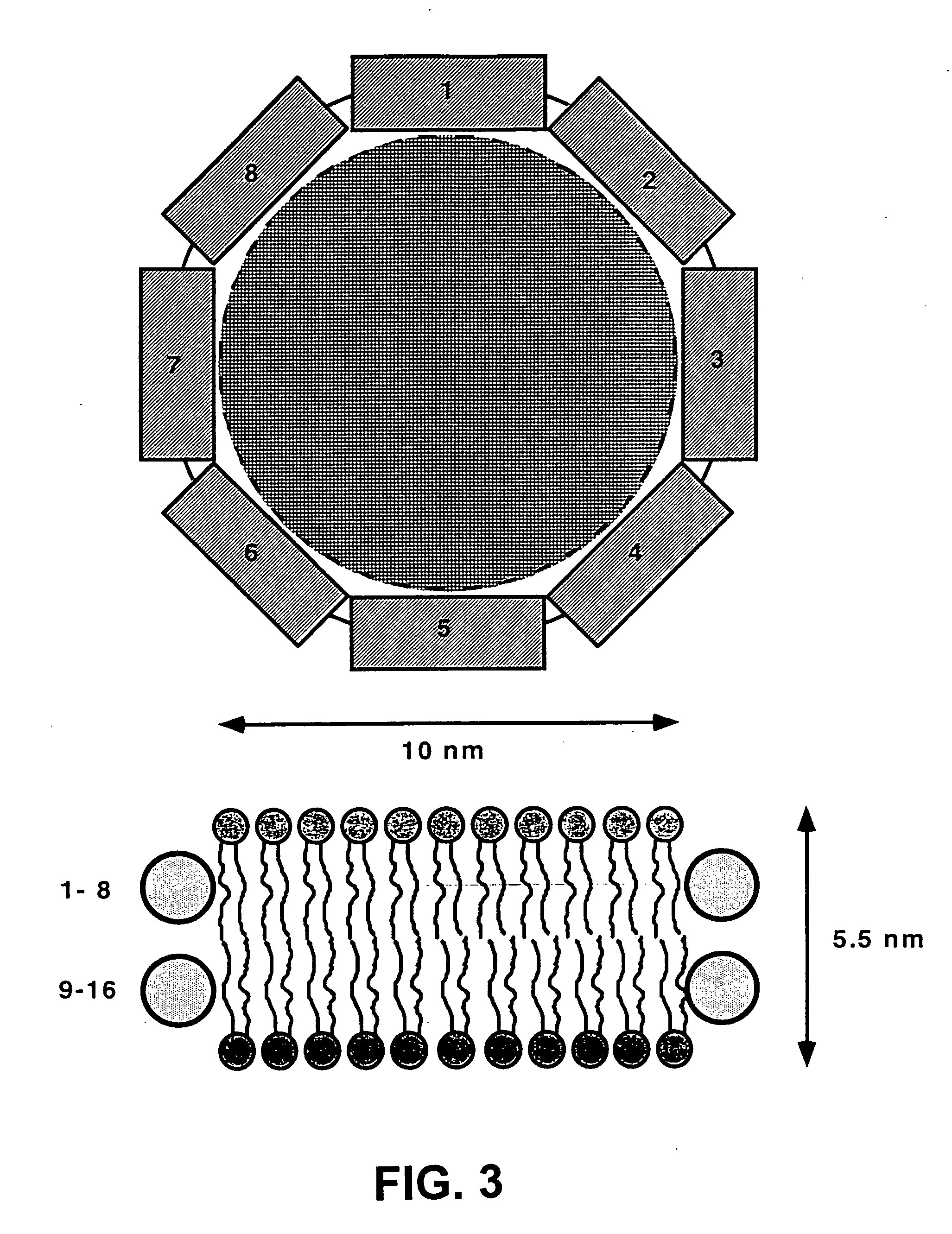
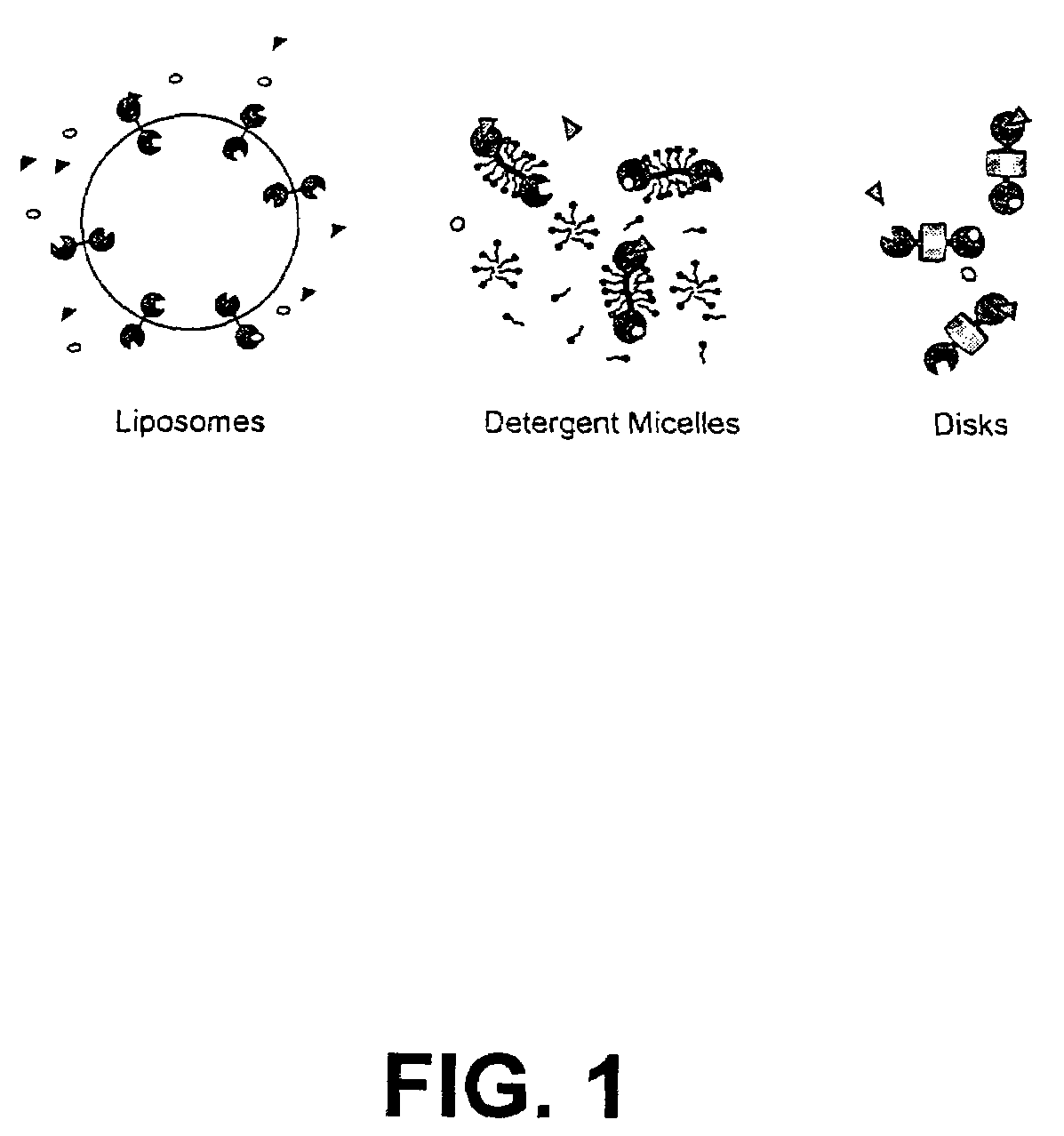
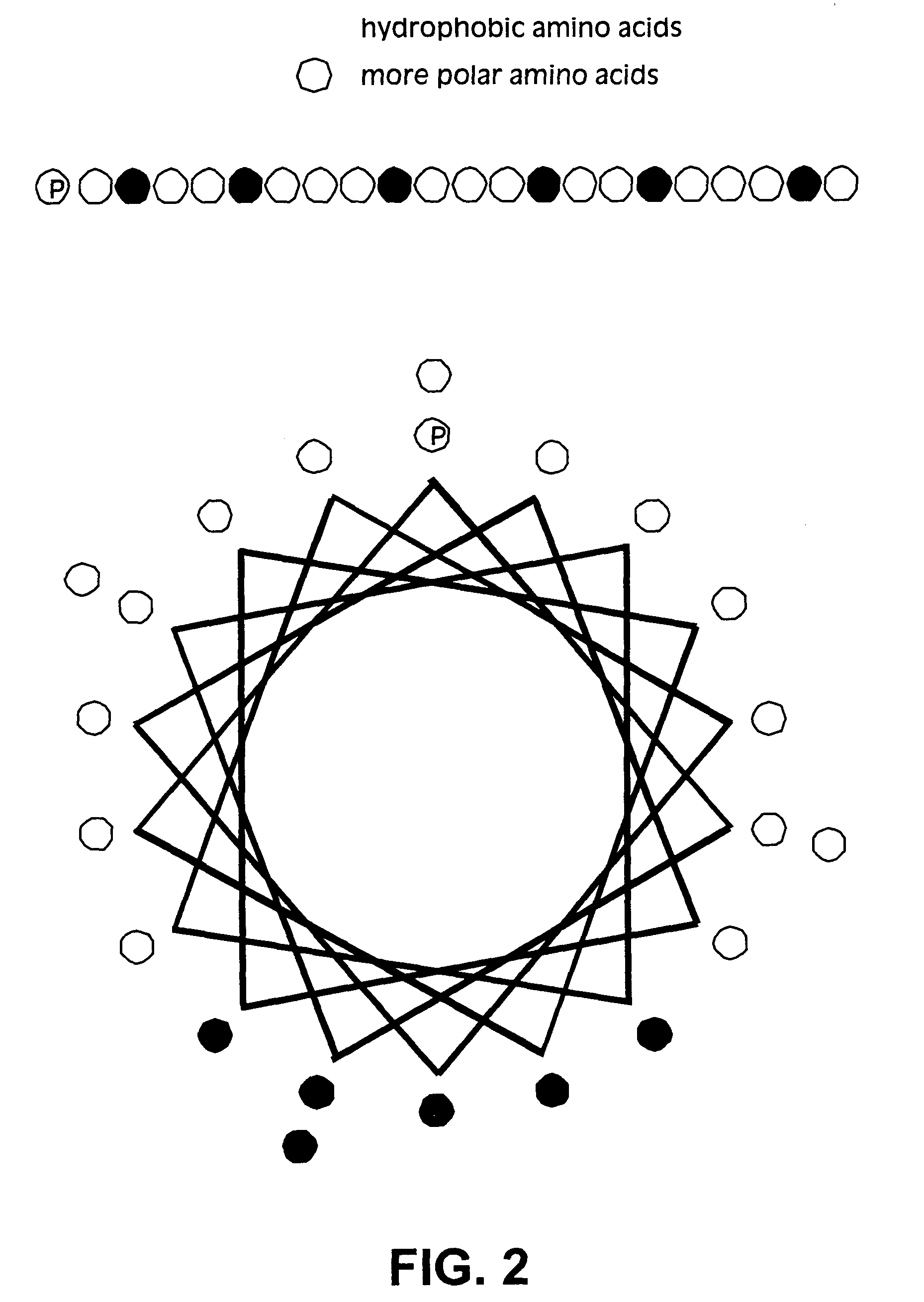
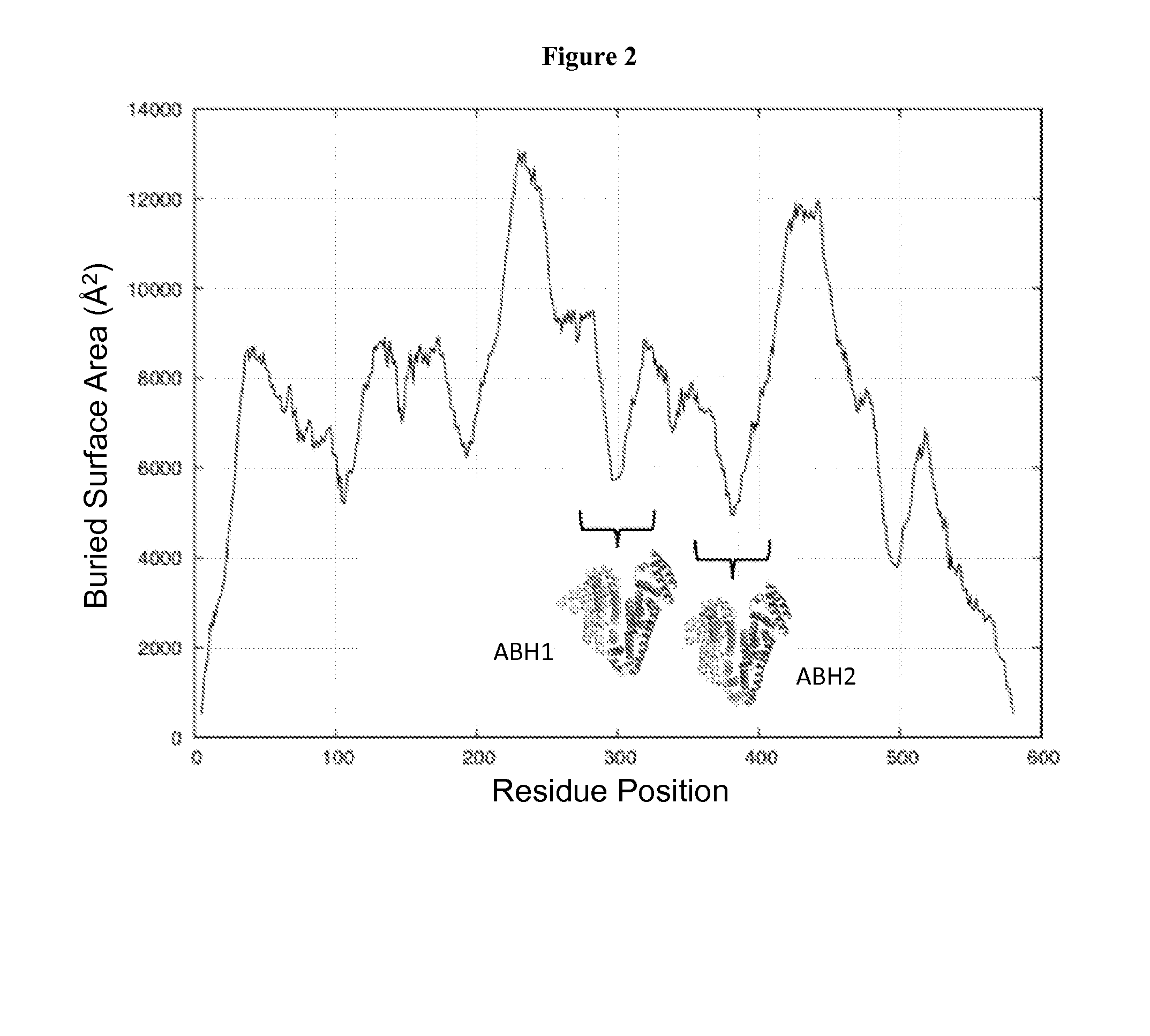
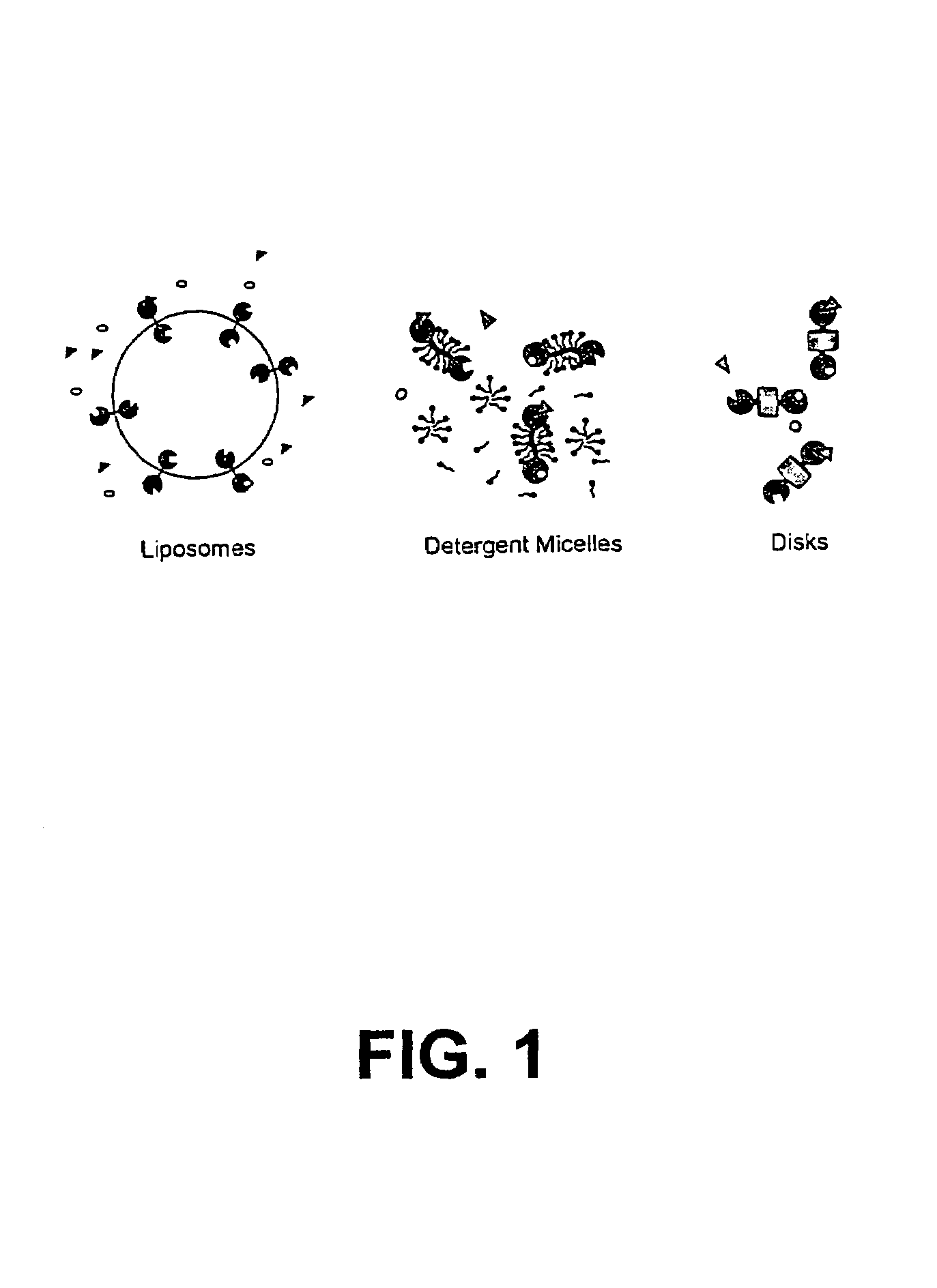
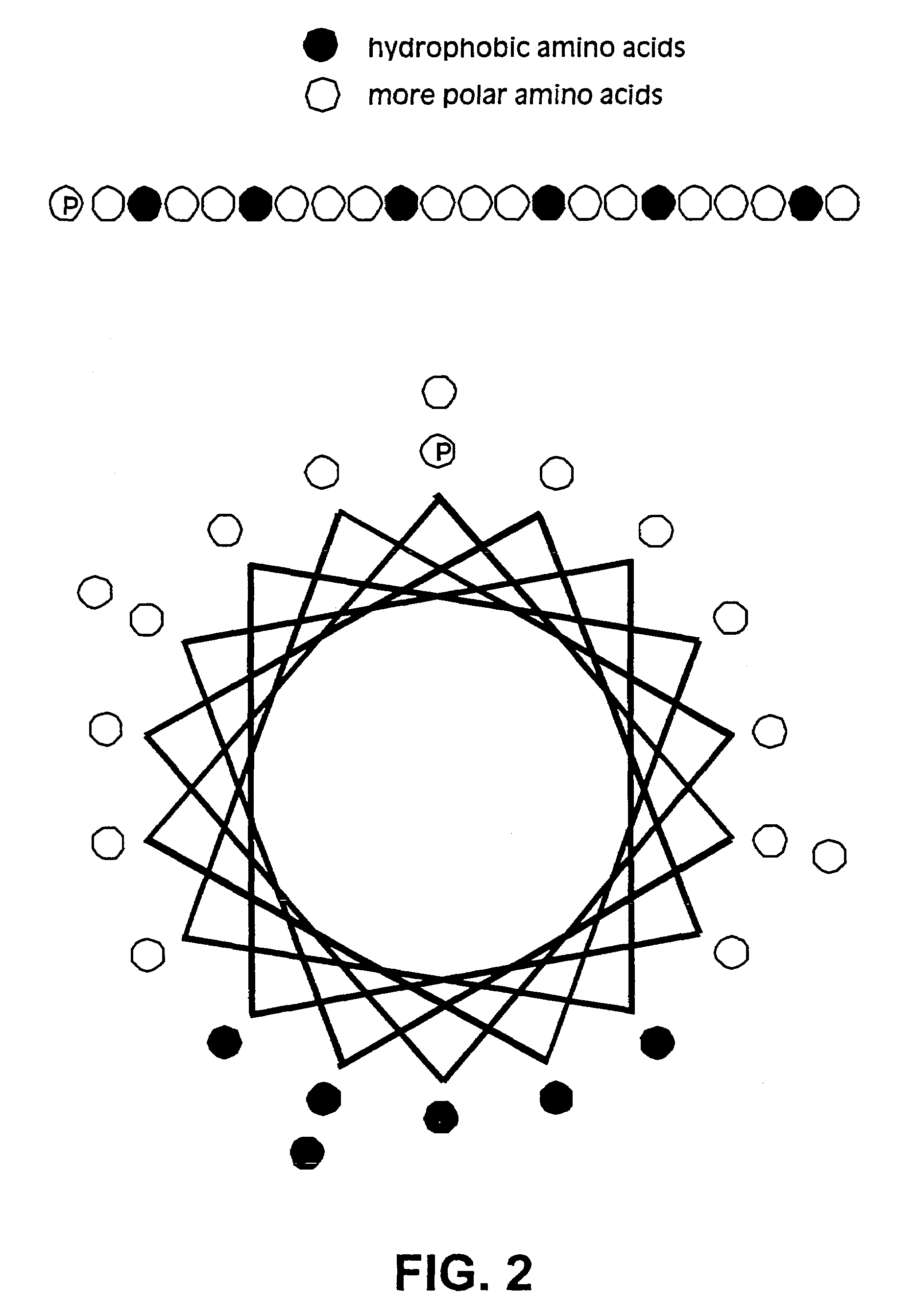
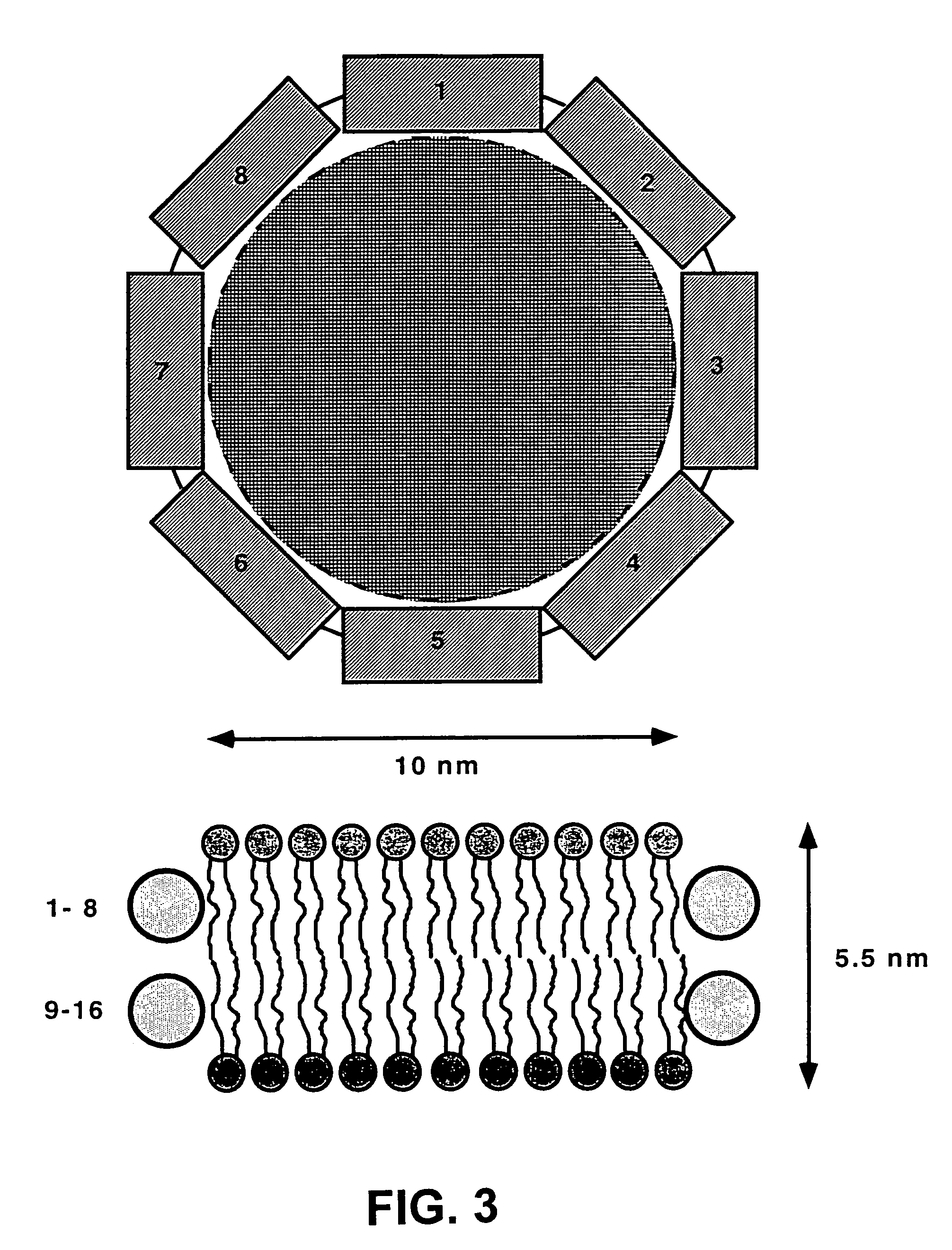
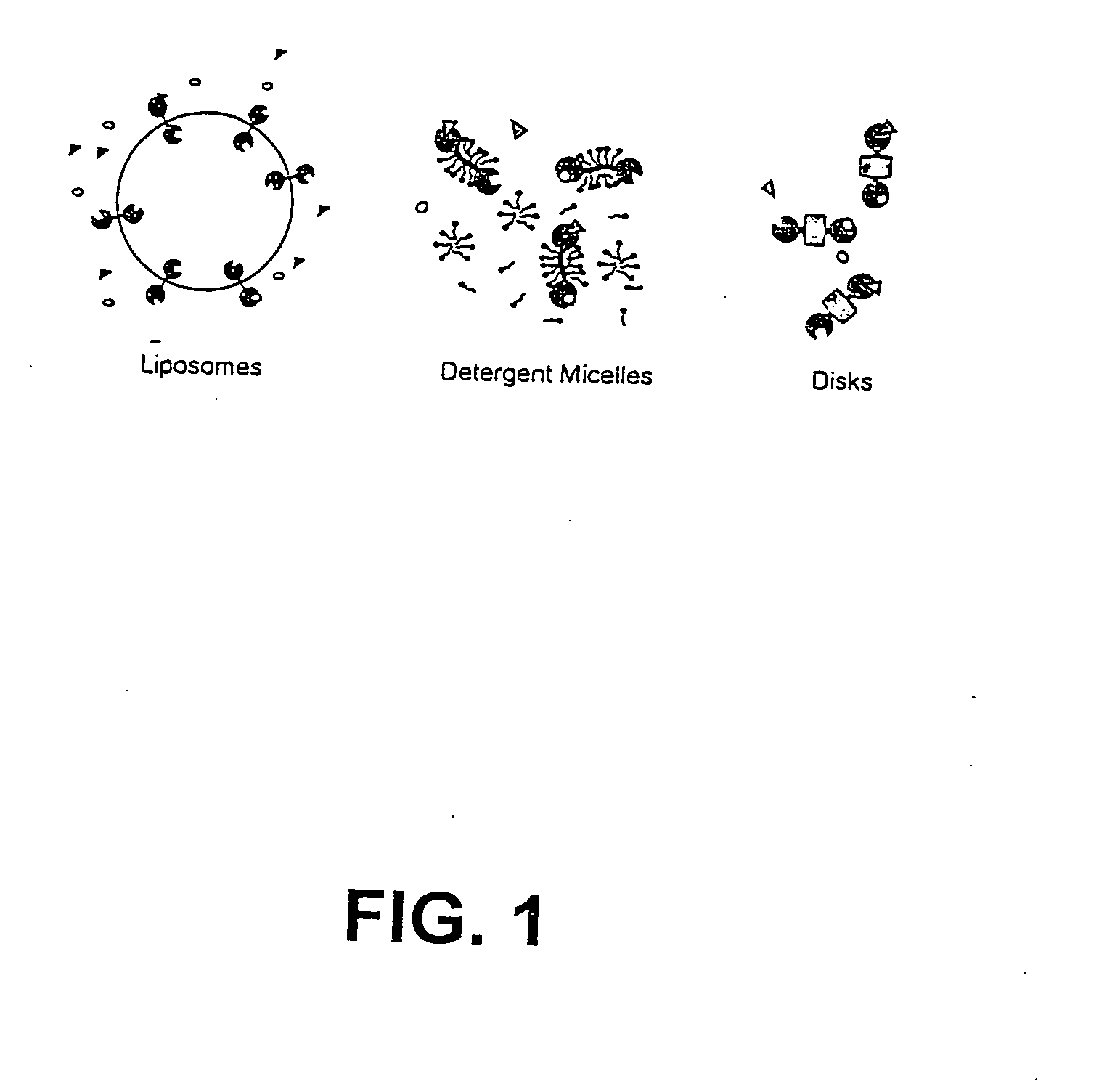
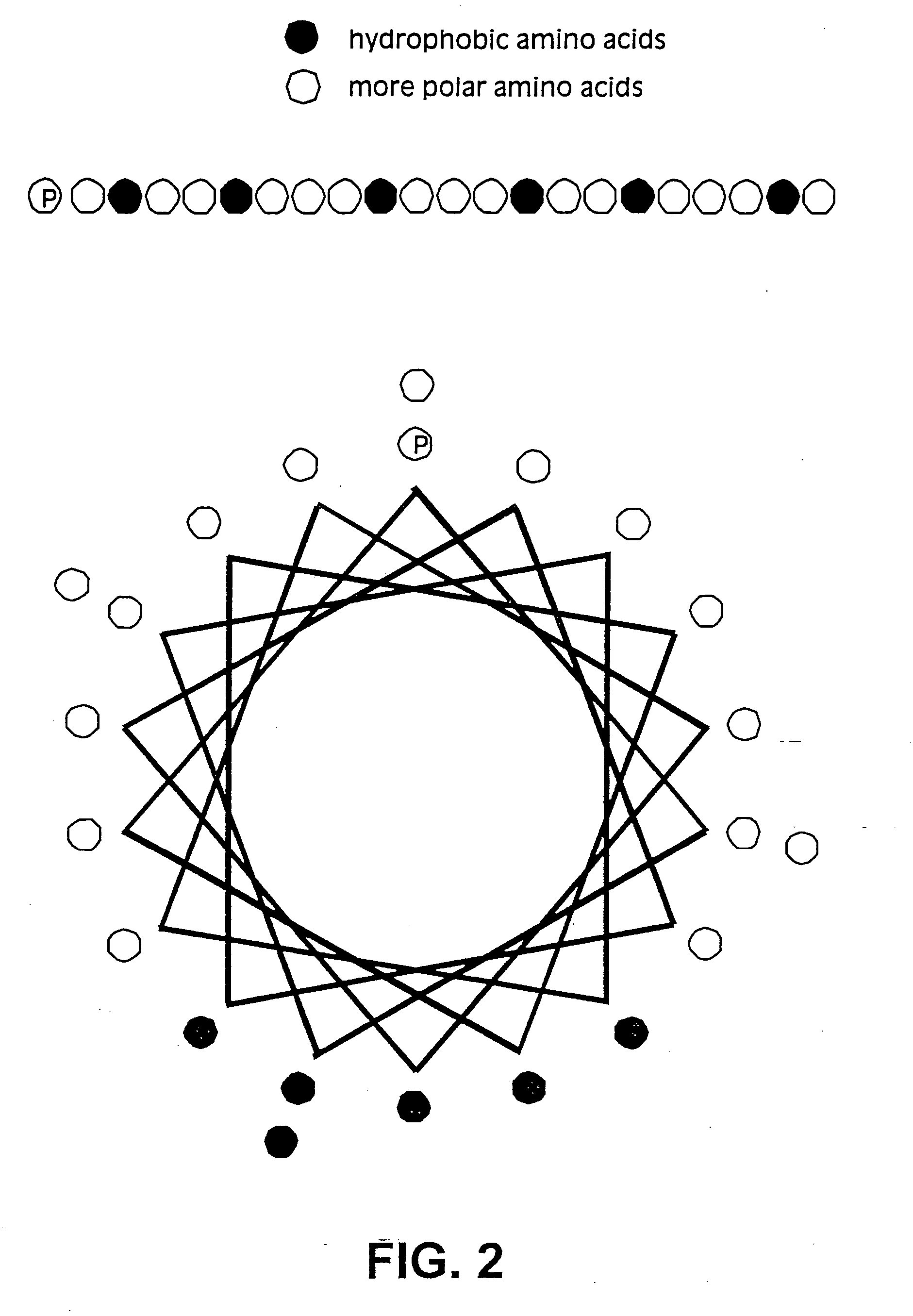
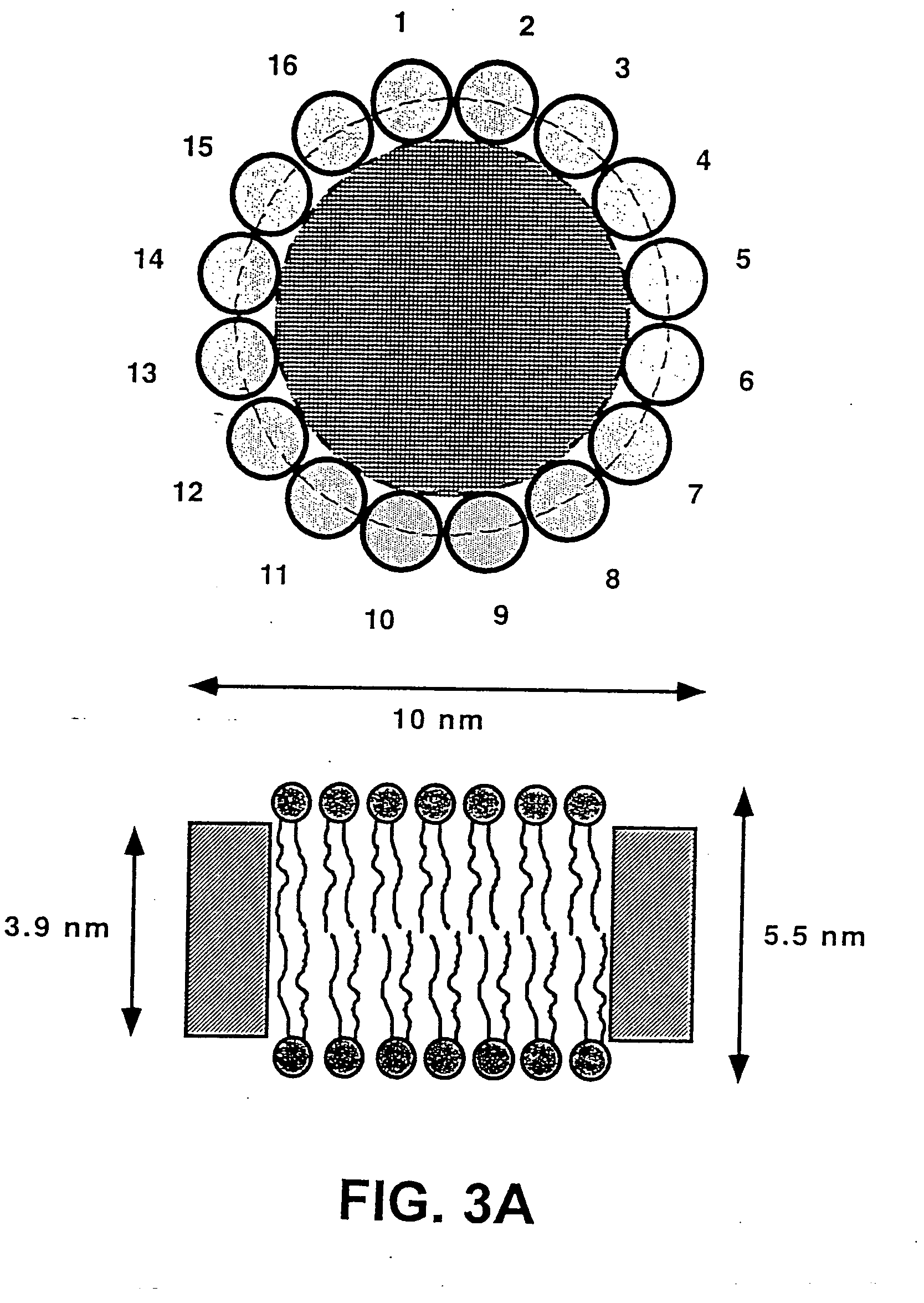
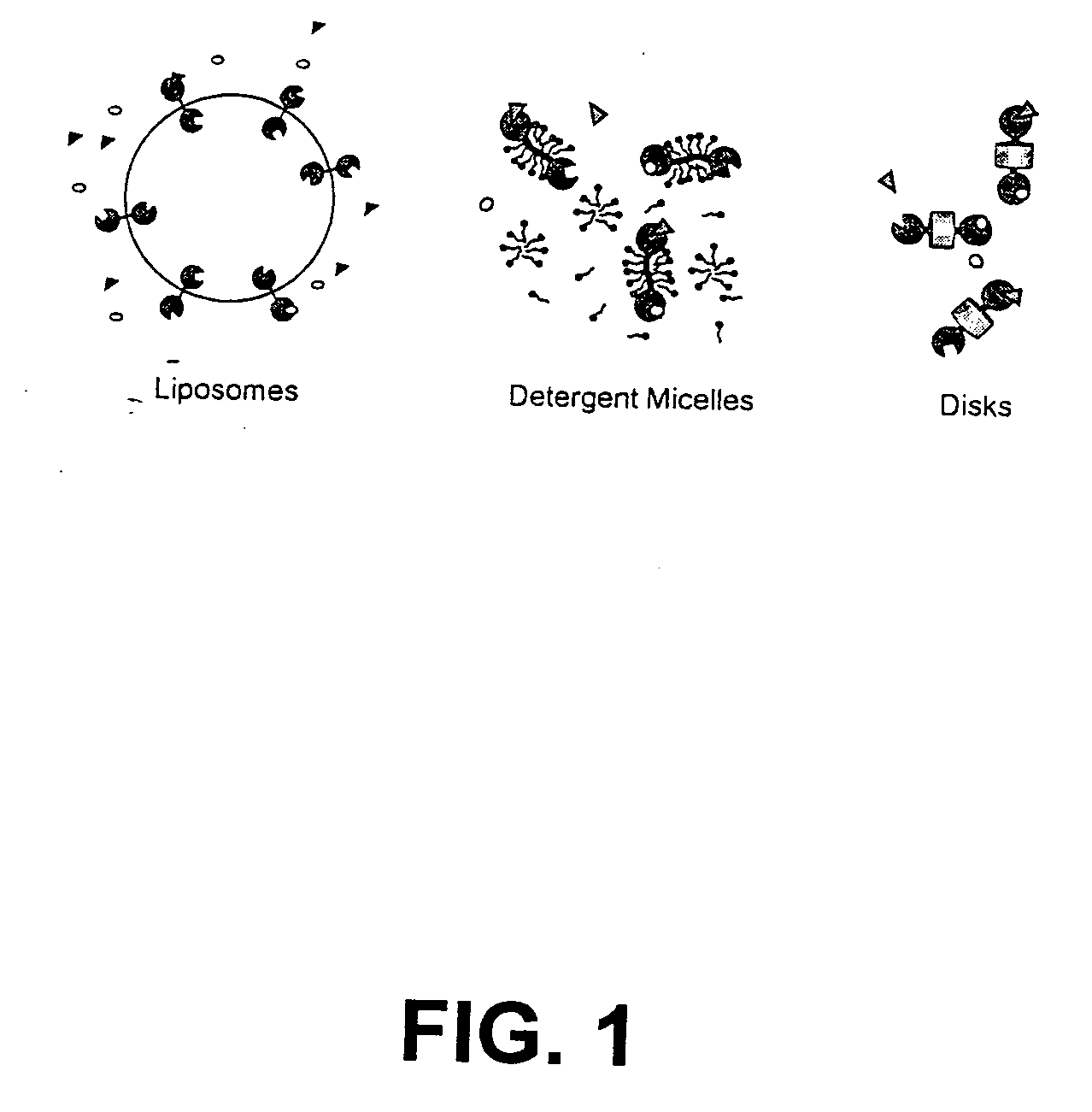
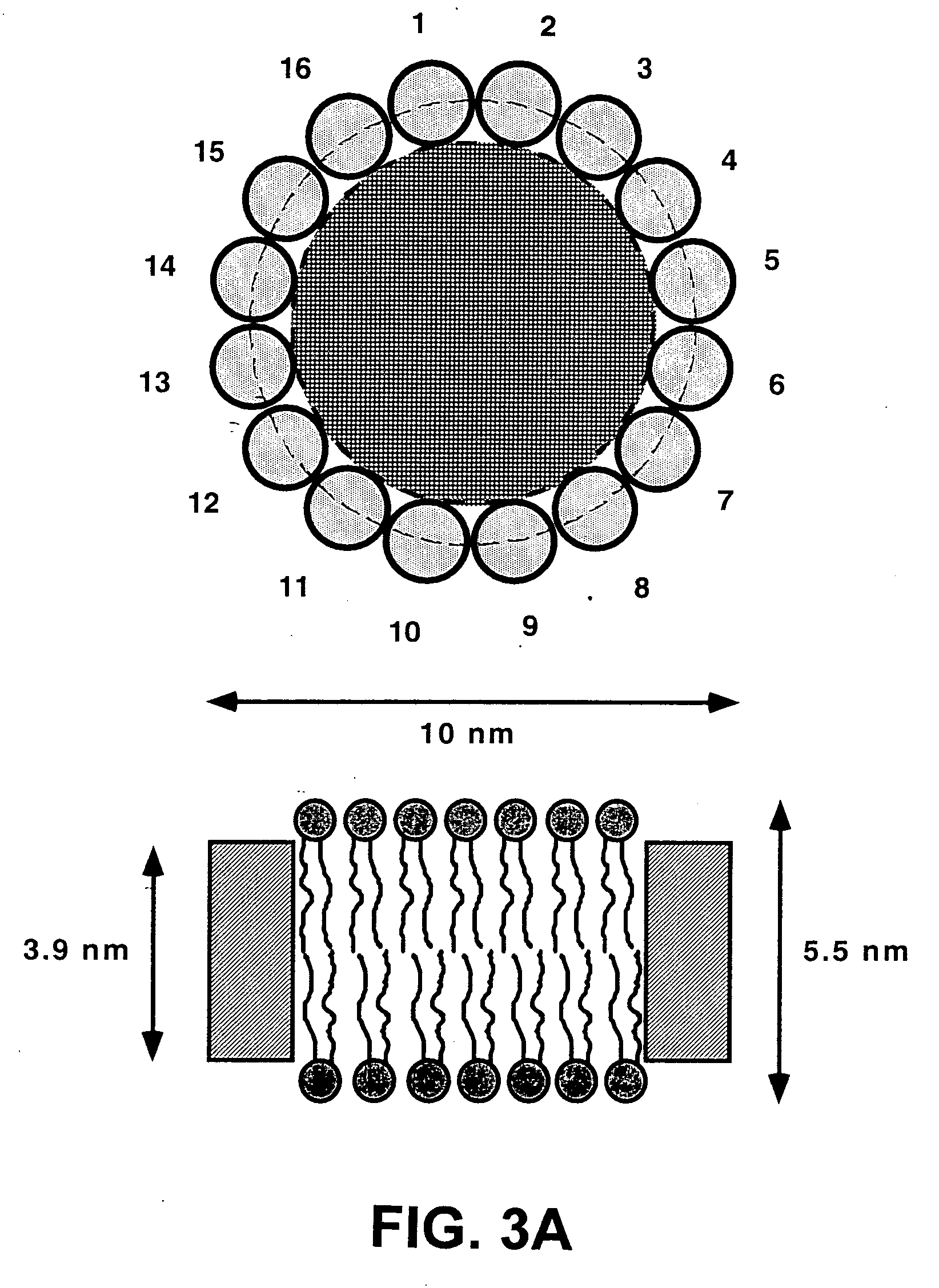
Membrane proteins are difficult to express in recombinant form, purify, and characterize, at least in part due to their hydrophobic or partially hydrophobic properties. The membrane scaffold proteins (MSP) of the present invention assemble with target membrane or other hydrophobic or partially hydrophobic proteins or membrane fragments to form soluble nanoscale particles which preserve their native structure and function; they are improved over liposomes and detergent micelles. In the presence of phospholipid, MSPs form nanoscopic phospholipid bilayer disks, with the MSP stabilizing the particle at the perimeter of the bilayer domain. The particle bilayer structure allows manipulation of incorporated proteins in solution or on solid supports, including for use with such surface-sensitive techniques as scanning probe microscopy or surface plasmon resonance. The nanoscale particles, which are robust in terms of integrity and maintenance of biological activity of incorporated proteins, facilitate pharmaceutical and biological research, structure / function correlation, structure determination, bioseparation, and drug discovery.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
Membrane scaffold proteins
InactiveUS20050182243A1Increase stability and monodispersityImprove purification effectBacterial antigen ingredientsProtozoa antigen ingredientsNative structurePhospholipid
The membrane scaffold proteins (MSP) of the present invention assemble with hydrophobic or partially hydrophobic proteins to form soluble nanoscale particles which preserve native structure and function; they are improved over liposomes and detergent micelles, in terms of stability and preservation of biological activity and native conformation. In the presence of phospholipid, MSPs form nanoscopic phospholipid bilayer disks, with the MSP stabilizing the particle at the perimeter of the bilayer domain. The particle bilayer structure allows manipulation of incorporated proteins in solution or on solid supports, including for use with such surface-sensitive techniques as scanning probe microscopy or surface plasmon resonance. The nanoscale particles, which are robust in terms of integrity and maintenance of biological activity of incorporated proteins, facilitate pharmaceutical and biological research, structure / function correlations, structure determinations, bioseparations, and drug discovery.
Owner:BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS THE A BODY & POLITIC OF THE STATE OF ILLINOIS
Membrane scaffold proteins
InactiveUS7048949B2Great stability and size homogeneity and useful functionalityEasy to understandPowder deliveryFungiNative structurePhospholipid
Membrane proteins are difficult to express in recombinant form, purify, and characterize, at least in part due to their hydrophobic or partially hydrophobic properties. Membrane scaffold proteins (MSP) assemble with target membrane or other hydrophobic or partially hydrophobic proteins or membrane fragments to form soluble nanoscale particles which preserve their native structure and function; they are improved over liposomes and detergent micelles. In the presence of phospholipid, MSPs form nanoscopic phospholipid bilayer disks, with the MSP stabilizing the particle at the perimeter of the bilayer domain. The particle bilayer structure allows manipulation of incorporated proteins in solution or on solid supports, including for use with such surface-sensitive techniques as scanning probe microscopy or surface plasmon resonance. The nanoscale particles facilitate pharmaceutical and biological research, structure / function correlation, structure determination, bioseparation, and drug discovery.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
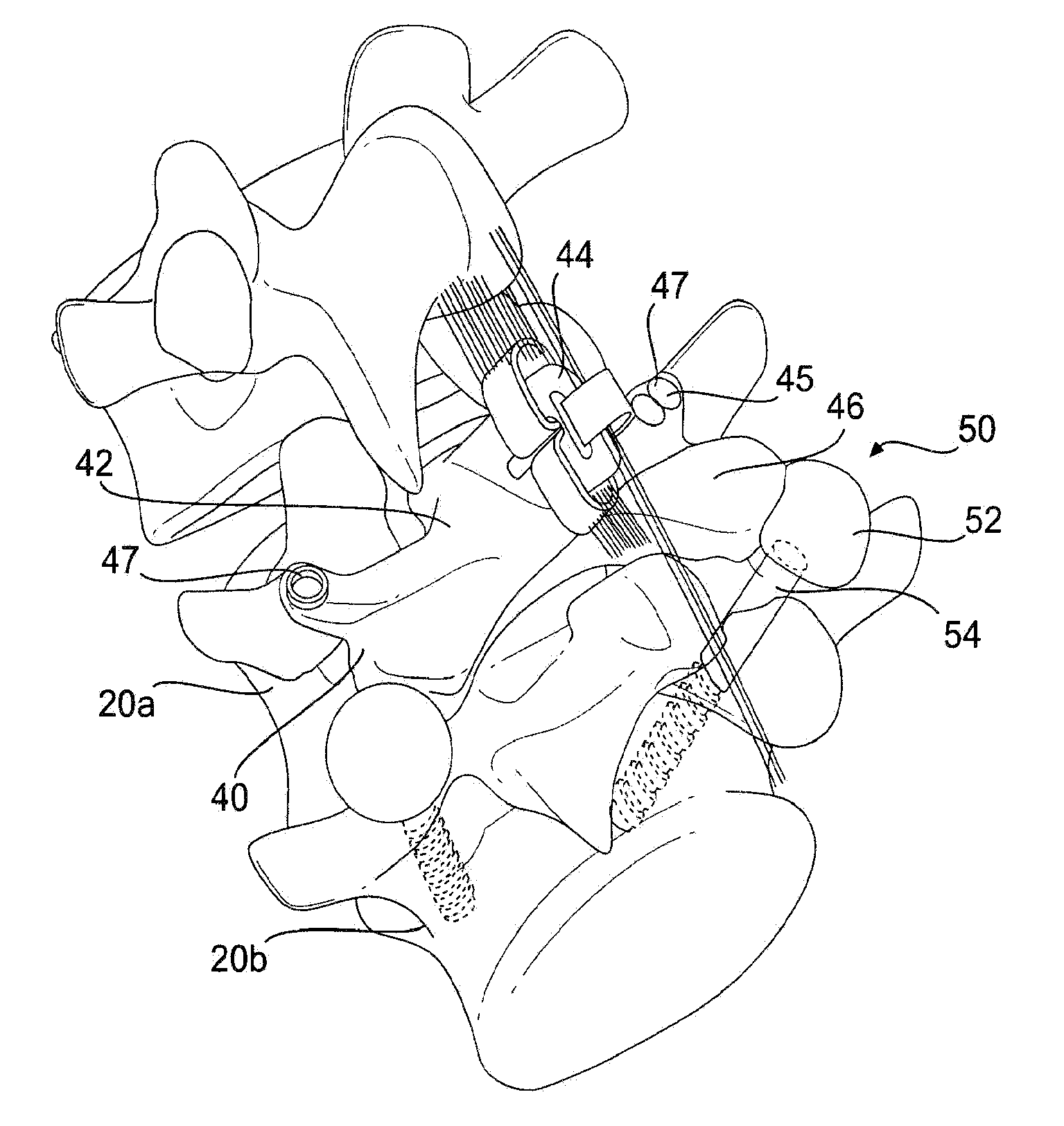
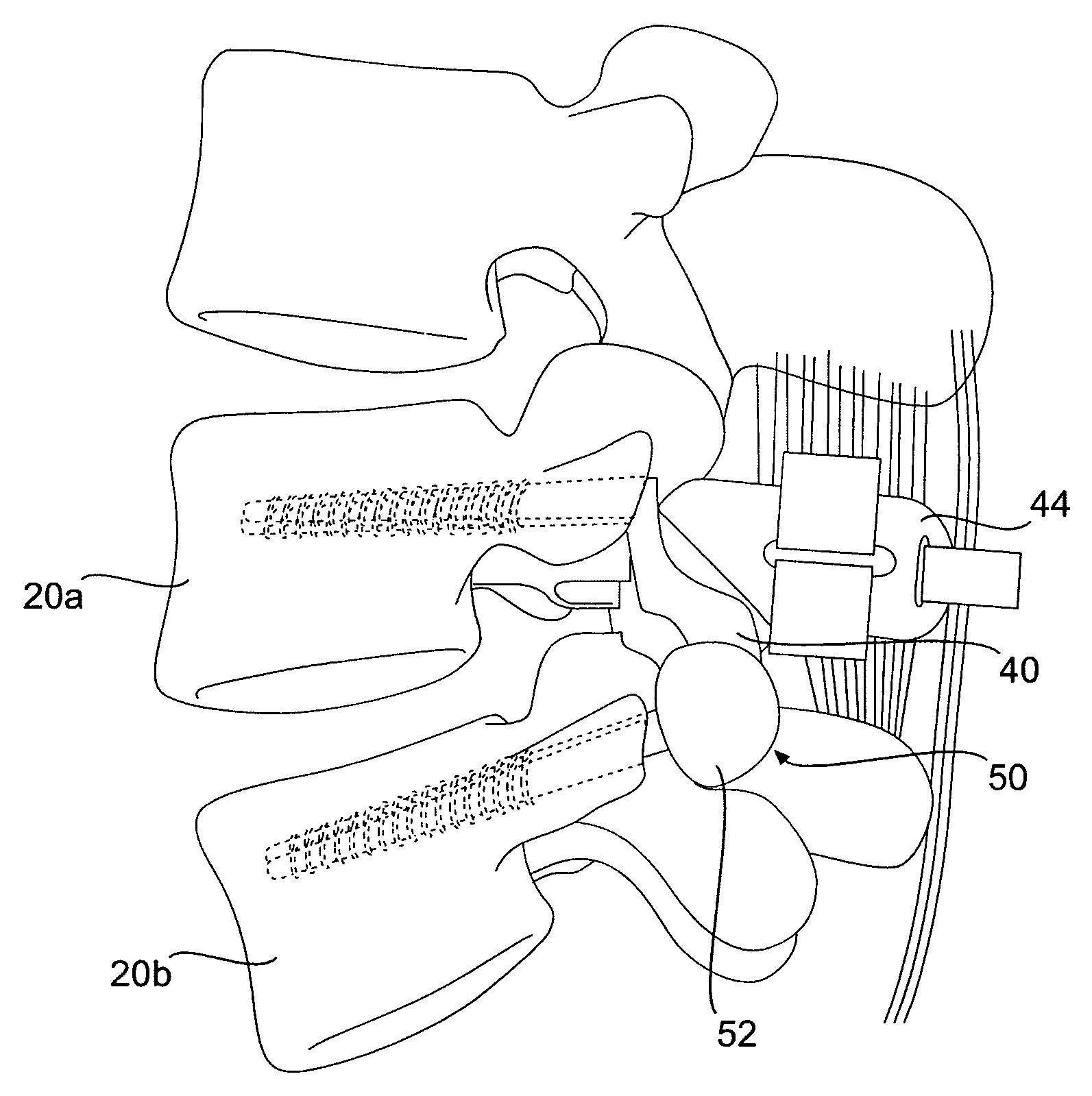
Prosthesis and Method for Replacing Degenerative Vertebral Portions
A posterior arch prosthesis according to the present invention includes a main body, two transverse protuberances extending transversely from either side of the main body, articulating surfaces formed on each of the transverse protuberances, and a posterior protuberance extending posteriorly from the main body. The posterior protuberance is attached to a first vertebra from which the natural posterior arch has been removed such that the articulating surfaces are arranged in articulating contact with surfaces on an adjacent, second vertebra. The posterior protuberance includes an attachment to which native structure may be reattached upon attachment of the prosthesis to the first vertebra.
Owner:WARSAW ORTHOPEDIC INC
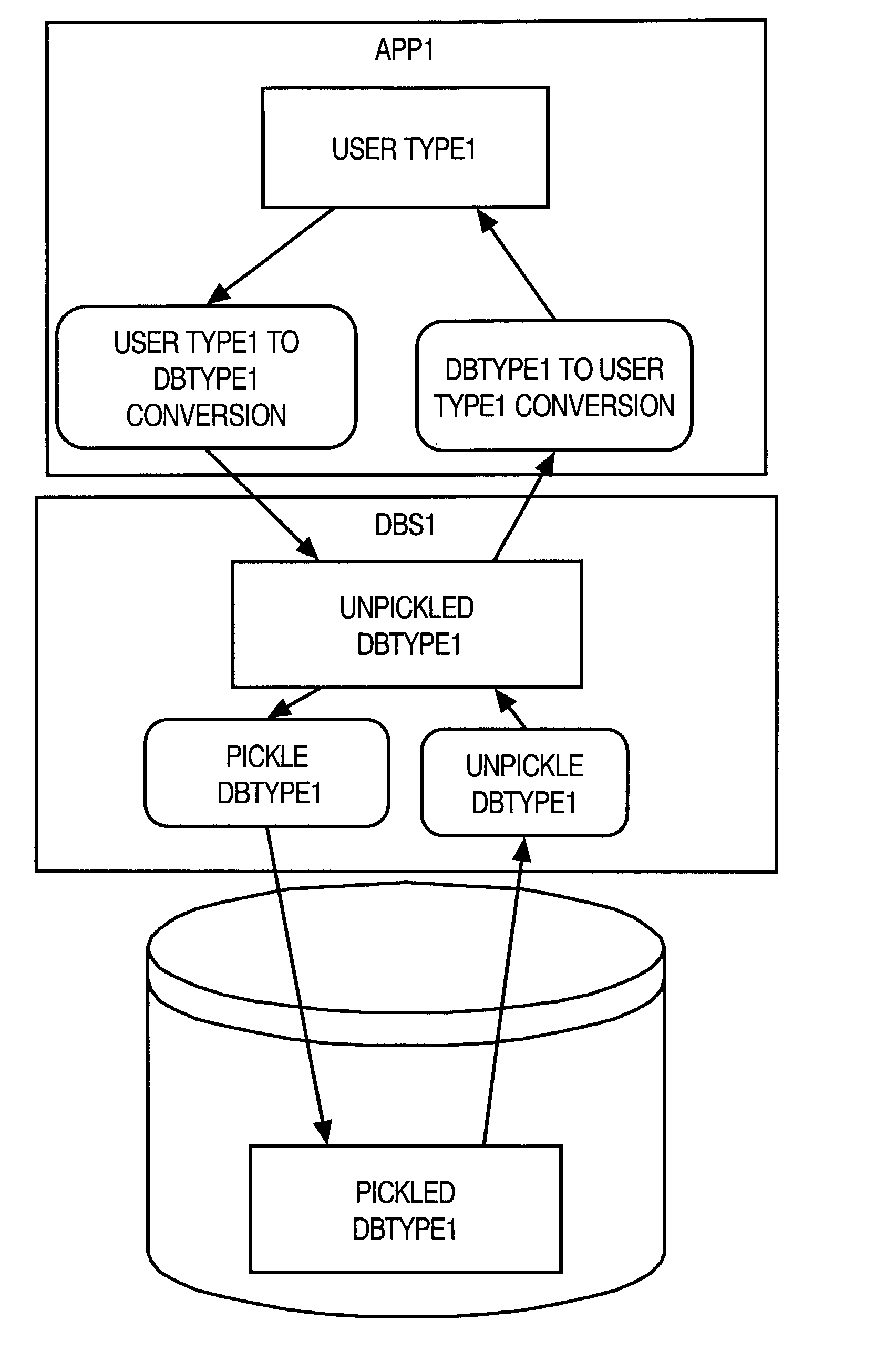
Opaque types
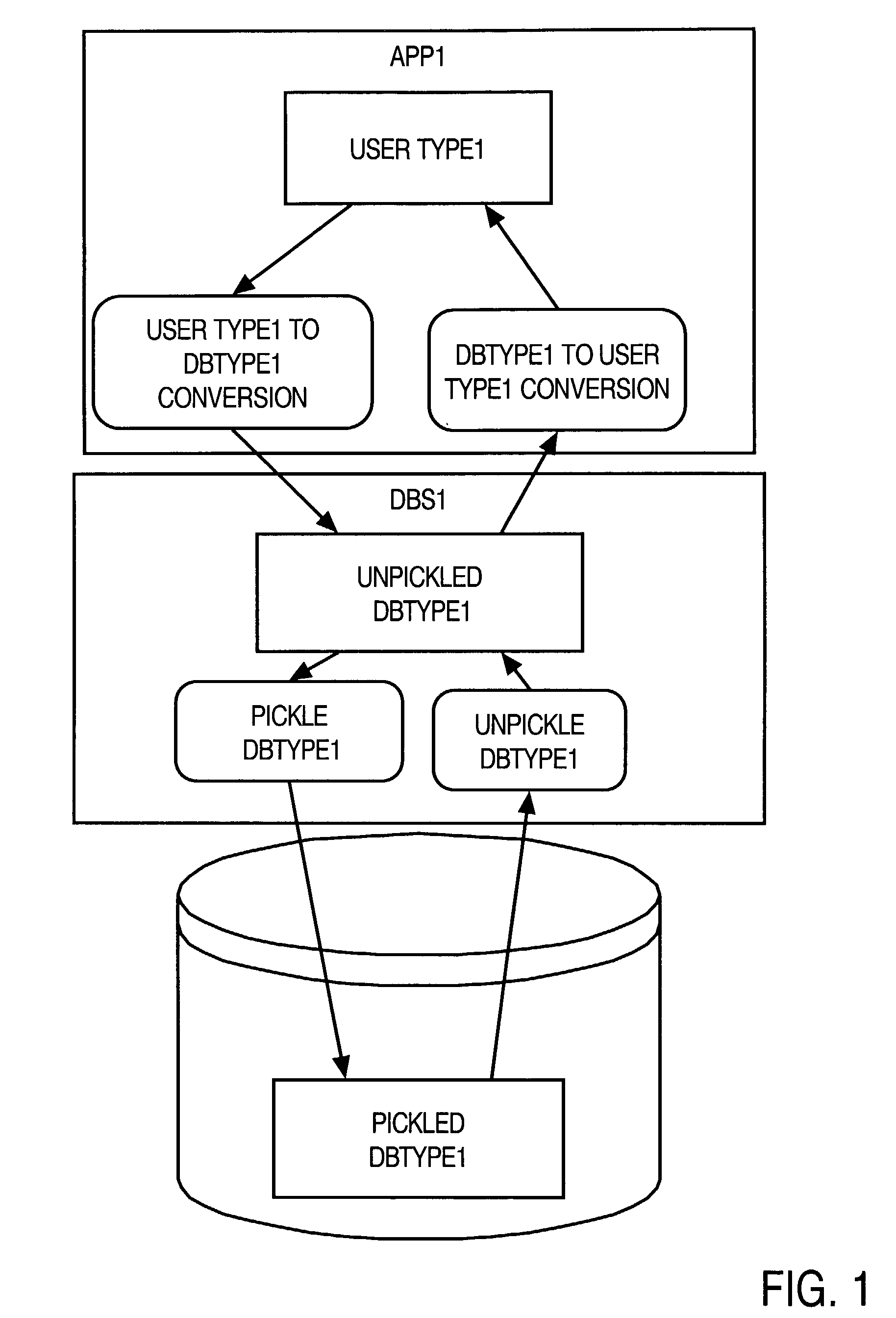
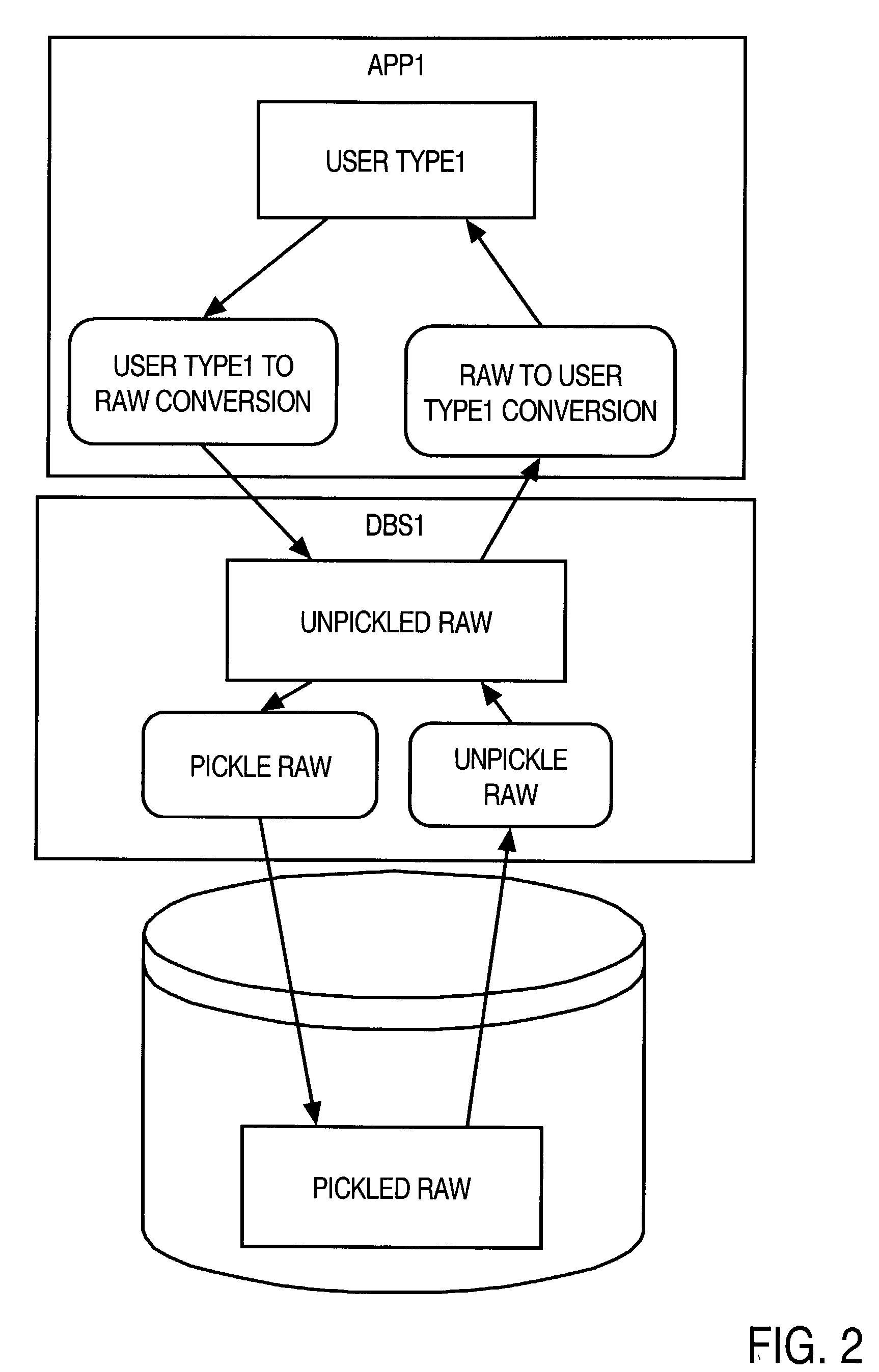
InactiveUS20020174128A1Data processing applicationsDigital data processing detailsNative structureSystem call
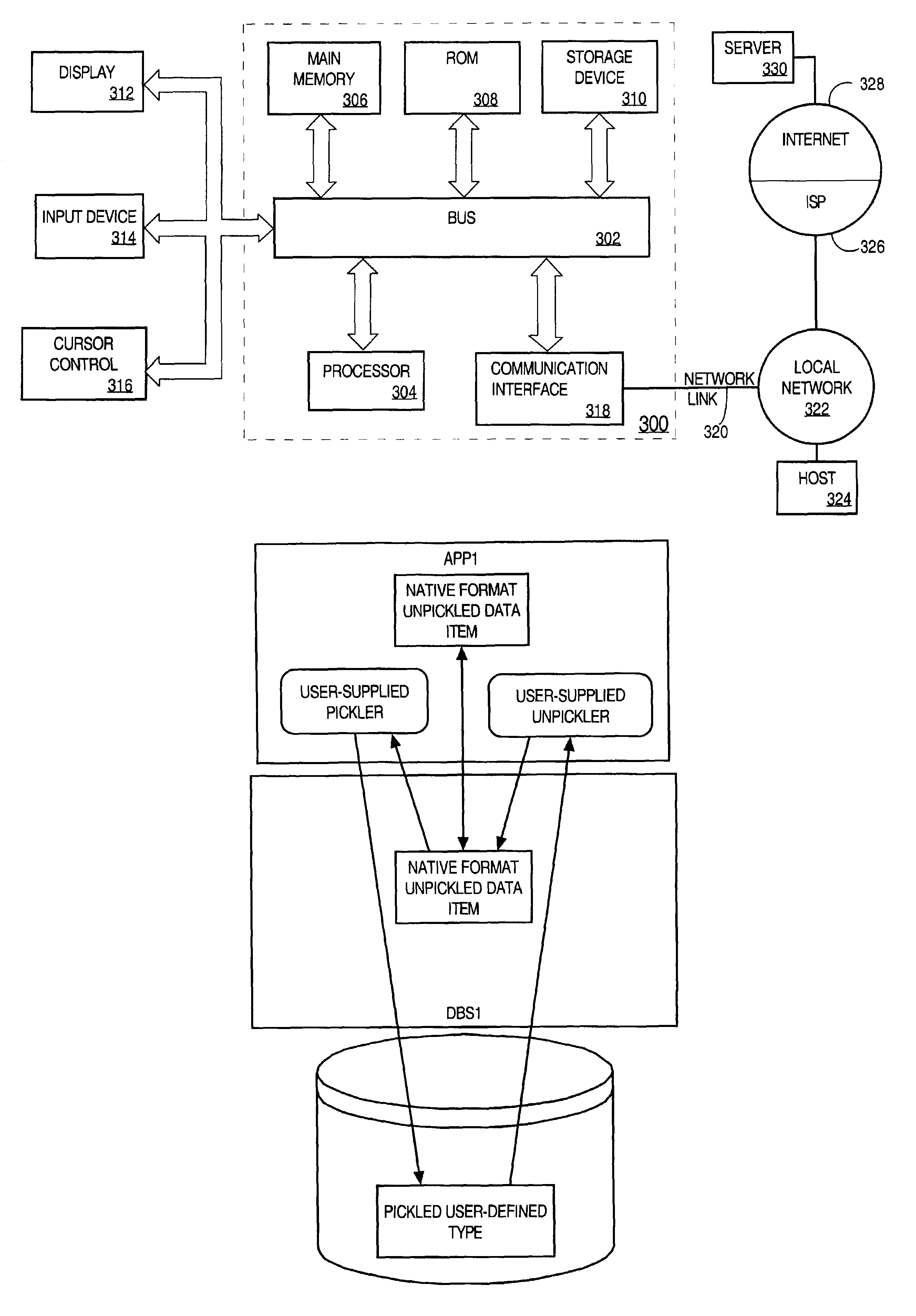
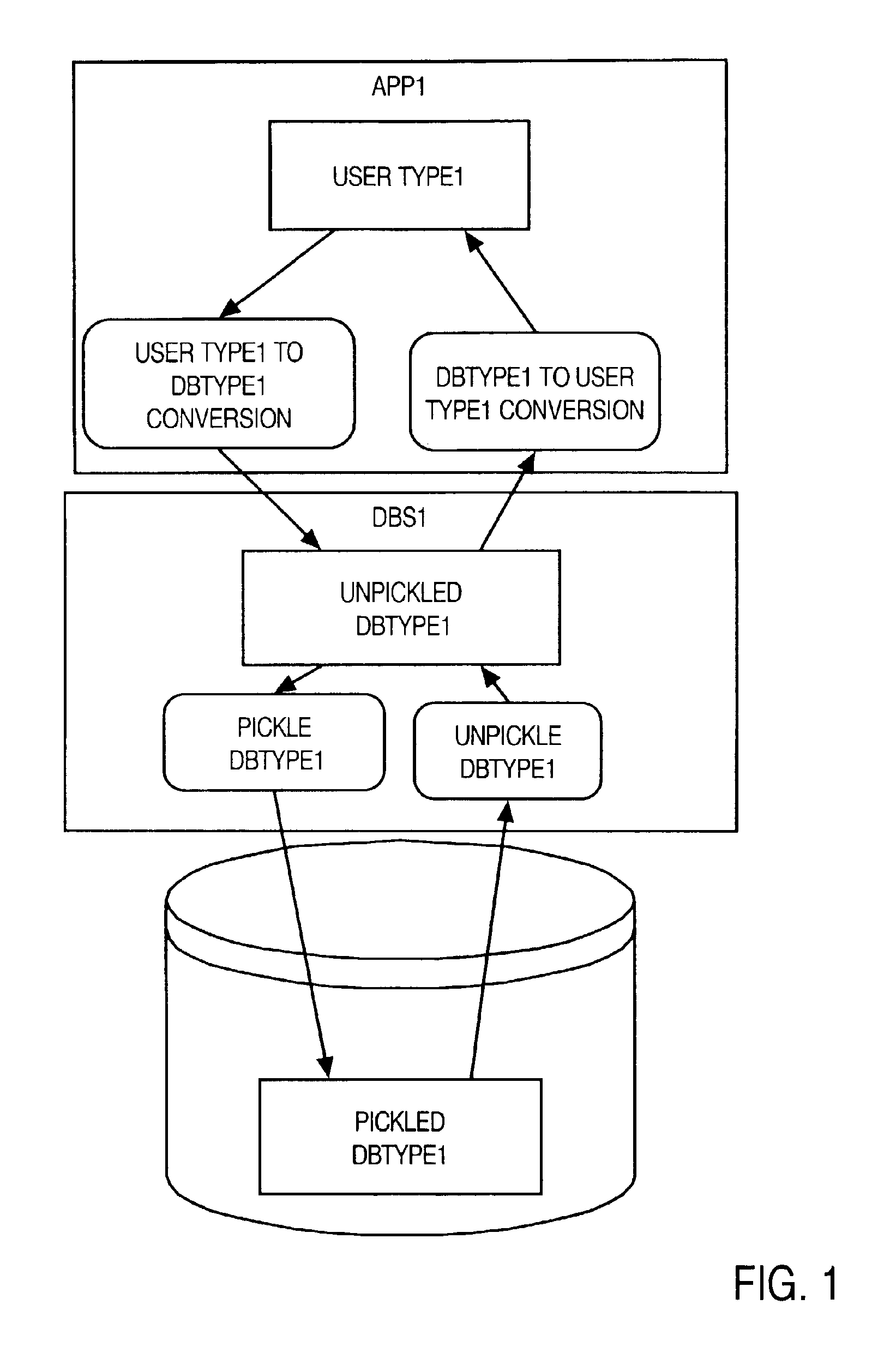
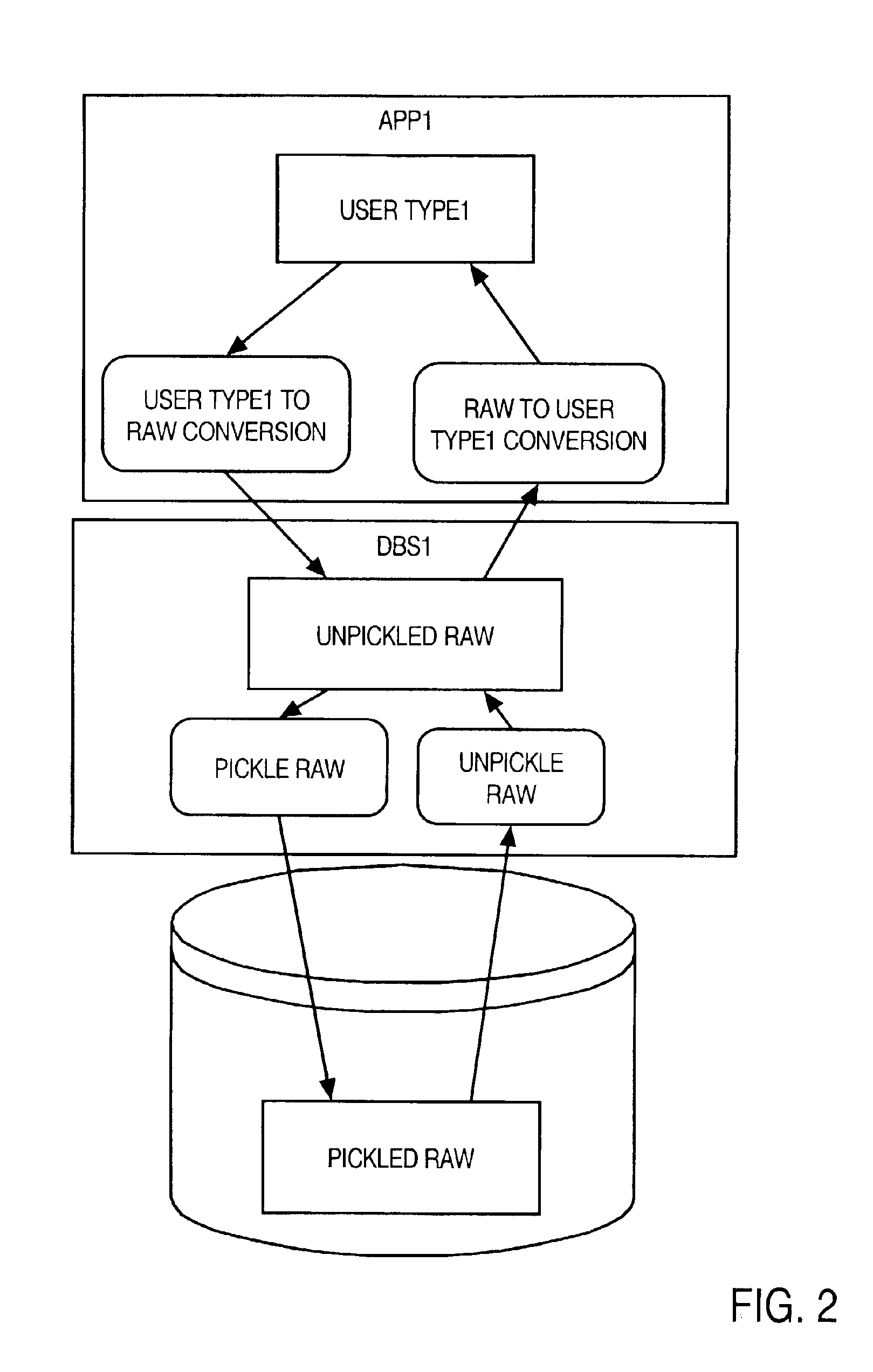
A method and apparatus are provided for handling within a database system data items that are associated with data types whose native structure is not known to the database system. The data items are stored within the database system in their native structure, even though it is not understood by the database system. To store the data items, the database system calls a pickling routine that is provided by the user, or by the runtime subsystem of the programming environment that is native to the data item. To retrieve the routine from storage, the database system calls an unpickling routine, also provided by the user or the appropriate runtime subsystem. Because the database maintains the data items in their native format, no conversions are required as the data items are passed between the database system and external routines that manipulate the data items.
Owner:ORACLE INT CORP
Systems, methods and apparatus for protein folding simulation
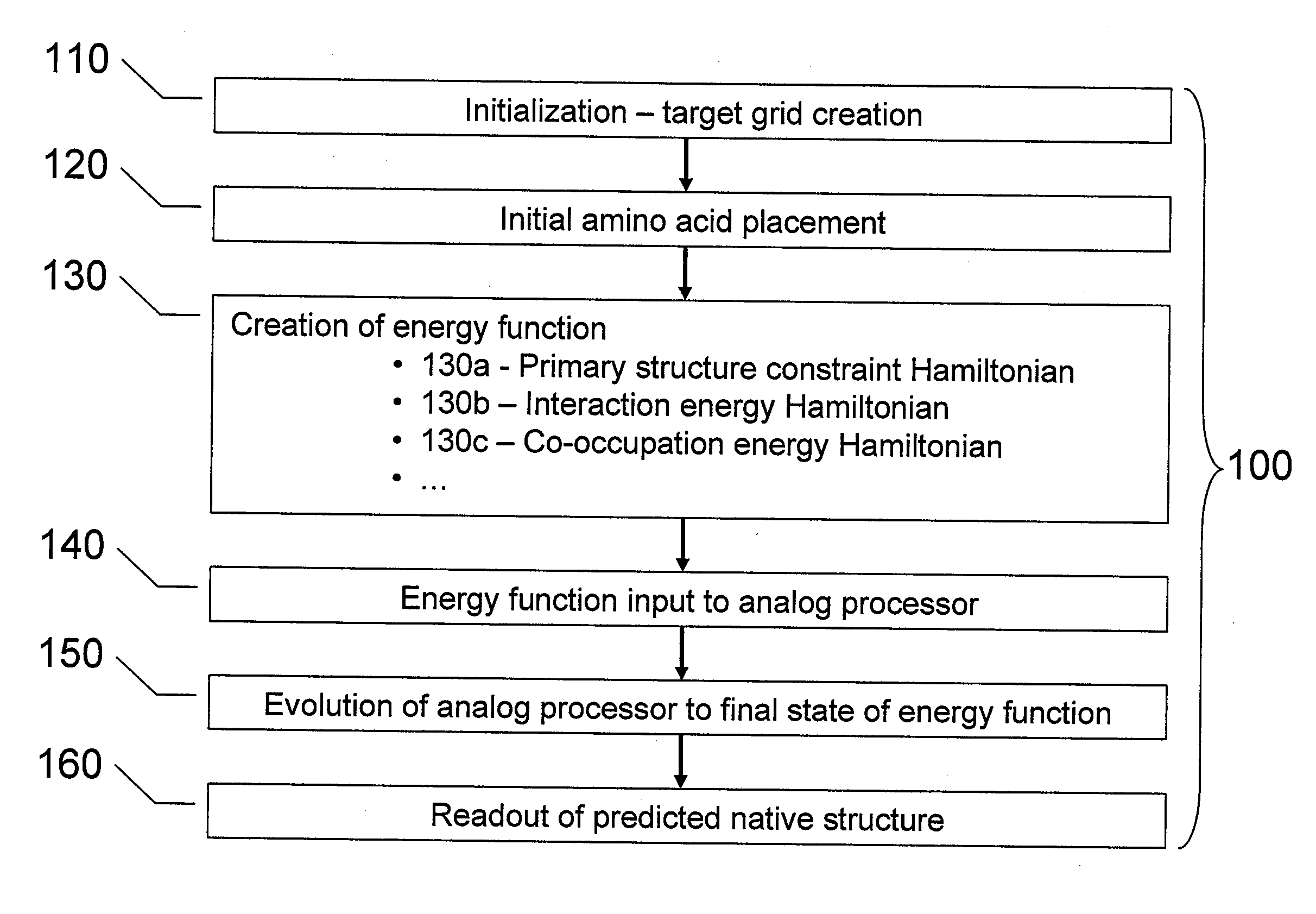
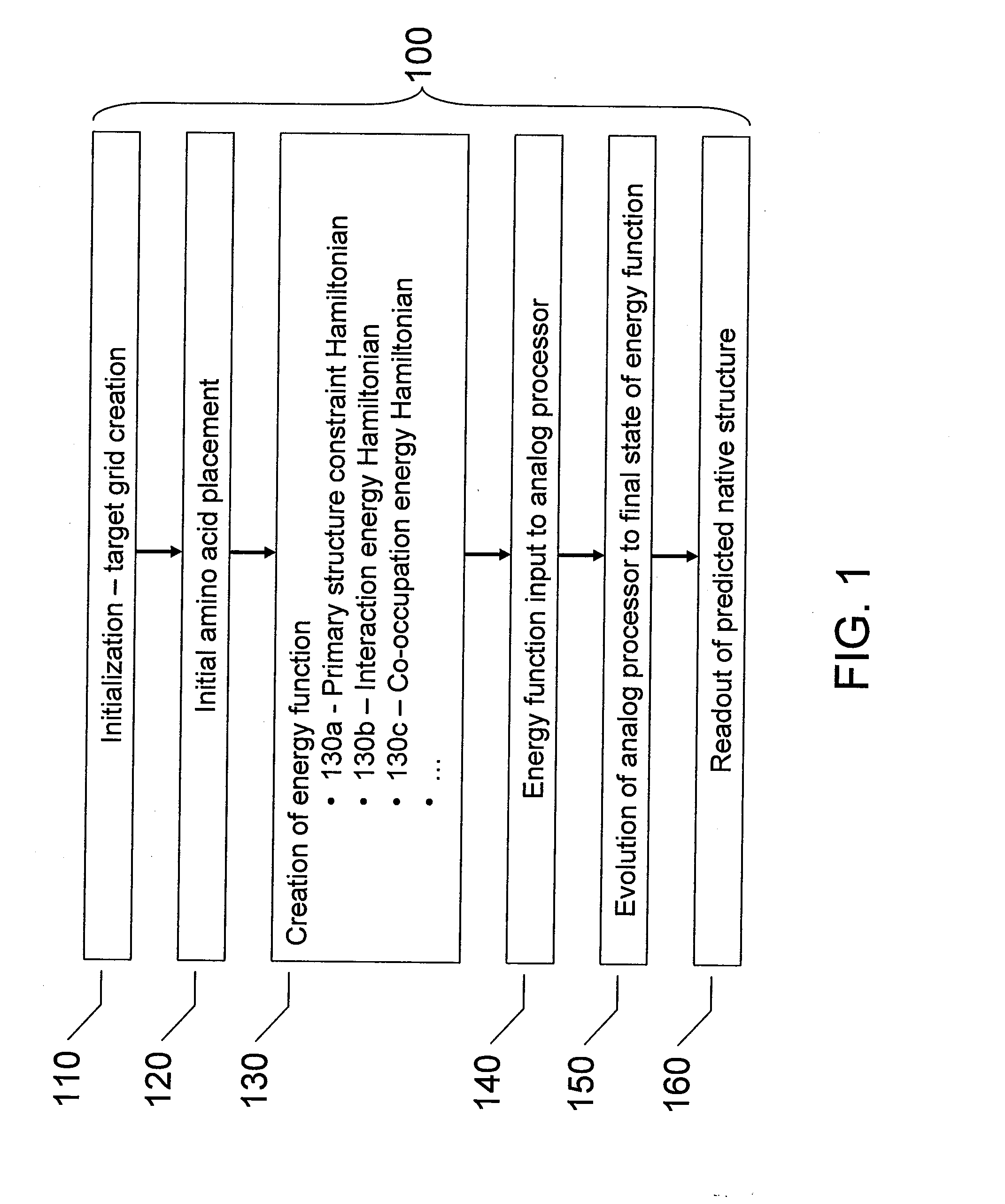
InactiveUS20080052055A1Improve legibilityAnalogue computers for chemical processesSpecial data processing applicationsNative structureAnalog processor
Analog processors such as quantum processors are employed to predict the native structures of proteins based on a primary structure of a protein. A target graph may be created of sufficient size to permit embedding of all possible native multi-dimensional topologies of the protein. At least one location in a target graph may be assigned to represent a respective amino acid forming the protein. An energy function is generated based assigned locations in the target graph. The energy function is mapped onto an analog processor, which is evolved from an initial state to a final state, the final state predicting a native structure of the protein.
Owner:D WAVE SYSTEMS INC
Prosthesis and method for replacing degenerative vertebral portions
A posterior arch prosthesis according to the present invention includes a main body, two transverse protuberances extending transversely from either side of the main body, articulating surfaces formed on each of the transverse protuberances, and a posterior protuberance extending posteriorly from the main body. The posterior protuberance is attached to a first vertebra from which the natural posterior arch has been removed such that the articulating surfaces are arranged in articulating contact with surfaces on an adjacent, second vertebra. The posterior protuberance includes an attachment to which native structure may be reattached upon attachment of the prosthesis to the first vertebra.
Owner:SDGI HLDG +1
Novel monoclonal antibody and use of the same
The present invention provides an anti-PAC1 monoclonal antibody capable of recognizing a PAC1 having a native structure, a PAC1 activity regulator (in particular, activity inhibitor) containing the antibody, a prophylactic / therapeutic agent for a disease associated with accentuation of a bioactivity of PAC1, containing the antibody, a diagnostic reagent for a disease associated with an abnormality of PAC1 activity, containing the antibody, and a screening method for a substance that regulates the expression of PAC1, using the antibody and a PAC1-expressing cell.
Owner:KYOTO UNIV +1
Multivalent Heteromultimer Scaffold Design and Constructs
ActiveUS20140066378A1Function increaseImprove stabilityBacteriaPeptide/protein ingredientsNative structureMolecular entity
Provided herein are multifunctional heteromultimer proteins. In specific embodiments is a heteromultimer comprising: at least two polypeptide constructs, each polypeptide construct comprising at least one cargo polypeptide attached to a transporter polypeptide, said transporter polypeptides derived from a monomeric native protein such that said monomeric constructs associate to form the heteromultimer and said transporter polypeptides associate to form a quasi-native structure of the monomeric native protein or analog thereof. These therapeutically novel molecules encompass heteromultimers comprising constructs that function as scaffolds for the conjugation or fusion of therapeutic molecular entities (cargo polypeptides) resulting in the creation of bispecific or multivalent molecular species. Provided herein is a method for creation of bispecific or multivalent molecular species.
Owner:ZYMEWORKS INC
Membrane scaffold proteins
InactiveUS7592008B2Increase stability and monodispersityImprove purification effectPeptide/protein ingredientsPharmaceutical delivery mechanismNative structurePhospholipid
The membrane scaffold proteins (MSP) of the present invention assemble with hydrophobic or partially hydrophobic proteins to form soluble nanoscale particles which preserve native structure and function; they are improved over liposomes and detergent micelles, in terms of stability and preservation of biological activity and native conformation. In the presence of phospholipid, MSPs form nanoscopic phospholipid bilayer disks, with the MSP stabilizing the particle at the perimeter of the bilayer domain. The particle bilayer structure allows manipulation of incorporated proteins in solution or on solid supports, including for use with such surface-sensitive techniques as scanning probe microscopy or surface plasmon resonance. The nanoscale particles, which are robust in terms of integrity and maintenance of biological activity of incorporated proteins, facilitate pharmaceutical and biological research, structure / function correlations, structure determinations, bioseparations, and drug discovery.
Owner:BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS THE A BODY & POLITIC OF THE STATE OF ILLINOIS
CRFR2 selective ligands
InactiveUS7141546B1Valid choicePharmaceutical formulations are facilitatedCompound screeningApoptosis detectionPeptide analogNative structure
CRF peptide analogs that bind to CRFR2 with an affinity far greater than they bind to CRFR1. These analogs exhibit CRF antagonist activity, and they can be based upon the native structures of sauvagine, CRF, and urocortin.
Owner:SALK INST FOR BIOLOGICAL STUDIES
Universal procedure for refolding recombinant proteins
A universal folding method that has been demonstrated to be effective in refolding a variety of very different proteins expressed in bacteria as inclusion bodies has been developed. Representative proteins that can be dissolved and refolded in biologically active form, with the native structure, are shown in Table I. The method has two key steps to unfold and then refold the proteins expressed in the inclusion bodies. The first step is to raise the pH of the protein solution in the presence of denaturing agents to pH greater than 9, preferably 10. The protein solution may be maintained at the elevated pH for a period of up to about 24 hours, or the pH immediately decreased slowly, in increments of about 0.2 pH units / 24 hours, until the solution reaches a pH of about 8.0, or both steps used. In the preferred embodiment, purified inclusion bodies are dissolved in 8 M urea, 0.1 M Tris, 1 mM glycine, 1 mM EDTA, 10 mM beta-mercaptoethanol, 10 mM dithiothreitol (DTT), 1 mM reduced glutathione (GSH), 0.1 mM oxidized glutathione (GSSG), pH 10. The absorbance at 280 nm (OD280) of the protein solution is 5.0. This solution is rapidly diluted into 20 volumes of 20 mM Tris base. The resulting solution is adjusted to pH 9.0 with 1 M HCl and is kept at 4° C. for 24 hr. The pH is adjusted to pH 8.8 and the solution is kept at 4° C. for another 24 hrs. This process is repeated until the pH is adjusted to 8.0. After 24 hr at pH 8.0, the refolded proteins can be concentrated by ultrafiltration and applied to a gel filtration column for purification.
Owner:OKLAHOMA MEDICAL RES FOUND
Membrane scaffold proteins
Membrane proteins are difficult to express in recombinant form, purify, and characterize, at least in part due to their hydrophobic or partially hydrophobic properties. Membrane scaffold proteins (MSP) assemble with target membrane or other hydrophobic or partially hydrophobic proteins or membrane fragments to form soluble nanoscale particles which preserve their native structure and function; they are improved over liposomes and detergent micelles. In the presence of phospholipid, MSPs form nanoscopic phospholipid bilayer disks, with the MSP stabilizing the particle at the perimeter of the bilayer domain. The particle bilayer structure allows manipulation of incorporated proteins in solution or on solid supports, including for use with such surface-sensitive techniques as scanning probe microscopy or surface plasmon resonance. The nanoscale particles facilitate pharmaceutical and biological research, structure / function correlation, structure determination, bioseparation, and drug discovery.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
Compositions and methods for proteomic investigations
Abstract of the Disclosure The present invention provides a variety of related proteomics analytical modalities that are open-ended, rapid, convenient and suitable for implementation in a high throughput parallel assay system. Specificity-determining compositions and methods are disclosed for use in proteomics. These compositions and methods provide a protein resolved from other protein species contained in a sample fluid, in its native, biologically functional conformation. The present invention provides a specificity-determining substrate that forms a complex with a protein molecule in a homogenous fashion. The specificity-determining substrate includes a specificity-determining ligand bound to a support, wherein optionally the substrate further includes a spacer bound between the ligand and the support. In addition a complex is provided that includes a specificity-determining substrate and a protein molecule. Furthermore, an array including a plurality of loci is provided, in which each locus includes a specificity-determining substrate of the invention. These substrates, complexes and arrays may be employed in a method of resolving a first protein from a fluid including one or more species of native, biologically active protein molecules, wherein the first protein retains its native structure and its biological activity; in a method of purifying one or more first proteins from a fluid including one or more species of native, biologically active protein molecules, wherein the purified first protein retains its native structure and its biological activity; in a method of characterizing one or more proteins in a fluid including one or more species of protein molecule; and in a method of identifying one or more proteins in a sample fluid wherein the concentration of the one or more proteins in the sample fluid differs from the concentration of the one or more proteins in a reference fluid.
Owner:ROY SWAPAN +2
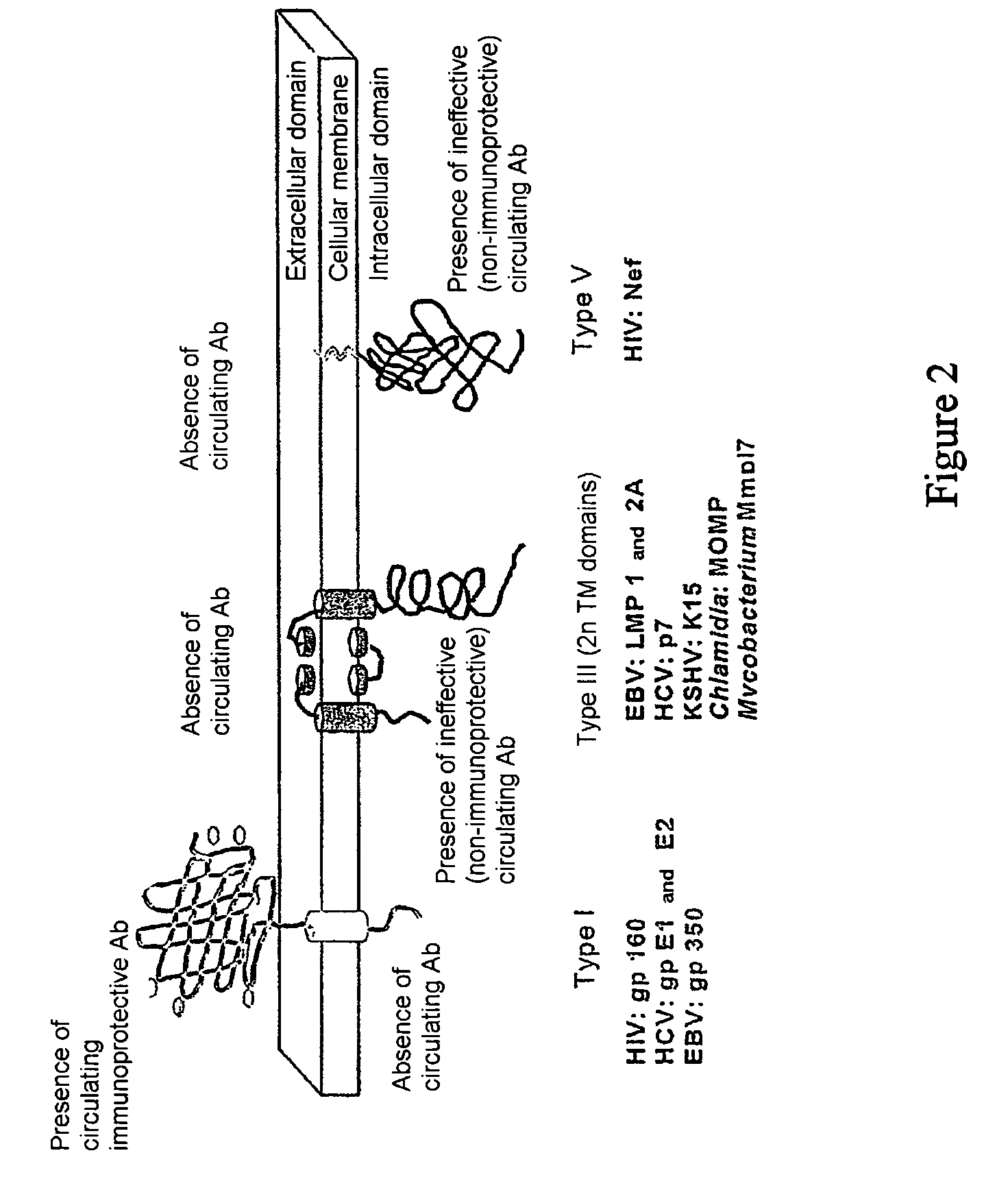
Antigen imitating extracellular areas of membrane proteins of type III produced from intracellular pathogenic micro-organisms, derived conformational antibodies and the use thereof
An antigen derived from an intracellular pathogenic micro-organism characterized in that it comprises at least on peptidic fragment which essentially consists of the concatenation of sequences of at least two extracellular adjacent areas in the native structure of a membrane protein of type III of said intracellular pathogenic micro-organism, derived conformational antibodies and the application hereof.
Owner:CENT NAT DE LA RECHERCHE SCI +2
Dihedral angle information auxiliary energy function selection based protein structure prediction method
ActiveCN109448784AReduce the impactEnhance local structureInstrumentsMolecular structuresNative structureEnergy functional
The invention discloses a dihedral angle information auxiliary energy function selection based protein structure prediction method. extracting dihedral angle information according to a Ramachandran plot corresponding to a protein residue; performing assessment on conformation by using the energy functions and the dihedral angle information in a Rosetta algorithm; giving different weights to two fractions, designing a new scoring function, and performing selection on the fragment assembled conformation by using the scoring function so that the influence on the three-dimensional structures of proteins due to inexact energy functions can be reduced; and performing global search and local search on the conformation, and performing enhancement on local structures on the basis of guaranteeing the global topological structure of the constellation so that more near-native structures can be obtained. Thus, the dihedral angle information auxiliary energy function selection based protein structure prediction method with good sampling capabilities and high prediction precision can be provided.
Owner:深圳新锐基因科技有限公司
Novel proteome analysis method and devices therefor
InactiveCN1496481AAvoid difficultiesMaintain structureMaterial analysis by observing effect on chemical indicatorLibrary screeningNative structureMembrane protein interactions
The present invention provides a proteome analysis method including grouping a proteome into membrane proteins and compounds capable of interacting with the membrane proteins, while retaining their native structure and function, and analyzing both the membrane proteins and the compounds based on biological affinity, and devices therefor.
Owner:PROTOSERA
Cellular And Viral Inactivation
The invention involves inactivation of viral populations by treating the viral populations with a compound to crosslink proteins in the viral membrane, UV irradiation and further inactivation of the viruses using detergent(s). According to the invention, this method preserves the native structure of viral epitopes so that the inactivated viral preparations can be used in immunological compositions that will inhibit and / or prevent viral infection when administered to an animal.
Owner:UNITED STATES OF AMERICA
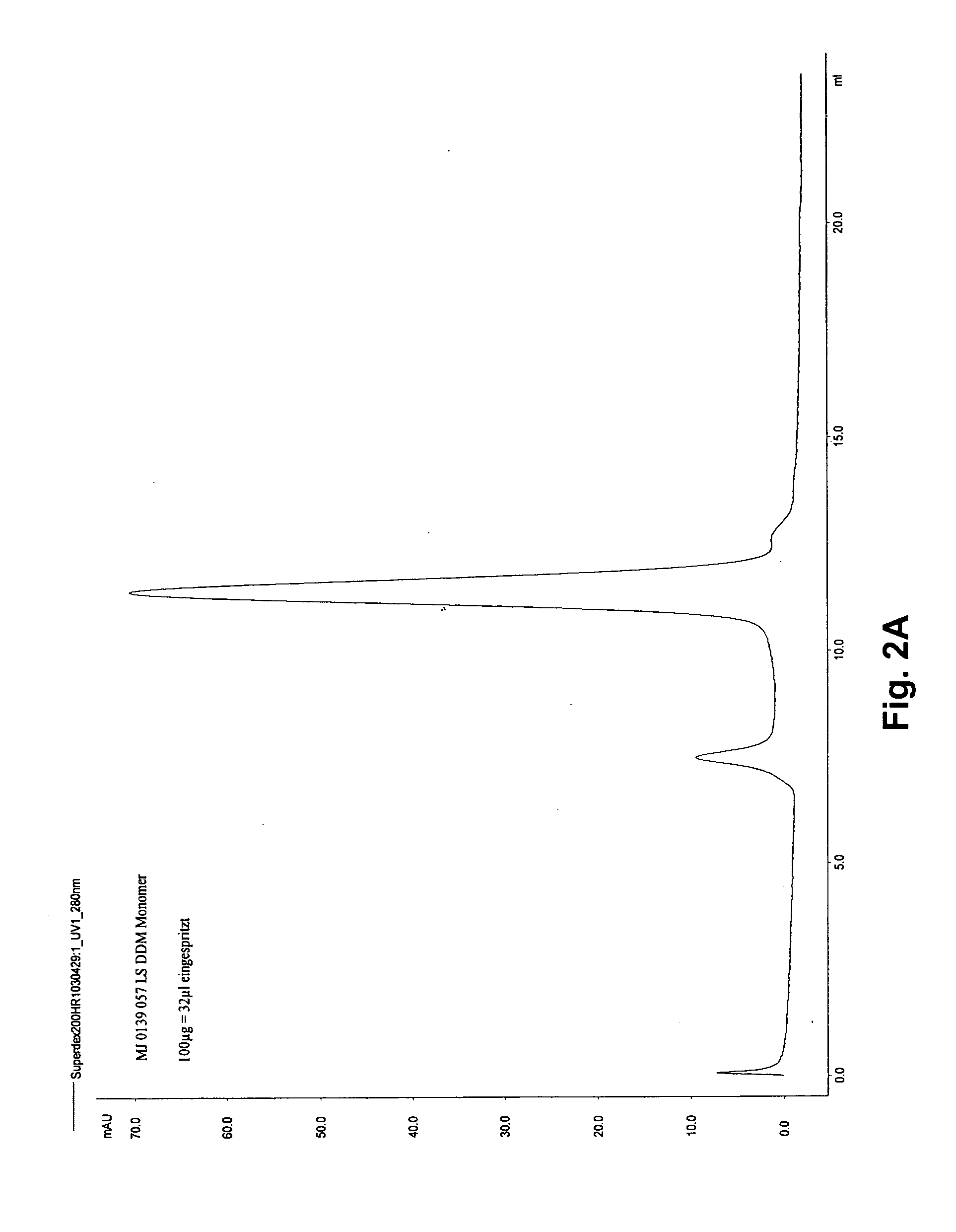
Refolding of membrane proteins
InactiveUS6939951B1High protein yieldCell receptors/surface-antigens/surface-determinantsDepsipeptidesNative structureCell Membrane Proteins
In a method for production of membrane proteins or receptors folded into their native structure, first, proteins solubilized in a first detergent are provided. To induce folding of proteins into their native form, the first detergent is exchanged for a second detergent. Both for the first and for the second detergent, examples are shown.
Owner:M PHASYS
Membrane scaffold proteins
InactiveUS20060057662A1Great stability and size homogeneity and useful functionalityEasy to understandFungiBacteriaNative structurePhospholipid
Membrane proteins are difficult to express in recombinant form, purify, and characterize, at least in part due to their hydrophobic or partially hydrophobic properties. Membrane scaffold proteins (MSP) assemble with target membrane or other hydrophobic or partially hydrophobic proteins or membrane fragments to form soluble nanoscale particles which preserve their native structure and function; they are improved over liposomes and detergent micelles. In the presence of phospholipid, MSPs form nanoscopic phospholipid bilayer disks, with the MSP stabilizing the particle at the perimeter of the bilayer domain. The particle bilayer structure allows manipulation of incorporated proteins in solution or on solid supports, including for use with such surface-sensitive techniques as scanning probe microscopy or surface plasmon resonance. The nanoscale particles facilitate pharmaceutical and biological research, structure / function correlation, structure determination, bioseparation, and drug discovery.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS
Opaque types
InactiveUS7007037B2Reduce the burden onReduce eliminateData processing applicationsDigital data processing detailsNative structureSystem call
A method and apparatus are provided for handling within a database system data items that are associated with data types whose native structure is not known to the database system. The data items are stored within the database system in their native structure, even though it is not understood by the database system. To store the data items, the database system calls a pickling routine that is provided by the user, or by the runtime subsystem of the programming environment that is native to the data item. To retrieve the routine from storage, the database system calls an unpickling routine, also provided by the user or the appropriate runtime subsystem. Because the database maintains the data items in their native format, no conversions are required as the data items are passed between the database system and external routines that manipulate the data items.
Owner:ORACLE INT CORP
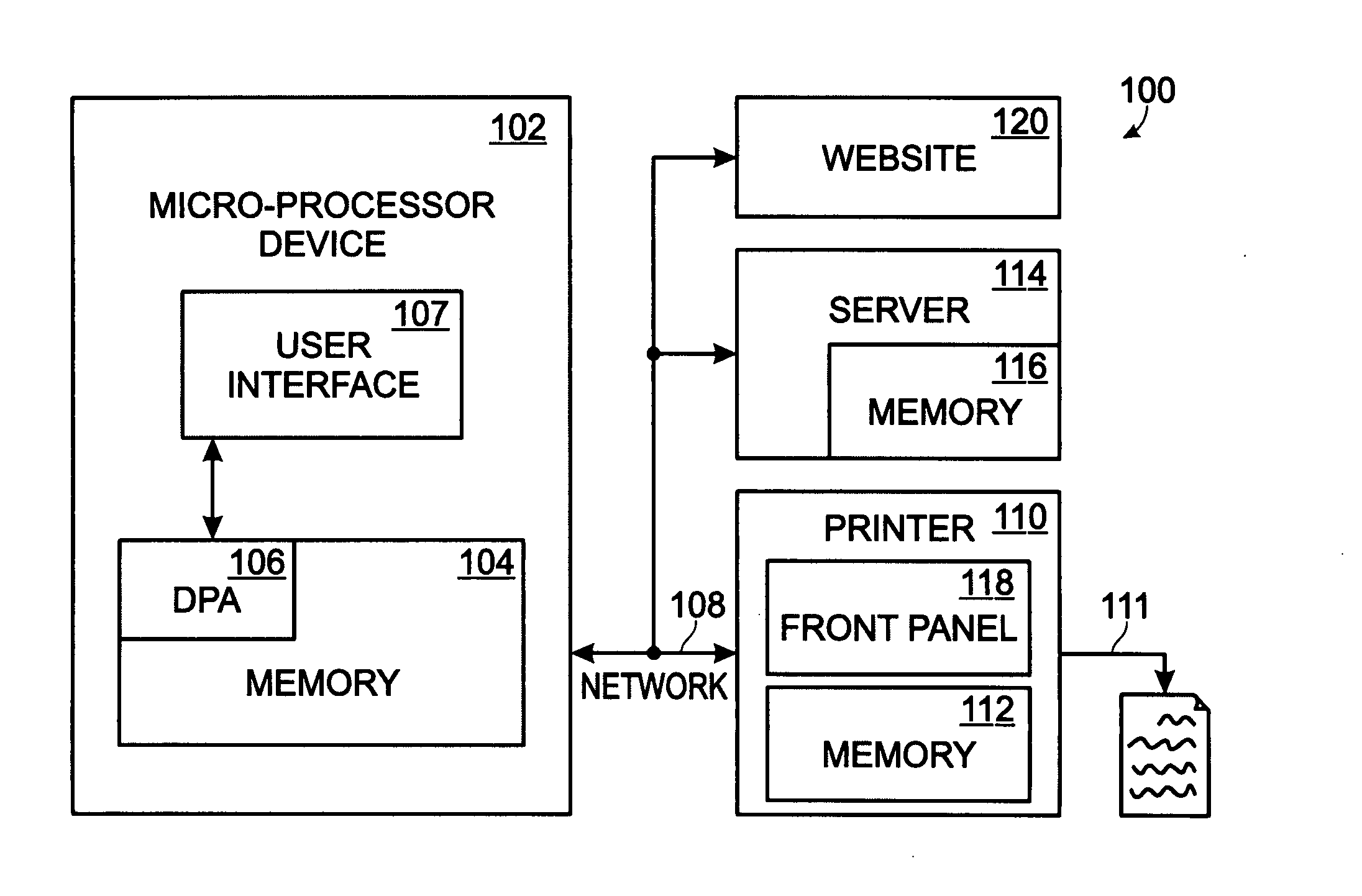
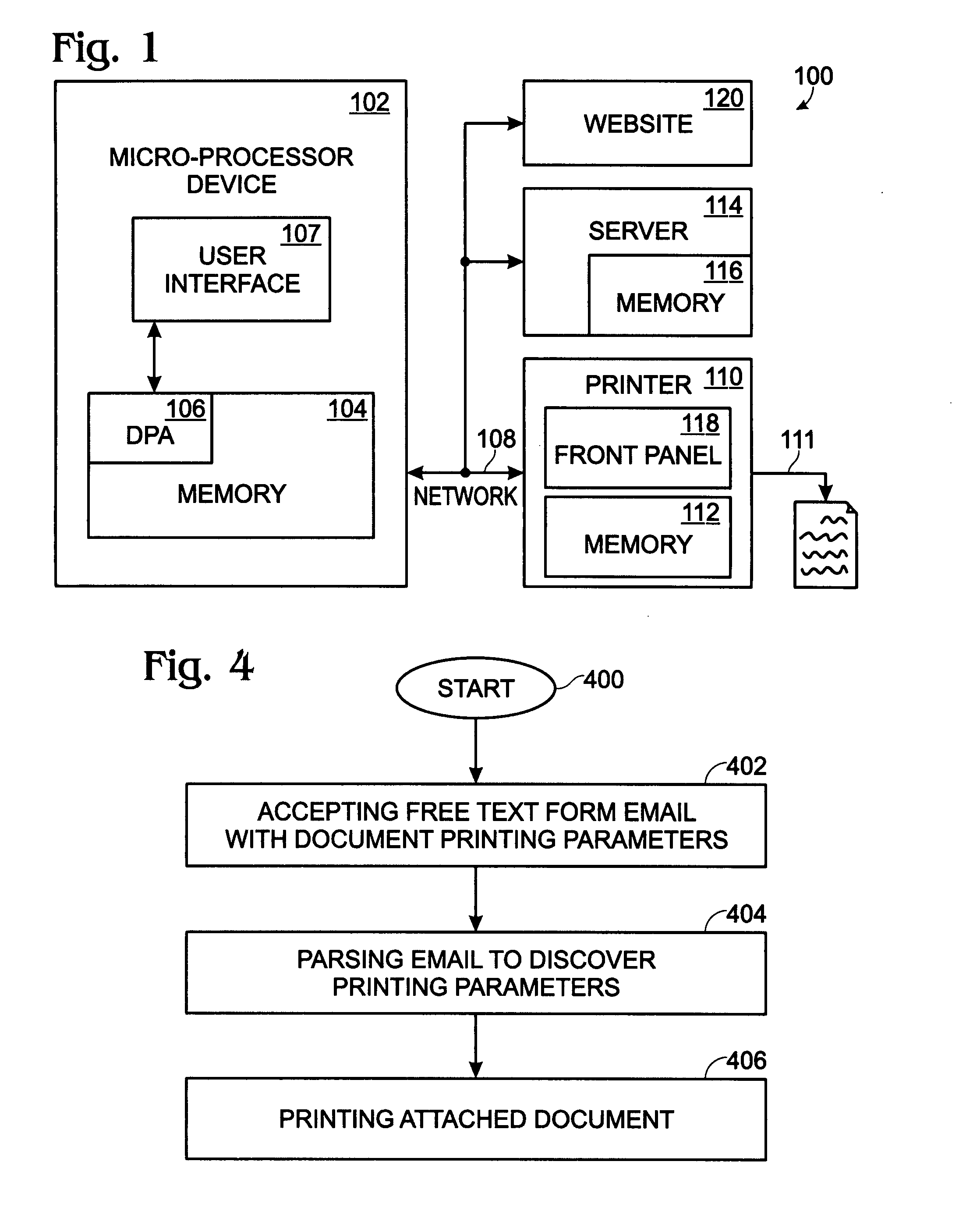
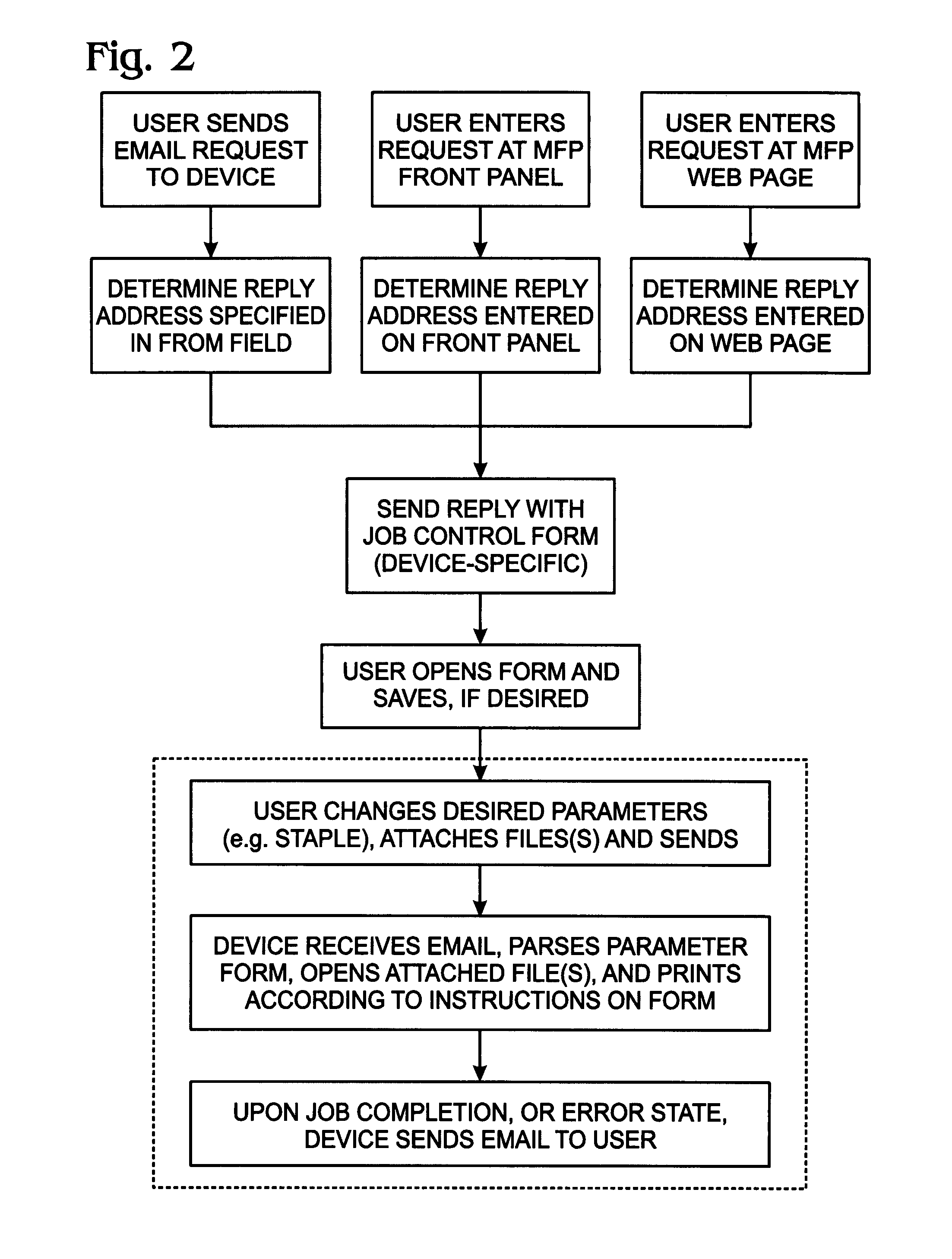
System and method for delivering native structure document printing instructions
ActiveUS8014006B2Multiple digital computer combinationsData switching networksEmail addressNative structure
A system and method are provided for delivering document print instructions in a system of network-connected devices. The method comprises: accepting printer control data at a network-connected email address; generating the printer-controller form in response to the printer control data; calling the printer-controller form; populating printer-controller form parameters; attaching a document to the printer-controller form; emailing the printer-controller form, with attachment, to a network-connected printer; and, printing the attached document in response to the printer-controller form parameters. Printing the attachment in response to the printer-controller form parameters includes the printer: opening the emailed printer-controller form; parsing the printer-controller form parameters; and, printing in response to the parsed parameters. In some aspects of the method, the printer-controller form is saved at a network-connected server, and calling a printer-controller form includes accessing the form from the server. Alternately, the printer-controller form is generated by the printer and delivered, via email.
Owner:SHARP KK
Ice breaker and snow blower
InactiveCN102268862AReduce operating temperatureImprove uniformitySnow cleaningNative structureCooling effect
The ice-breaking and snow-cleaning machine belongs to urban environmental protection machinery; on the lower side of the frame assembly, between the ice-breaking shaft assembly and the ice and snow scraper, a natural cooling heat dissipation pipe is equipped, and the electric air-cooled radiator is installed on the frame assembly. On the upper side, the oil pipe connects the distributor with the electric air-cooled radiator, the electric air-cooled radiator and the hydraulic oil tank in turn, and the oil pipe connects the hydraulic motor with the natural cooling heat dissipation pipe, and the natural cooling heat dissipation pipe with the distributor in turn; The structure design is reasonable, the cooling effect of hydraulic oil is good, the use is reliable, the failure is less, the adjustment is simple and convenient, the operation efficiency is high, the quality is good, and the service life is long.
Owner:王发祥
Refolding of membrane proteins
InactiveUS20060058509A1High protein yieldHigh yieldCell receptors/surface-antigens/surface-determinantsPeptide preparation methodsNative structureCell Membrane Proteins
In a method for production of membrane receptors folded into their native structure, first, receptors solubilized in a first detergent are provided. To induce folding of receptors into their native form, the first detergent is exchanged for a second detergent. Both for the first and for the second detergent, examples are shown.
Owner:M PHASYS
Method for producing a membrane protein
ActiveUS20100291189A1Improve hydrophobicityPromote formationAntibacterial agentsPeptide/protein ingredientsNative structureNucleotide
A method is provided for producing a membrane protein folded to its native structure or active structure in a lipid disc or a liposome. The method comprises: (a) preparing a reaction solution for cell-free protein synthesis containing a polynucleotide encoding a membrane protein, a steroidal detergent, and a phospholipid, wherein the steroidal detergent is contained at a concentration higher than its critical micelle concentration; (b) decreasing the concentration of said steroidal detergent in the reaction solution; and (c) synthesizing the membrane protein simultaneously with formation of a lipid disk or liposome into which the synthesized membrane protein is integrated.
Owner:RIKEN
Coal and gas outburst simulation test method
Belonging to the field of coal-mine gas disaster control, the invention puts forward an outburst simulation test method. Heating and pressurization are carried out on lignite and other low metamorphic non-outburst test coal samples in an outburst simulation test device to let the substances deform, break and metamorphose to generate gas so as to form outburst coal with outburst potential, and then coal and gas outburst simulation test study can be carried out. The specific test steps include: A. collecting lignite and other low metamorphic non-outburst coal samples on the spot, conducting laboratory vacuum drying and degassing for 48h, on the basis of maintaining a coal sample primary structure, preparing a coal sample according to the size of the test device; B. under an airtight condition, carrying out temperature (20-390DEG C), pressure (a confining pressure of 20-220MPa), time (1h-120d), moisture (moisture percentage) and other single parameter and multi-parameter coupled outburst potential inoculation evolution simulation test; and C. using a quick opening device to open an outburst cover suddenly, and simulating a dynamic emergence process of coal and gas outburst.
Owner:HENAN POLYTECHNIC UNIV
Peptides and recombinant proteins mimicking interferons
InactiveUS20060165654A1AvoidingUndesirable side-effectPeptide/protein ingredientsAntibody mimetics/scaffoldsNative structureSide effect
The present invention provides peptide and recombinant protein mimetics specifically representing the antiviral and antiproliferative activities of interferons with avoiding interferons' undesirable side effects during treatment of a variety of diseases. The basic embodiment of the present invention was to link together, through a short artificial spacer, two primary protein sequences locating distant to each other in the interferon's native structure.
Owner:ZAVYALOV VLADIMIR +5
Refolding of ion channel proteins
InactiveUS20050255556A1Avoid disadvantagesCell receptors/surface-antigens/surface-determinantsSugar derivativesIon Channel ProteinNative structure
A method of ion channels folded into their active structure, or functional subunits thereof, is described. For this purpose, initially expressed subunits, which are solubilized and denatured in a first detergent, of an ion channel are provided, and subsequently the first detergent is replaced by a second detergent which induces folding of the subunits of an ion channel into their native structure. The subunits of the ion channel are then assembled into its active structure.
Owner:M PHASYS
A simulation test method for coal and gas outburst
Belonging to the field of coal-mine gas disaster control, the invention puts forward an outburst simulation test method. Heating and pressurization are carried out on lignite and other low metamorphic non-outburst test coal samples in an outburst simulation test device to let the substances deform, break and metamorphose to generate gas so as to form outburst coal with outburst potential, and then coal and gas outburst simulation test study can be carried out. The specific test steps include: A. collecting lignite and other low metamorphic non-outburst coal samples on the spot, conducting laboratory vacuum drying and degassing for 48h, on the basis of maintaining a coal sample primary structure, preparing a coal sample according to the size of the test device; B. under an airtight condition, carrying out temperature (20-390DEG C), pressure (a confining pressure of 20-220MPa), time (1h-120d), moisture (moisture percentage) and other single parameter and multi-parameter coupled outburst potential inoculation evolution simulation test; and C. using a quick opening device to open an outburst cover suddenly, and simulating a dynamic emergence process of coal and gas outburst.
Owner:HENAN POLYTECHNIC UNIV
Acellular corneas, methods of producing the same and uses thereof
A method of producing an acellular cornea includes steps of subjecting a cornea of an animal to a decellularization process, and has not the step of treating the cornea with a protease, a chelating agent, a detergent, a glycerol, or a combination thereof. When a native cornea is processed by the method, the native structure and conformation of the native cornea are preserved while immunogenic matters are reduced to a level that the thus produced cornea may serve as a three-dimensional scaffold for host cells to grow thereon after transplantation.
Owner:ACRO BIOMEDICAL CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com