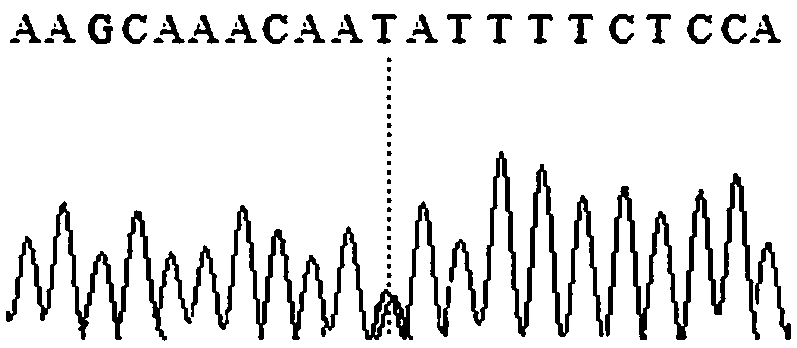
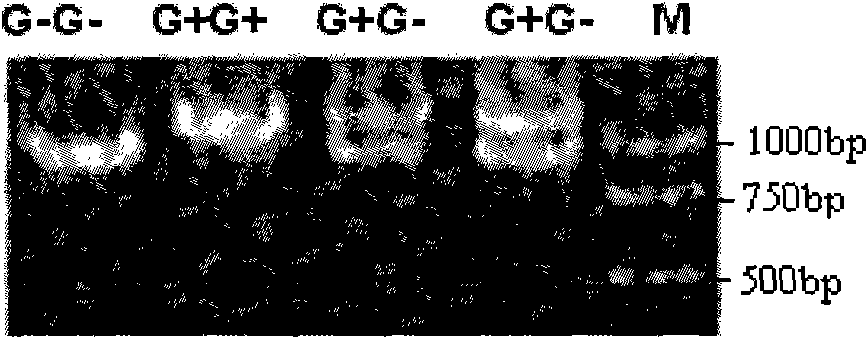Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
37results about How to "Achieve early selection" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Molecular marker related with diameter character of sheep wool fibers as well as specific primer and application thereof
ActiveCN107604078AHigh precisionSpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceFiber diameter
The invention relates to the technical field of animal molecular markers, in particular to a molecular marker related with a diameter character of sheep wool fibers as well as a specific primer and application thereof in selection of diameter character of Chinese Merino sheep wool fibers. The molecular marker related with the diameter character of the sheep wool fibers is located at the No.30 exonof the No.21 chromosome of FAT3 gene. The molecular marker has the advantages that the function of using rs425144268 SNP site as the molecular marker of the diameter of the sheep wool fibers is disclosed for the first time, and the function of applying the rs425144268 SNP site to the selection of the diameter character of the Chinese Merino sheep wool fibers is disclosed for the first time; whenthe excellent diameter character of the wool fibers is identified by the molecular marker related with the diameter character of the sheep wool fibers, the operation is simple, and the sheep with smaller fiber diameter can be screened in an assisting way; because the detection level of the molecular marker is started, the accuracy of variety selection is improved, and the detection efficiency is improved.
Owner:新疆畜牧科学院畜牧研究所 +1
Molecular marker capable of predicting and identifying sheep wool length and specific primer pair and application
ActiveCN107619870ASpeed up the genetic selection processAchieve early selectionMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceIFNAR2 Gene
The invention relates to the technical field of animal molecular markers, in particular to a molecular marker capable of predicting and identifying a sheep wool length and application of a specific primer pair in predicting and identifying wool length character of Chinese Merino lambs. The molecular marker capable of predicting and identifying the sheep wool length is located at a first exon of aNo. 1 chromosome of IFNAR2 genes. The invention firstly discloses an rs407032027 SNP locus serving as a molecular maker capable of predicting and identifying the sheep wool length, and firstly discloses the application of the rs407032027 SNP locus in the selection of the wool length character of the Chinese Merino lamb; and when the molecular marker capable of predicting and identifying the sheepwool length is adopted to select a sheep variety with excellent wool length character, the molecular marker has the advantages of simple operation, low expense, high accuracy and capability of automatic detection.
Owner:新疆畜牧科学院畜牧研究所 +1
Molecular marker and specific primers for assisting in test of wilt disease resistance in brassica oleracea and use thereof
InactiveCN102108407AAchieve early selectionMicrobiological testing/measurementAgricultural scienceSheath blight
The invention discloses a molecular marker and specific primers for assisting in test of wilt disease resistance in brassica oleracea and use thereof. The invention provides a reagent for assisting in the test of wilt disease resistance in brassica oleracea and / or assisting in screening brassica oleracea with wilt disease resistance, which is a specific primer pair formed by nucleotides represented by a sequence 2 and a sequence 3 in a sequence table. The invention also provides a specific gene fragment which is at a genetic distance about 2.87cM to the wilt disease resistance gene in brassica oleracea and is formed by nucleotides represented by a sequence 1 in a sequence table. The result of the identification of wilt disease resistance in brassica oleracea and / or screening of the brassica oleracea with wilt disease resistance with assistance from the reagent (primer pair) or sequence characterized amplified region (SCAR) marker, which are provided by the invention, is 97 percent consistent with that of field identification. When used in breeding, the reagent, molecular marker or method has the advantages of accuracy, quickness, capability of realizing early breeding and the like and has a bright application prospect.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES +1
Molecular identification method of peanut hybrid
ActiveCN105525012AAccurate identificationRapid identificationMicrobiological testing/measurementEnzyme digestionAgricultural science
The invention discloses a molecular identification method of a peanut hybrid. The method comprises the following steps: conducting reduced-representation genome sequencing on the male parent and female parent of the peanut hybrid, and analyzing a sequencing result so as to identify an SNP molecular marker between the male parent and the female parent and design a universal primer for the male parent and the female parent; and extracting the genome DNA of the peanut hybrid, and verifying whether the peanut hybrid simultaneously has the common characteristic segments of the male parent and the female parent through PCR reaction and enzyme digestion. By virtue of the method, the peanut hybrid can be rapidly and accurately identified.
Owner:BIOTECH RES CENT SHANDONG ACADEMY OF AGRI SCI
Molecular marker relevant to laying duck egg laying performance and application thereof in breeding
ActiveCN108546764AProper extractionFast wayMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceMutation
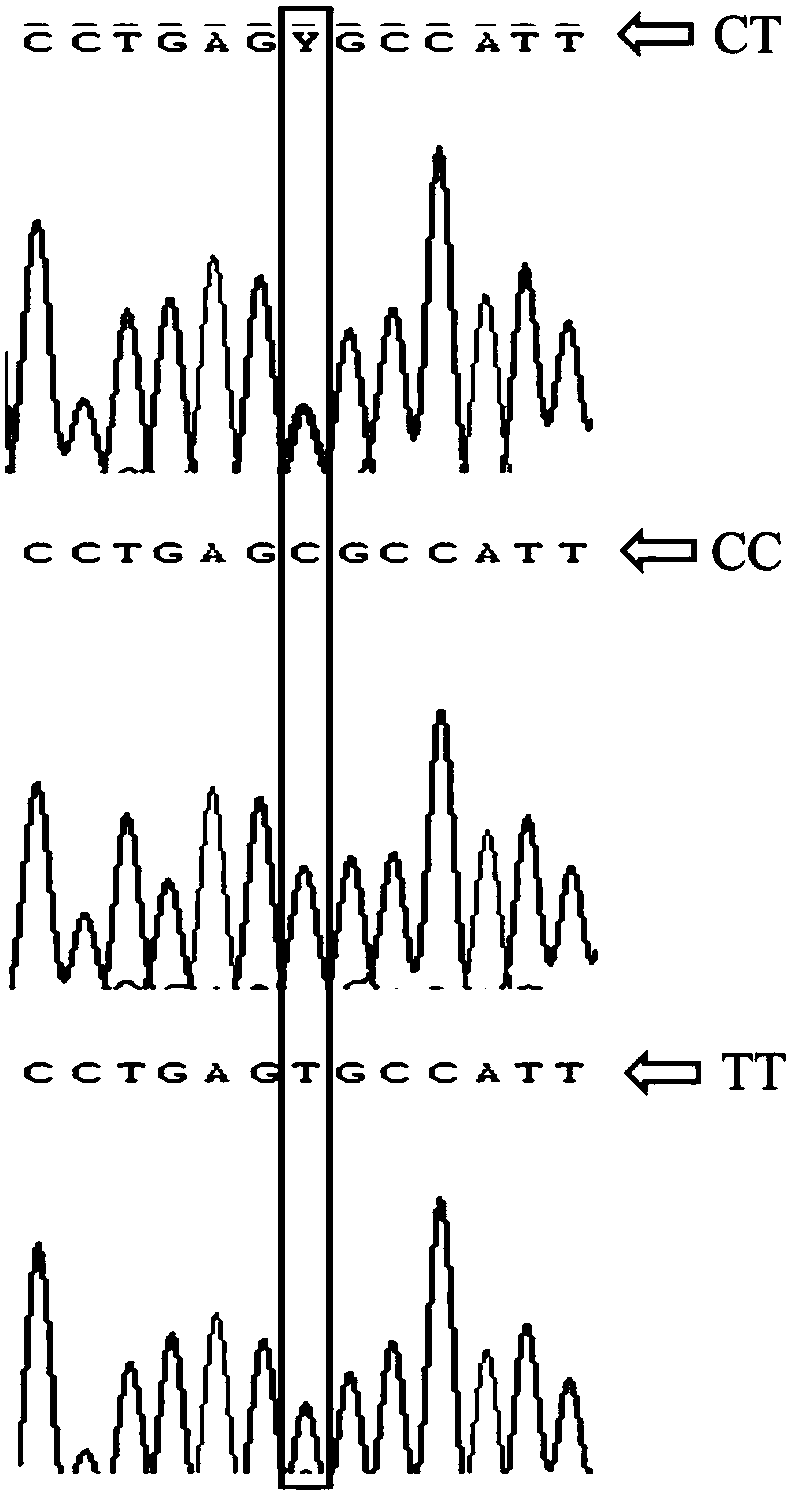
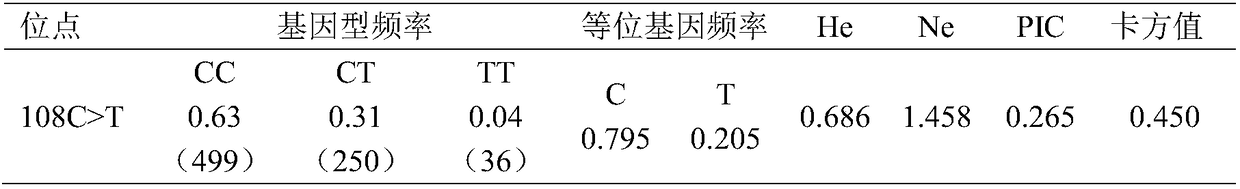
The invention relates to a molecular marker relevant to the laying duck egg laying performance and application thereof in breeding, and belongs to the technical field of molecular markers. The nucleotide sequence of the molecular marker is shown as SEQ ID NO.1; the 108-th site position in the sequence has a C>T mutation; the site has the obvious relevance with the duck egg laying starting day age,the 34-week-age egg laying quantity and the 72-week-age egg laying quantity; the egg laying starting day age of the TT gene type is obviously lower than that of the CC and CT gene type; the 34-week-age egg laying quantity of the TT gene type is obviously higher than that of the CC gene type; the 72-week-age egg laying quantity of the TT gene type is obviously higher than that of the CC and CT gene type; through PCR amplification and product sequencing typing, the TT gene type duct can be selected. The molecular marker realizes the early stage breeding on the duck egg laying performance; the powerful help is provided for the work of the egg laying type duck breeding work.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
CYP3A88-molecular-marker breeding method for sorting porcine reproductive and respiratory syndrome (PRRS)-resistant pigs and application thereof
ActiveCN103131772AReduced activitySimple methodMicrobiological testing/measurementDiseasePromoter activity
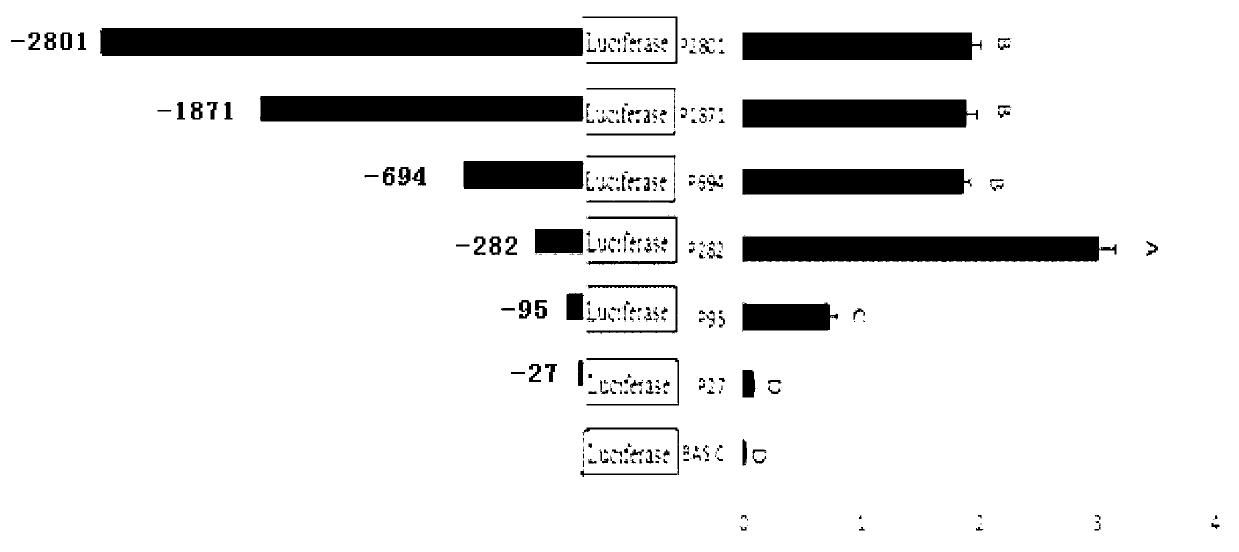
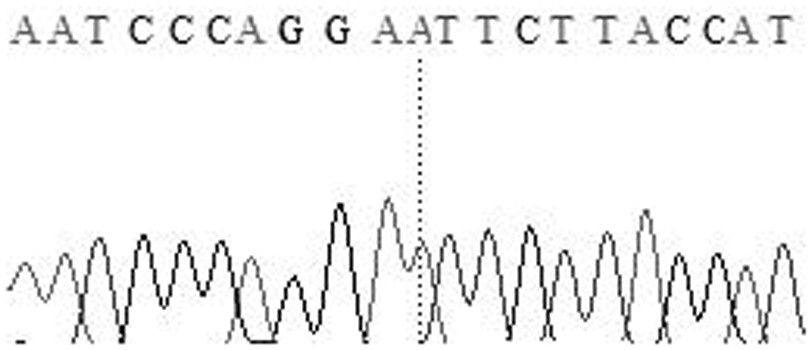
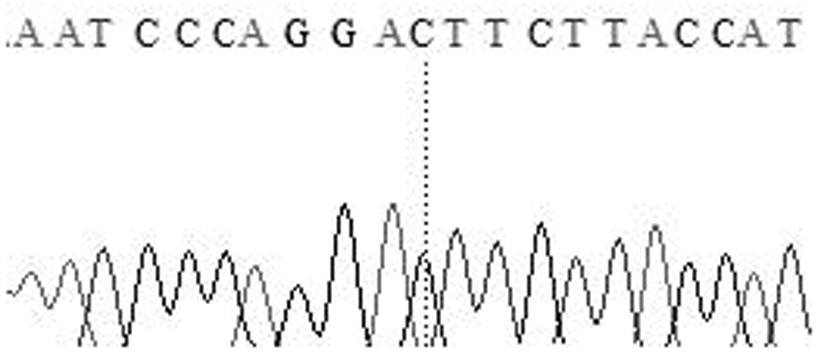
The invention relates to the field of molecular genetics, in particular to the application of a molecular marker method in pig breeding for disease resistance, wherein according to the molecular marker method, the molecule at a mutation site in a CYP3A88 gene 5' regulatory region of a pig is marked. The inventor of the method discovers that the CYP3A885' regulatory region of a large Chinese streaky-head pig and the CYP3A885' regulatory region of a Duroc long hybrid pig have a plurality of differences, wherein an A-to-T mutation exists at the -78 site, through a luciferase reporter gene system, the fact that promoter activity of the -78 site is lowered remarkably after an A at the -78 site is mutated into a T in a site-directed mutagenesis mode is found, and the promoter activity of the -78 site is the same as the low level of messenger ribonucleic acid (mRNA) expression of a CYP3A88 gene of the porcine reproductive and respiratory syndrome (PRRS)-resistant large Chinese streaky-head pig. Therefore, through genotype detection of the -78 site of the CYP3A88 regulatory region in a pig genome, the genotype of the -78 site of the CYP3A88 regulatory region can be used as a modular marker associated with traits of the PPRS, the molecular marker method not only is simple, convenient and rapid, but also cannot be affected by the environment, and early selection for breeding can be realized.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecular marking method for predicting and identifying length of sheep wool
InactiveCN103276098ASpeed up the genetic selection processSpeed up the breeding processMicrobiological testing/measurementEnzyme digestionGenotype
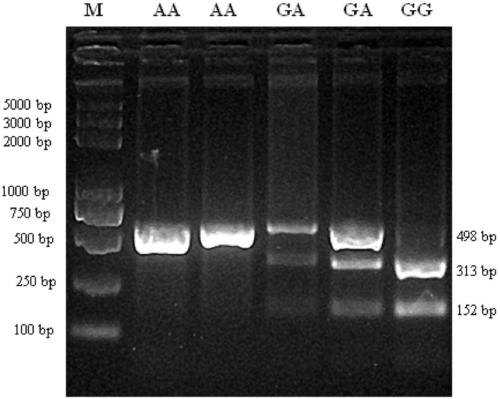
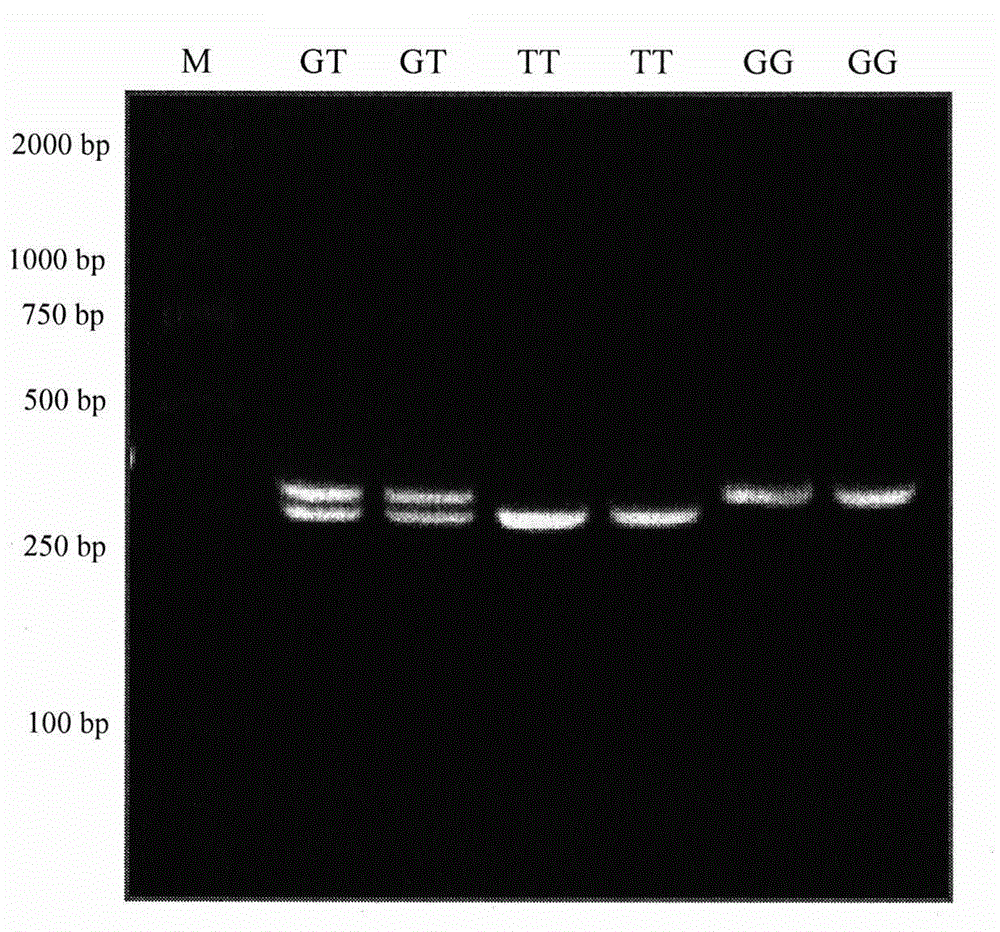
The invention relates to a molecular marking method, in particular to a molecular marking method for predicting and identifying the length of sheep wool. The method comprises the steps as follows: 1), extracting sheep genome DNA, designing a primer, performing PCR (polymerase chain reaction) amplification, performing enzyme digestion, and obtaining an enzyme digestion product; 2), subjecting the enzyme digestion product to electrophoretic separation, and judging genotypes according to an electrophoretic separation result; 3), performing association analysis and estimating the least square mean value of characters, and obtaining a result that the length of wool of a GG genotype individual is remarkably higher than that of wool of AA genotype and AG genotype individuals in the three genotypes; and 4), constructing wool length character breeding population taking the GG genotype individual as the primary so that the length of the sheep wool is predicated and identified. According to the method, the operation is simple, the cost is low, the accuracy is high, and automatic detection can be performed. Selective breeding of the sheep wool length is performed with the molecular marking method, so that a genetic breeding process of the length characters of the sheep wool can be accelerated, breeding sheep can be early selected, the sheep can be selected and left after birth, and a sheep breeding process is accelerated.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marker primer for aided identification of red pepper gray mold resistance and application of molecular marker primer
InactiveCN105296625AAchieve early selectionMajor application prospectsMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceRed peppers
The invention discloses a molecular maker primer for aided identification of red pepper gray mold resistance and an application of the molecular marker primer. A reagent for aided identification of red pepper gray mold resistance and / or aided identification of red pepper gray mold resistance, provided by the invention, is a molecular maker primer pair CARpi-1-200, and is a specific primer pair consisting of nucleotides. By using the primer pair, provided by the invention, for aided identification of red pepper gray mold resistance and / or aided screening of red peppers with gray mold resistance, compared with a field identification result, the disease-resistant coincidence rate reaches 90%. During red pepper seed cultivation, by using the molecular maker disclosed by the invention, the genes for resisting gray molds, in red pepper breeding materials, can be subjected to molecular maker aided selection before the attack of the gray molds in a seedling stage, thereby improving the breeding efficiency and accelerating the breeding progress.
Owner:崔青青
Molecular marker method for two mutation sites of chicken MMP13 gene 5' control region and application of molecular marking method in chicken breeding
ActiveCN105420352AIncrease egg productionAchieve early selectionMicrobiological testing/measurementAnimal husbandryBreeding chickenGenetic linkage disequilibrium
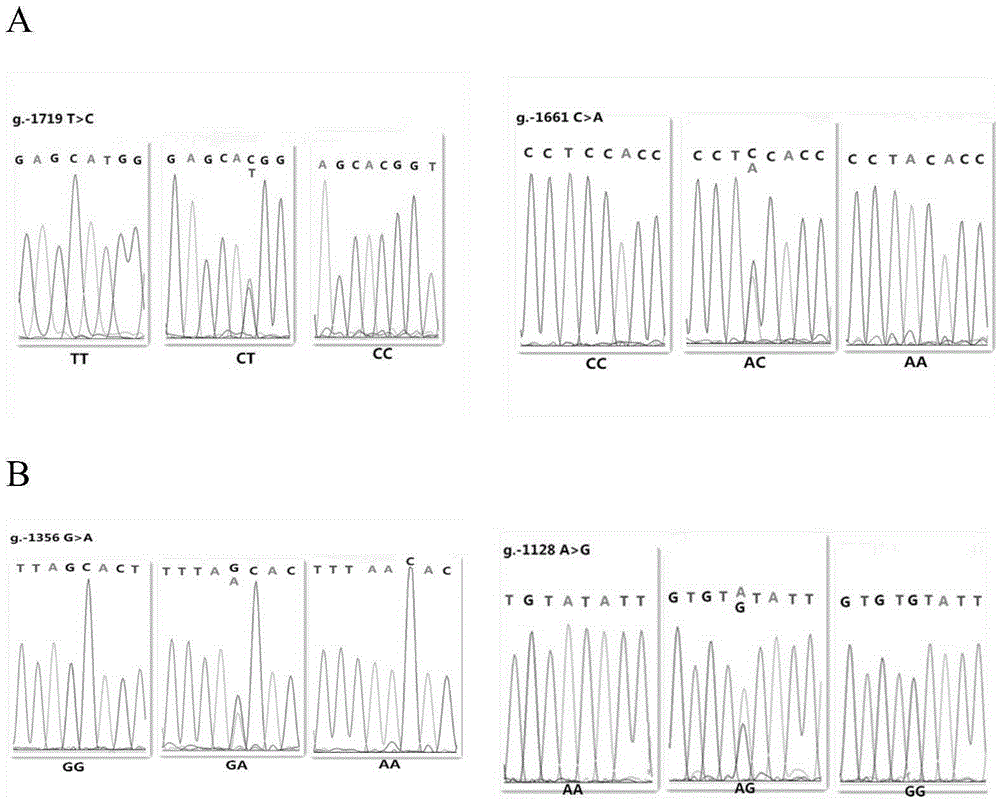
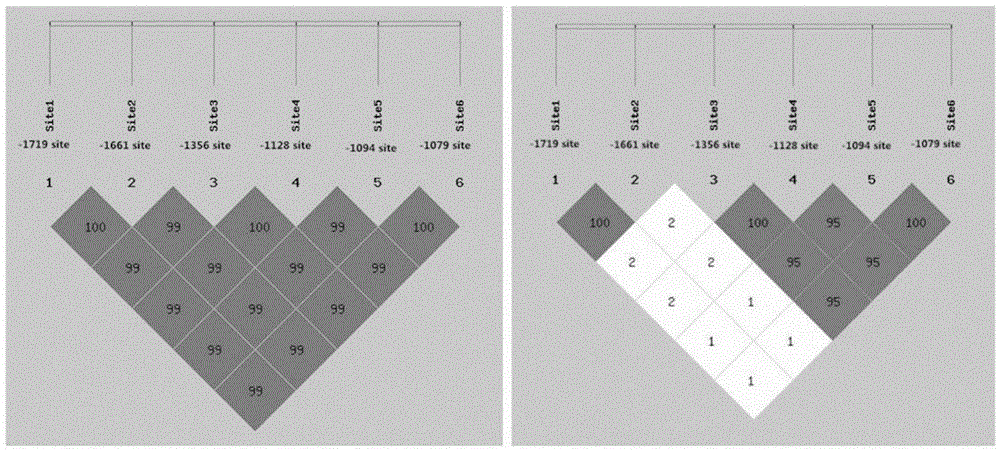
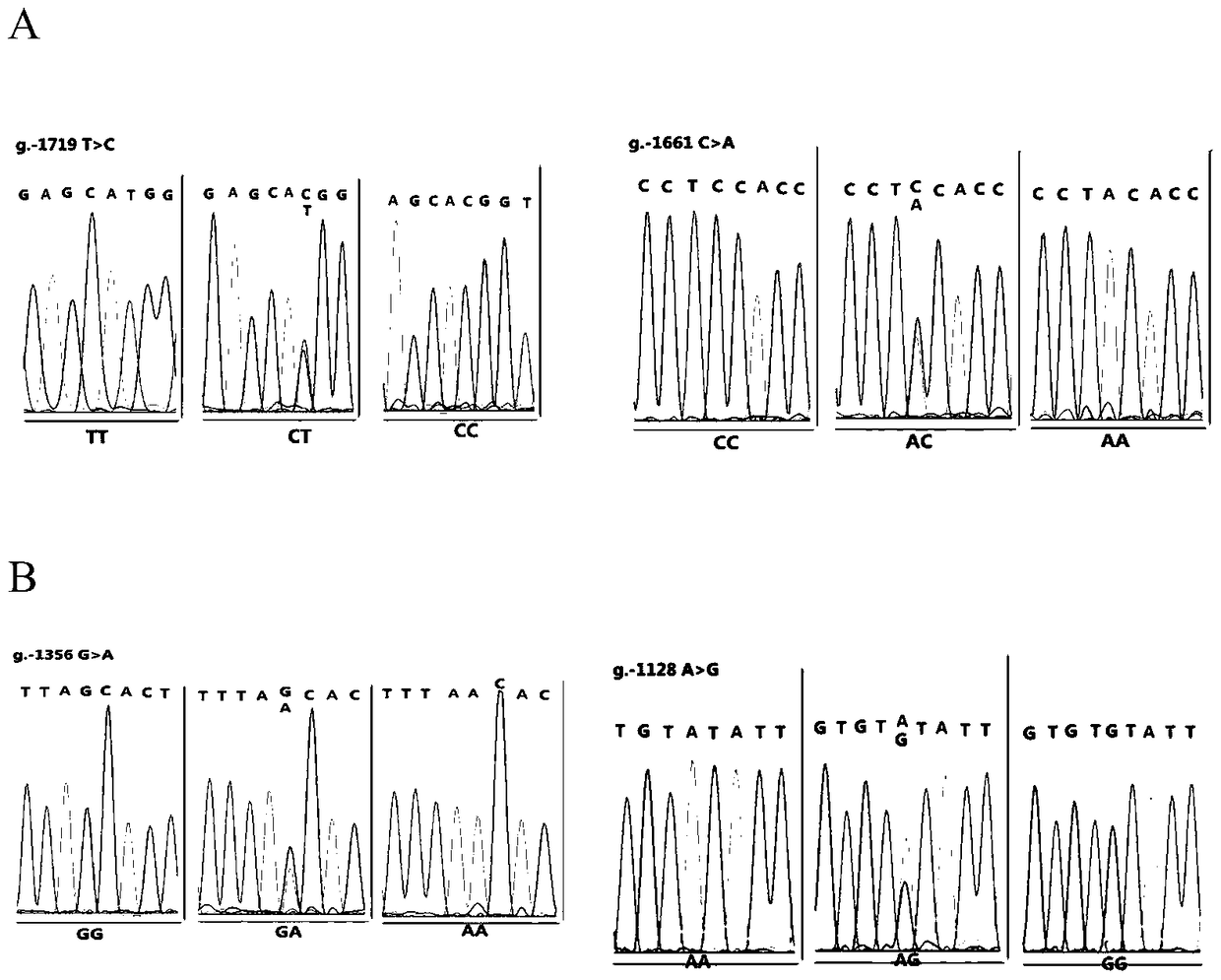
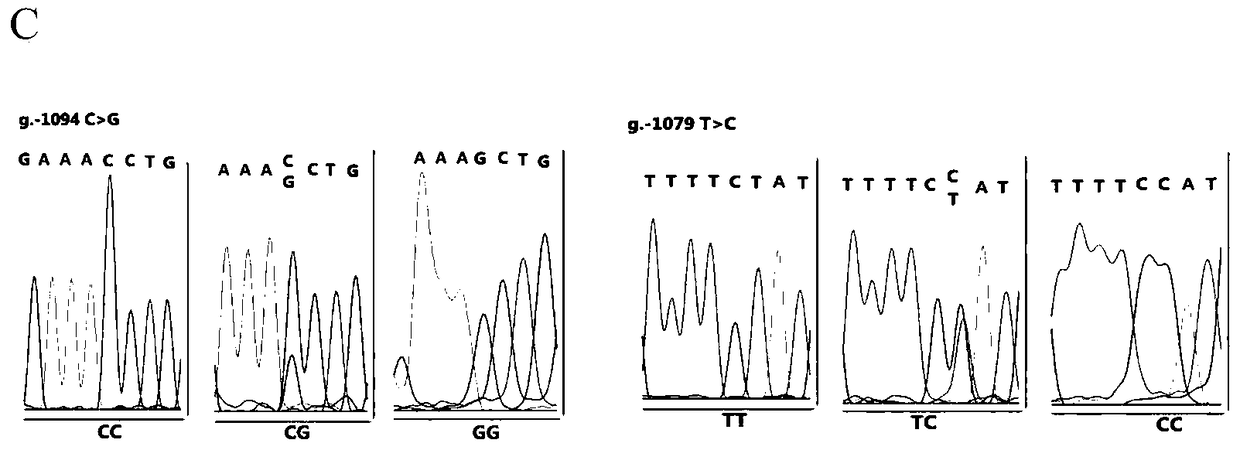
The invention relates to the field of molecular genetics, in particular to a molecular marker method for two mutation sites of a chicken MMP13 gene 5' control region and application of the molecular marker method in chicken breeding. It is found by the inventor that six mutation sites exist in the chicken MMP13 5' control region, namely, -1719 (T>C), -1661 (C>A), -1356 (G>A), -1128 (A>G), -1094 (C>A) and -1079 (T>C); linkage disequilibrium analysis is performed through the SHEsis online software, and it is found through the result that the sites -1719 and -1661, the sites -1356 and -1128 and the sites -1094 and -1079 are respectively in a completely-linked state in White Recessive Rock. The method is easy, convenient and fast to implement, beneficial for breeding chicken breeds laying eggs early at a high yield and capable of providing favorable help for the marker-assisted breeding work.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
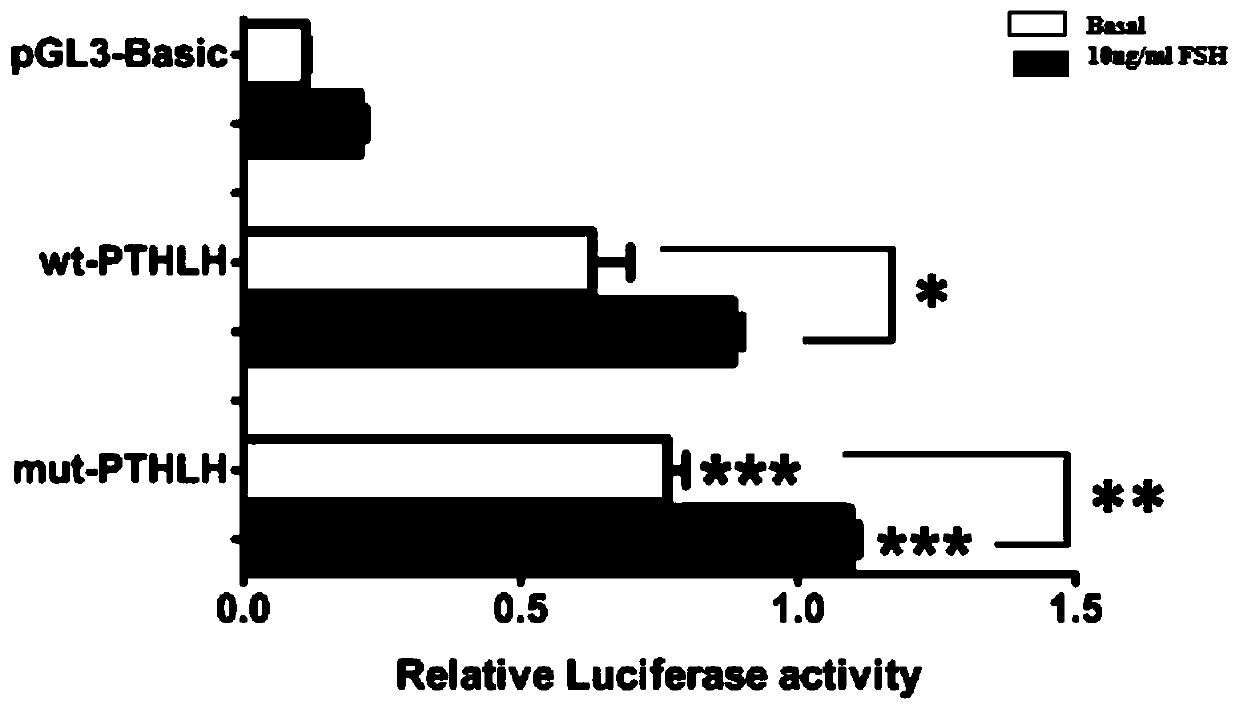
Molecular marking method for two mutation sites in chicken PTHLH gene 5' regulatory region and application thereof in chicken breeding
ActiveCN106048043AIncrease egg productionAchieve early selectionMicrobiological testing/measurementAnimal husbandryBreeding chickenGenetic linkage disequilibrium
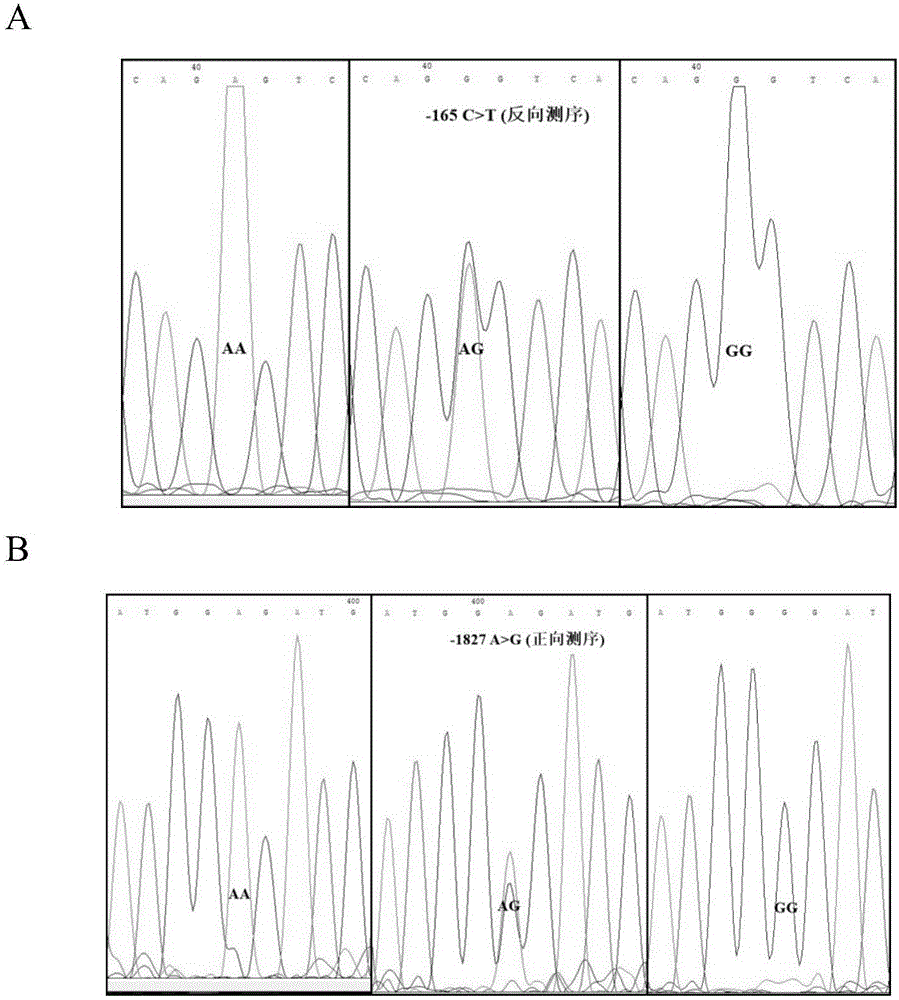
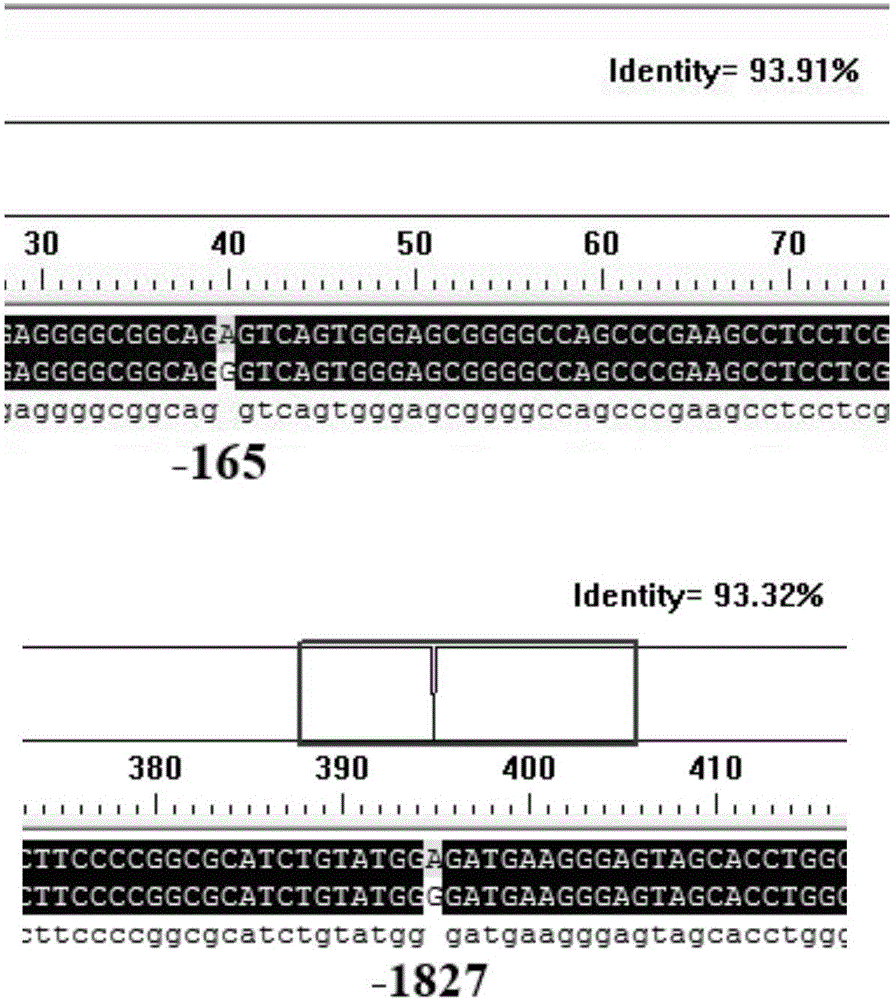
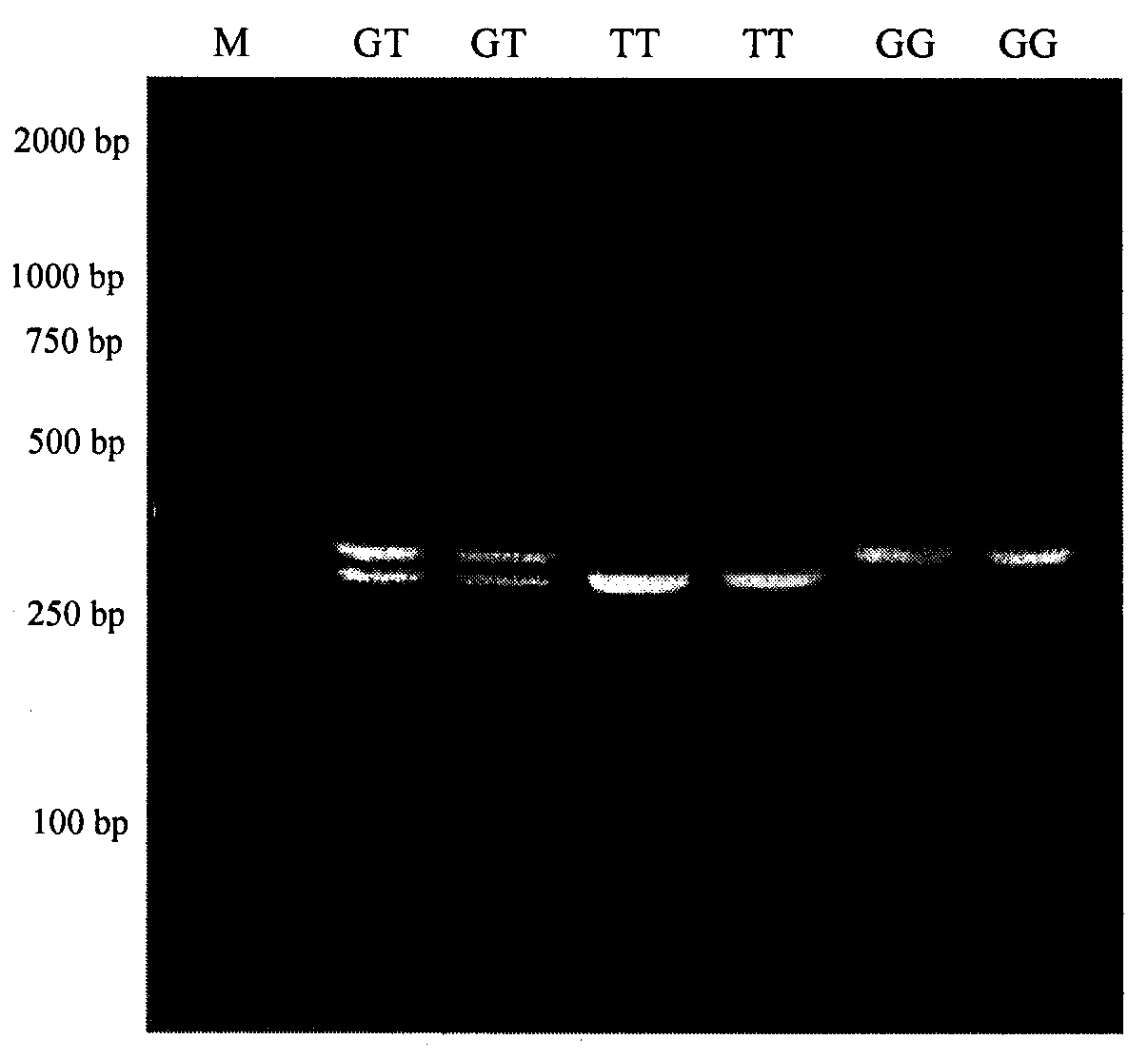
The invention relates to a molecular marking method for two mutation sites in a chicken PTHLH gene 5' regulatory region and application thereof in chicken breeding. Two mutation sites (namely the -1827 (A>G) site and the -165(C>T) site) exist in the chicken PTHLH gene 5' regulatory region, SHEsis online software is used for linkage disequilibrium analysis, and the result shows that the two sites in recessive white plymouth rock are in a complete linkage state. The two sites are related with the first-egg age and egg laying quantity at the 32th week, the first-egg age of AC / GT gene type is remarkably earlier than that of the GT / GT type and the AC / AC type, and meanwhile the egg laying quantity at the 32th week is remarkably larger than that of the other two types. The method is simple, fast and beneficial for breeding chicken variety with the early first-egg age and the large egg laying quantity and provides favorable help for marker-assisted breeding work.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecular marker primer pair capable of predicting and identifying the natural length of sheep and wool and application thereof
ActiveCN109182558ASpeed up the breeding processAchieve early selectionMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceEnzyme digestion
The invention discloses a molecular marker primer pair capable of predicting and identifying the natural length of sheep and wool and application thereof, wherein the pair of primers is shown in sequence lists Seq ID No. 1 and Seq ID No. 2. The primer pairs were designed and the sheep genomic DNA was extracted for amplification, and then digested with enzyme to obtain digested products. Enzyme digestion product electrophoresis separation, according to the electrophoresis separation result carries on the genotype determination; The results showed that the natural wool length of GG genotype population was significantly higher than that of AA genotype and GA genotype population. Finally, the ewes with GG genotype were selected as breeding population. The invention can predict and identify thenatural length of sheep wool, provides a more effective molecular marker method for improving the quality traits of sheep wool, and is applied to the field of screening sheep molecular assisted breeding with long natural length of wool fiber. The invention is simple in operation and low in cost.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
Molecular marker relevant to reproductive performance of chicken and application of molecular marker in breeding
InactiveCN103789307ASimple methodNot affected by the environmentMicrobiological testing/measurementDNA preparationBiotechnologyNucleotide
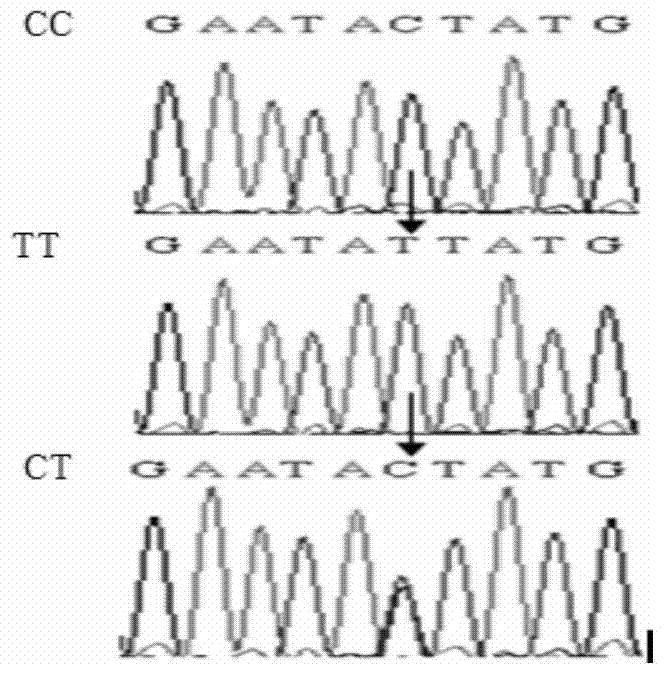
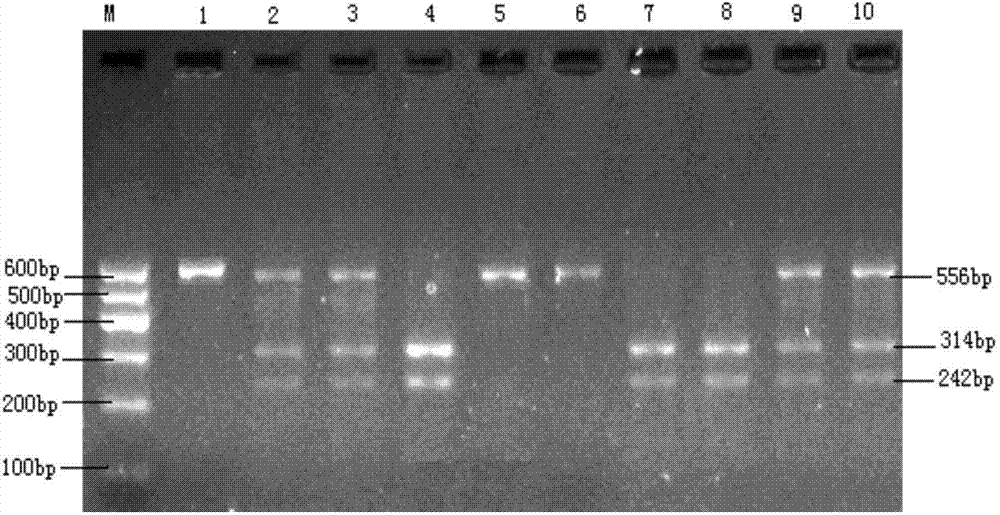
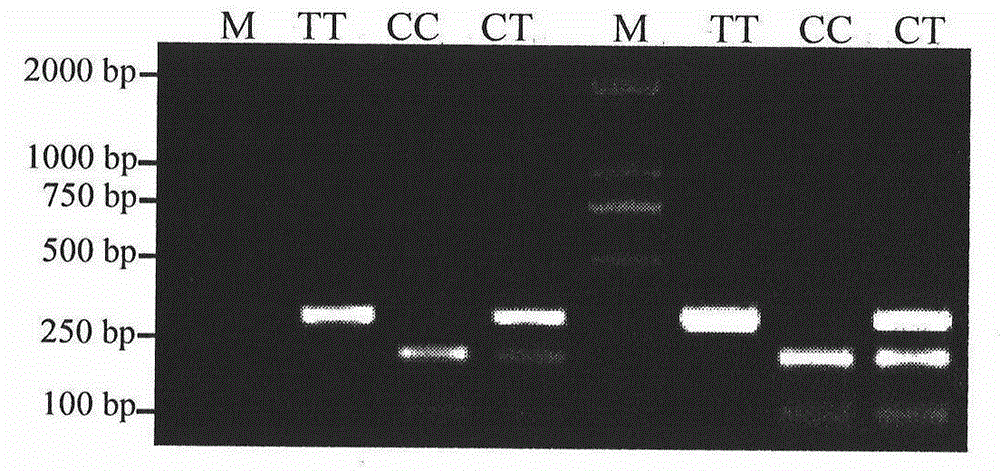
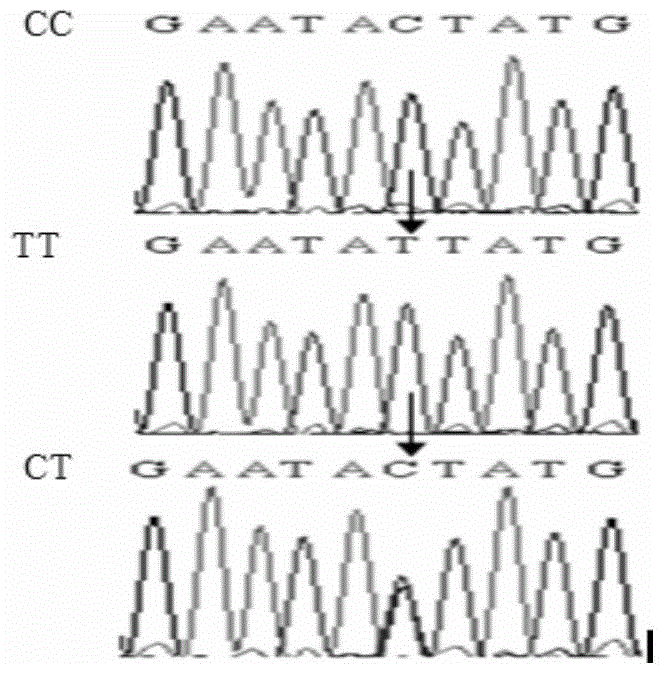
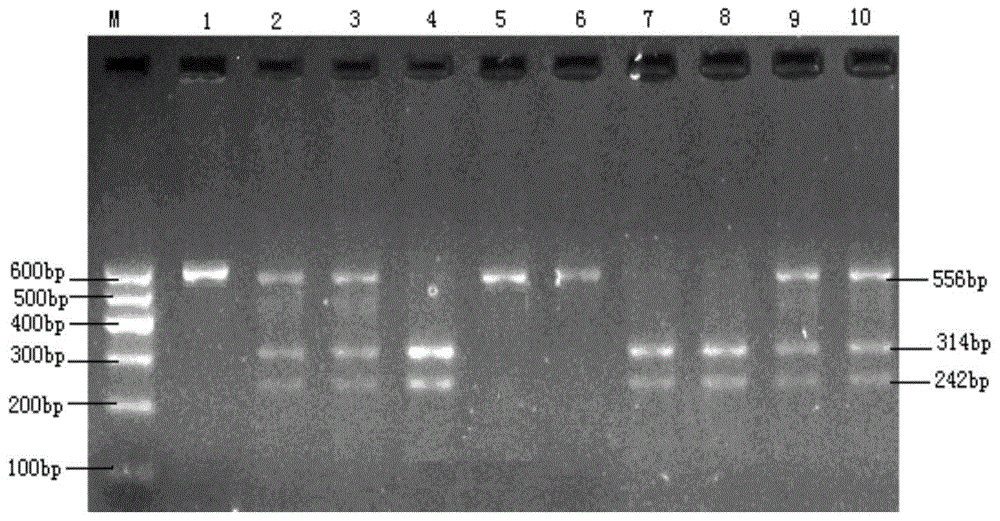
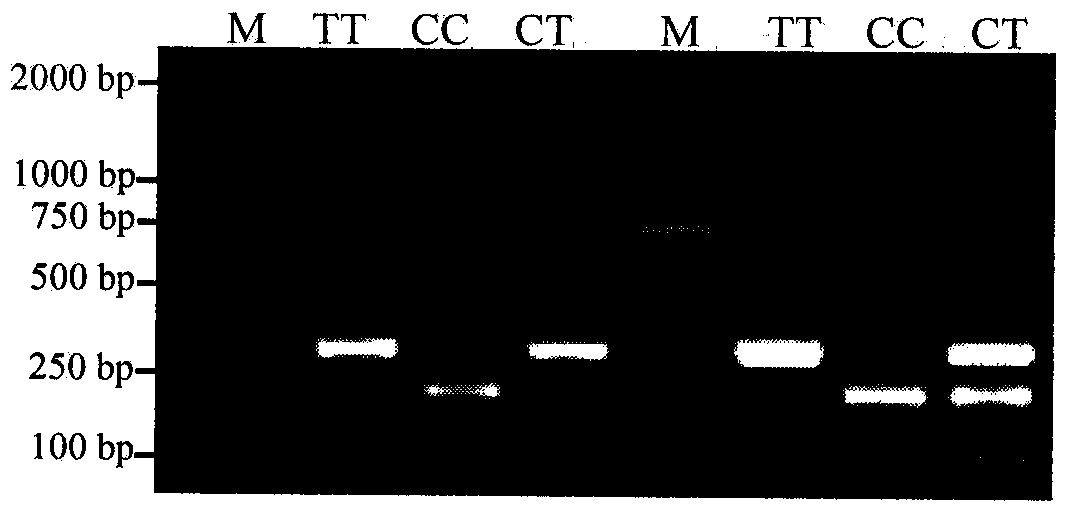
The invention discloses a molecular marker relevant to the reproductive performance of chicken and application of the molecular marker in breeding, belonging to the field of molecular genetics. The nucleotide sequence of the molecular marker is shown in SEQ ID No.1; a C->T base substitution exists at the 315th bp (base pair) of the sequence, thereby leading polymorphism of SspI-RFLP. The genome DNA of chicken is taken as a template, and primers shown in SEQ IDNo.2-3 are used for amplifying to obtain a PCR amplification product of 556bp; the PCR product is detected by using 1.5 percent agarose gel electrophoresis after being digested by an SspI restriction enzyme to obtain three banding patterns of CC, CT and TT; in local varieties with small egg weights and poor egg laying performance, CC genetype individuals are for reserved. A method for detecting the molecular marker relevant to the reproductive performance of chicken is simple and rapid and is not influenced by the environment, and early strain selection can be realized.
Owner:POULTRY INST SHANDONG ACADEMY OF AGRI SCI
Molecular marking method and primer pair for predicting and identifying wool fineness of sheep
InactiveCN104894253ASpeed up the breeding processAchieve early selectionMicrobiological testing/measurementDNA/RNA fragmentationForward primerEnzyme digestion
The invention provides a molecular marking method and a primer pair for predicting and identifying the wool fineness of sheep. The primer pair includes a forward primer ICT1F as shown in Seq No.1 in a sequence table and a reverse primer ICT1R as shown in Seq No.2 in the sequence table. The molecular marking method concretely comprises the following steps of designing the primer pair, extracting sheep genome DNA to amplify, and carrying out enzyme digestion to obtain an enzyme-digested product; carrying out electrophoretic separation on the enzyme-digested product, and judging the genetype according to an electrophoretic separation result; carrying out correlation analysis, and estimating the least square average value of a character to obtain a result that the wool fineness of a colony with a CT genetype in three genetypes is remarkably lower than that of a colony with a TT genetype; and dividing experimental colonies into three kinds of types. By using the molecular marking method, the wool fineness of the sheep can be predicted and identified; a more effective molecular marking method is provided for quality character improvement of sheep wool and marker assisted selection; and the molecular marking method can be effectively applied to the field of molecular-assisted breeding of sheep with superfine wool.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marker method of two mutation sites in chicken FSHR (Follicle-Stimulating Hormone Receptor) gene 5' control region and applications thereof in children breeding
ActiveCN102080126AIncrease egg productionAchieve early selectionMicrobiological testing/measurementHaplotypeFollicle-stimulating hormone receptor
The invention relates to the field of molecular genetics, in particular to a molecular marker method of two mutation sites in a chicken FSHR (Follicle-Stimulating Hormone Receptor) gene 5' control region and the applications thereof in children breeding. The inventor finds that chicken FSHR 5' control region -237 and -868 sites are not related to properties when single site marker is analyzed but finds that the two sites have remarkable difference (P=0.016) by corresponding a TTG+G- type to a small first laying age (123d) and corresponding a TTG+G+ type to a great first laying age (134d) when using the two site haplotypes to construct doubling types for multiple comparison. Accordingly, the invention can detect the two molecular markers related to the egg laying properties, and the method not only is simple, convenient and rapid, but also avoids environmental influence and can realize early seed section.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecular marker method capable of indicating and identifying curling degree of sheep wools and primer pair for molecular marker method
InactiveCN105063213AAchieve early selectionEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationElectrophoresisMolecular breeding
The invention provides a molecular marker method capable of indicating and identifying curling degree of sheep wools and a primer pair for the molecular marker method. The primer pair comprises an upper primer SHCBP1-P1-F shown in the sequence chart Seq No.1 and a lower primer SHCBP1-P1-R shown in the sequence chart Seq No.2. The molecular marker method comprises the following steps: designing a primer pair, extracting a sheep genome DNA for amplification, and then performing digestion to obtain a digestion product; performing electrophoretic separation on the digestion product, and judging the genotypes according to the electrophoretic separation result; performing correlation analysis, estimating the least squares means of the character, wherein the result is that the wool fineness of a GG genotype group and a GT genotype group in three genotypes is obviously higher than that of a TT genotype group; dividing the test group into three types to complete the method. The molecular marker method can indicate and identify the curling degree of sheep wools, and can be effectively applied to the auxiliary molecular breeding field of high-curling-degree sheep.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marking method of fat deposition and meat quality traits of lean meat type Dingyuan pig strain
ActiveCN110982908AAchieve early selectionEarly selection is accurateFood processingMicrobiological testing/measurementBiotechnologyEnzyme digestion
The invention discloses a molecular marker of fat deposition and meat quality traits of a lean meat type Dingyuan pig strain. The nucleotide sequence of the molecular marker is shown in SEQ ID NO:1, a398th base in the above sequence is a base mutation site, and the base mutation site is G or C. The invention discloses a primer pair for detecting the above molecular marker. The invention disclosesa molecular marking method for the fat deposition and meat quality traits of the lean meat type Dingyuan pig strain. The method comprises the following steps: extracting genomic DNA containing the above molecular marker in pigs; performing a polymerase chain reaction by using the above primer pair, the genomic DNA and a PCR reaction buffer solution to obtain an in-vitro amplification product; performing an enzyme digestion reaction on the in-vitro amplification product by using a restriction enzyme to obtain an enzyme-digested product; and performing polymorphism detection on the enzyme-digested product to obtain genotypes of each individual. The invention discloses an application of the above molecular marker and / or the above primer pair in the breeding of live pigs of the lean meat typeDingyuan pig strain.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Molecular marker method of beef cattle fat color trait
InactiveCN107354216AAchieve early selectionShorten the generation intervalMicrobiological testing/measurementAgricultural scienceBeta-Carotene
The invention discloses a molecular marker method of a beef cattle fat color trait. The method comprises steps as follows: DNA extraction of bovine genome, design of a third exon primer of a BCO2 (beta-carotene oxygenase 2) gene, in-vitro amplification and genotype detection. The method can be used for assistant selection of the fat color trait in cattle breeding and can realize early selection of breeder cattle; with the adoption of the gene pyramiding method, the good genotype of the BCO2 gene can be stabilized after one generation, that is, homozygosis of a good gene of the fat color trait of a beef cattle strain is realized through selection of one generation, the generation interval is greatly shortened, and the selection process is accelerated. The molecular marker method is simple and convenient to operate, condition requirements in the polymerase chain reaction process are low, the length of amplified fragments is short (525bp), amplification is easier, and the amplification efficiency and the judgement accuracy of the genotype are improved.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Molecular marker and specific primers for assisting in test of wilt disease resistance in brassica oleracea and use thereof
InactiveCN102108407BAchieve early selectionMicrobiological testing/measurementBiotechnologyDisease resistant
The invention discloses a molecular marker and specific primers for assisting in test of wilt disease resistance in brassica oleracea and use thereof. The invention provides a reagent for assisting in the test of wilt disease resistance in brassica oleracea and / or assisting in screening brassica oleracea with wilt disease resistance, which is a specific primer pair formed by nucleotides represented by a sequence 2 and a sequence 3 in a sequence table. The invention also provides a specific gene fragment which is at a genetic distance about 2.87cm to the wilt disease resistance gene in brassica oleracea and is formed by nucleotides represented by a sequence 1 in a sequence table. The result of the identification of wilt disease resistance in brassica oleracea and / or screening of the brassica oleracea with wilt disease resistance with assistance from the reagent (primer pair) or sequence characterized amplified region (SCAR) marker, which are provided by the invention, is 97 percent consistent with that of field identification. When used in breeding, the reagent, molecular marker or method has the advantages of accuracy, quickness, capability of realizing early breeding and the like and has a bright application prospect.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES +1
Genome of low porcine backfat thickness and molecular marking method and breeding application
ActiveCN111763749ANot affectedSimple methodMicrobiological testing/measurementFermentationNucleotideMedicine
The invention discloses a genome relates to a character of porcine backfat thickness and a molecular marking method and a breeding application. The genome is characterized in that a gene sequence SeqID No: 4 is a nucleotide sequence molecule of haplotype H4 present in a D-Loop region of a porcine mitochondrial genome, and pigs with the haplotype H4 are marked and determined by the molecular marking method. The beneficial effect of genome is as follows, 6-month-old sows with the haplotype H4 in the D-Loop region of the porcine mitochondrial gene have significantly lower backfat thickness thanthose with other haplotypes, indicating that the haplotype H4 is relevant to the character of low porcine backfat thickness. The haplotype H4 in the D-Loop region of the porcine mitochondrial genome is detected as a molecular breeding marker associated with low porcine backfat thickness, so that the method is simple and rapid, insusceptible to environment, and capable of achieving early breeding.
Owner:济宁安鑫养殖有限公司
A molecular marker related to chicken reproductive performance and its application in breeding
InactiveCN103789307BSimple methodNot affected by the environmentMicrobiological testing/measurementDNA preparationAgricultural scienceNucleotide
The invention discloses a molecular marker relevant to the reproductive performance of chicken and application of the molecular marker in breeding, belonging to the field of molecular genetics. The nucleotide sequence of the molecular marker is shown in SEQ ID No.1; a C->T base substitution exists at the 315th bp (base pair) of the sequence, thereby leading polymorphism of SspI-RFLP. The genome DNA of chicken is taken as a template, and primers shown in SEQ IDNo.2-3 are used for amplifying to obtain a PCR amplification product of 556bp; the PCR product is detected by using 1.5 percent agarose gel electrophoresis after being digested by an SspI restriction enzyme to obtain three banding patterns of CC, CT and TT; in local varieties with small egg weights and poor egg laying performance, CC genetype individuals are for reserved. A method for detecting the molecular marker relevant to the reproductive performance of chicken is simple and rapid and is not influenced by the environment, and early strain selection can be realized.
Owner:POULTRY INST SHANDONG ACADEMY OF AGRI SCI
A Molecular Marker Related to Laying Duck Laying Performance and Its Application in Breeding
InactiveCN108546764BProper extractionFast wayMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceMedicine
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
Molecular markers related to fiber diameter traits of sheep wool and their specific primer pairs and applications
ActiveCN107604078BHigh precisionSpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationChromosome 21Fiber diameter
The invention relates to the technical field of animal molecular markers, and relates to a molecular marker related to the diameter traits of sheep wool fibers and its specific primer pair and its application in the selection of Chinese Merino wool fiber diameter traits, which is related to the diameter traits of sheep wool fibers Molecular markers associated with traits, which are located at exon 30 of chromosome 21 of the FAT3 gene. The present invention discloses for the first time that the rs425144268 SNP site is used as a molecular marker of sheep wool fiber diameter, and discloses for the first time that the rs425144268 SNP site is applied to the selection of Chinese Merino sheep wool fiber diameter traits; When the molecular marker is used for the selection of good wool fiber diameter traits, it has the advantage of simple operation and can assist in the screening of sheep with small fiber diameter. Since it starts from the molecular marker detection level, it can improve the accuracy of breed selection and improve the detection efficiency.
Owner:新疆畜牧科学院畜牧研究所 +1
A Molecular Identification Method for Peanut Hybrids
ActiveCN105525012BRapid identificationAccurate identificationMicrobiological testing/measurementGenomic sequencingMolecular identification
The invention discloses a molecular identification method of a peanut hybrid. The method comprises the following steps: conducting reduced-representation genome sequencing on the male parent and female parent of the peanut hybrid, and analyzing a sequencing result so as to identify an SNP molecular marker between the male parent and the female parent and design a universal primer for the male parent and the female parent; and extracting the genome DNA of the peanut hybrid, and verifying whether the peanut hybrid simultaneously has the common characteristic segments of the male parent and the female parent through PCR reaction and enzyme digestion. By virtue of the method, the peanut hybrid can be rapidly and accurately identified.
Owner:BIOTECH RES CENT SHANDONG ACADEMY OF AGRI SCI
Method for assisting in identifying resistance of soybeans to soybean mosaic viruses
InactiveCN103320427BAchieve early selectionEfficient selectionMicrobiological testing/measurementDNA/RNA fragmentationSoybean mosaic virusDNA fragmentation
The invention discloses a method for assisting in identifying the resistance of soybeans to soybean mosaic viruses. The invention protects a primer pair A consisting of a DNA fragment shown by a sequence 1 in the sequence table and a DNA fragment shown by a sequence 2 in the sequence table; the primer pair A have functions of (a) or (b) as follow: (a) assisting in identifying the resistance of soybeans to soybean mosaic viruses, and (b) assisting in selecting soybeans capable of resisting to soybean mosaic viruses. By adopting the method of the invention, molecular-assisted selective breeding of soybeans capable of resisting to soybean mosaic virus SMV-N1 strains and soybean mosaic virus SMV-N3 strains is realized, early-stages (such as a cotyledon stage) selective breeding with efficiency over 98% can be realized comparing with conventional disease-resistant phenotype identification method, and experiment time and experiment cost are saved greatly. The method of the invention will play an important role in selective breeding of soybeans.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marking method for predicting and identifying length of sheep wool
InactiveCN103276098BEasy to separateSpeed up the genetic selection processMicrobiological testing/measurementEnzyme digestionGenotype
The invention relates to a molecular marking method, in particular to a molecular marking method for predicting and identifying the length of sheep wool. The method comprises the steps as follows: 1), extracting sheep genome DNA, designing a primer, performing PCR (polymerase chain reaction) amplification, performing enzyme digestion, and obtaining an enzyme digestion product; 2), subjecting the enzyme digestion product to electrophoretic separation, and judging genotypes according to an electrophoretic separation result; 3), performing association analysis and estimating the least square mean value of characters, and obtaining a result that the length of wool of a GG genotype individual is remarkably higher than that of wool of AA genotype and AG genotype individuals in the three genotypes; and 4), constructing wool length character breeding population taking the GG genotype individual as the primary so that the length of the sheep wool is predicated and identified. According to the method, the operation is simple, the cost is low, the accuracy is high, and automatic detection can be performed. Selective breeding of the sheep wool length is performed with the molecular marking method, so that a genetic breeding process of the length characters of the sheep wool can be accelerated, breeding sheep can be early selected, the sheep can be selected and left after birth, and a sheep breeding process is accelerated.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marker method of two mutation sites in the 5′ regulatory region of chicken pthlh gene and its application in chicken breeding
ActiveCN106048043BIncrease egg productionAchieve early selectionMicrobiological testing/measurementAnimal husbandryBreeding chickenComplete linkage
The invention relates to a molecular marking method for two mutation sites in a chicken PTHLH gene 5' regulatory region and application thereof in chicken breeding. Two mutation sites (namely the -1827 (A>G) site and the -165(C>T) site) exist in the chicken PTHLH gene 5' regulatory region, SHEsis online software is used for linkage disequilibrium analysis, and the result shows that the two sites in recessive white plymouth rock are in a complete linkage state. The two sites are related with the first-egg age and egg laying quantity at the 32th week, the first-egg age of AC / GT gene type is remarkably earlier than that of the GT / GT type and the AC / AC type, and meanwhile the egg laying quantity at the 32th week is remarkably larger than that of the other two types. The method is simple, fast and beneficial for breeding chicken variety with the early first-egg age and the large egg laying quantity and provides favorable help for marker-assisted breeding work.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Molecular marker method of two mutation sites in the 5′ regulatory region of chicken mmp13 gene and its application in chicken breeding
ActiveCN105420352BIncrease egg productionAchieve early selectionMicrobiological testing/measurementAnimal husbandryBreeding chickenGenetic linkage disequilibrium
The invention relates to the field of molecular genetics, in particular to a molecular marker method for two mutation sites of a chicken MMP13 gene 5' control region and application of the molecular marker method in chicken breeding. It is found by the inventor that six mutation sites exist in the chicken MMP13 5' control region, namely, -1719 (T>C), -1661 (C>A), -1356 (G>A), -1128 (A>G), -1094 (C>A) and -1079 (T>C); linkage disequilibrium analysis is performed through the SHEsis online software, and it is found through the result that the sites -1719 and -1661, the sites -1356 and -1128 and the sites -1094 and -1079 are respectively in a completely-linked state in White Recessive Rock. The method is easy, convenient and fast to implement, beneficial for breeding chicken breeds laying eggs early at a high yield and capable of providing favorable help for the marker-assisted breeding work.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
A molecular marker method and its primer pair that can predict and identify the curl degree of sheep wool
InactiveCN105063213BAchieve early selectionEasy to operateMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenotype
The invention provides a molecular marker method capable of indicating and identifying curling degree of sheep wools and a primer pair for the molecular marker method. The primer pair comprises an upper primer SHCBP1-P1-F shown in the sequence chart Seq No.1 and a lower primer SHCBP1-P1-R shown in the sequence chart Seq No.2. The molecular marker method comprises the following steps: designing a primer pair, extracting a sheep genome DNA for amplification, and then performing digestion to obtain a digestion product; performing electrophoretic separation on the digestion product, and judging the genotypes according to the electrophoretic separation result; performing correlation analysis, estimating the least squares means of the character, wherein the result is that the wool fineness of a GG genotype group and a GT genotype group in three genotypes is obviously higher than that of a TT genotype group; dividing the test group into three types to complete the method. The molecular marker method can indicate and identify the curling degree of sheep wools, and can be effectively applied to the auxiliary molecular breeding field of high-curling-degree sheep.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Molecular marker method and primer pair for predicting and identifying sheep wool fineness
InactiveCN104894253BSpeed up the breeding processAchieve early selectionMicrobiological testing/measurementDNA/RNA fragmentationForward primerEnzyme digestion
The invention provides a molecular marking method and a primer pair for predicting and identifying the wool fineness of sheep. The primer pair includes a forward primer ICT1F as shown in Seq No.1 in a sequence table and a reverse primer ICT1R as shown in Seq No.2 in the sequence table. The molecular marking method concretely comprises the following steps of designing the primer pair, extracting sheep genome DNA to amplify, and carrying out enzyme digestion to obtain an enzyme-digested product; carrying out electrophoretic separation on the enzyme-digested product, and judging the genetype according to an electrophoretic separation result; carrying out correlation analysis, and estimating the least square average value of a character to obtain a result that the wool fineness of a colony with a CT genetype in three genetypes is remarkably lower than that of a colony with a TT genetype; and dividing experimental colonies into three kinds of types. By using the molecular marking method, the wool fineness of the sheep can be predicted and identified; a more effective molecular marking method is provided for quality character improvement of sheep wool and marker assisted selection; and the molecular marking method can be effectively applied to the field of molecular-assisted breeding of sheep with superfine wool.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
PCR primer set for identification or screening of cabbage hybrid lethal parent type 2 and its application
ActiveCN112980986BShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyGenetics
The invention belongs to the technical field of molecular assisted breeding, in particular to a PCR primer set for identifying or screening cabbage hybrid lethal parent type 2 and its application. The PCR primer set includes primers KBoHL2-X, KBoHL2-Y, and KBoHL2-Common. The present invention detects whether the genotype at positions 46, 333, and 244 of the C04 chromosome of the cabbage to be tested is T / T or C / C, and determines whether the cabbage has the characteristics of hybrid lethal parent type 2 according to the genotype of the cabbage to be tested: T / T genotype of the cabbage to be tested It is the hybrid lethal parent type 2, and the tested cabbage of the C / C genotype is not the hybrid lethal parent type 2. Using the PCR primers, kit or method of the present invention for assisted breeding has the advantages of simple operation, strong specificity, good stability, and early breeding can be realized, and the coincidence rate of identification results and field phenotypes reaches 100%. It has great application prospect.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com