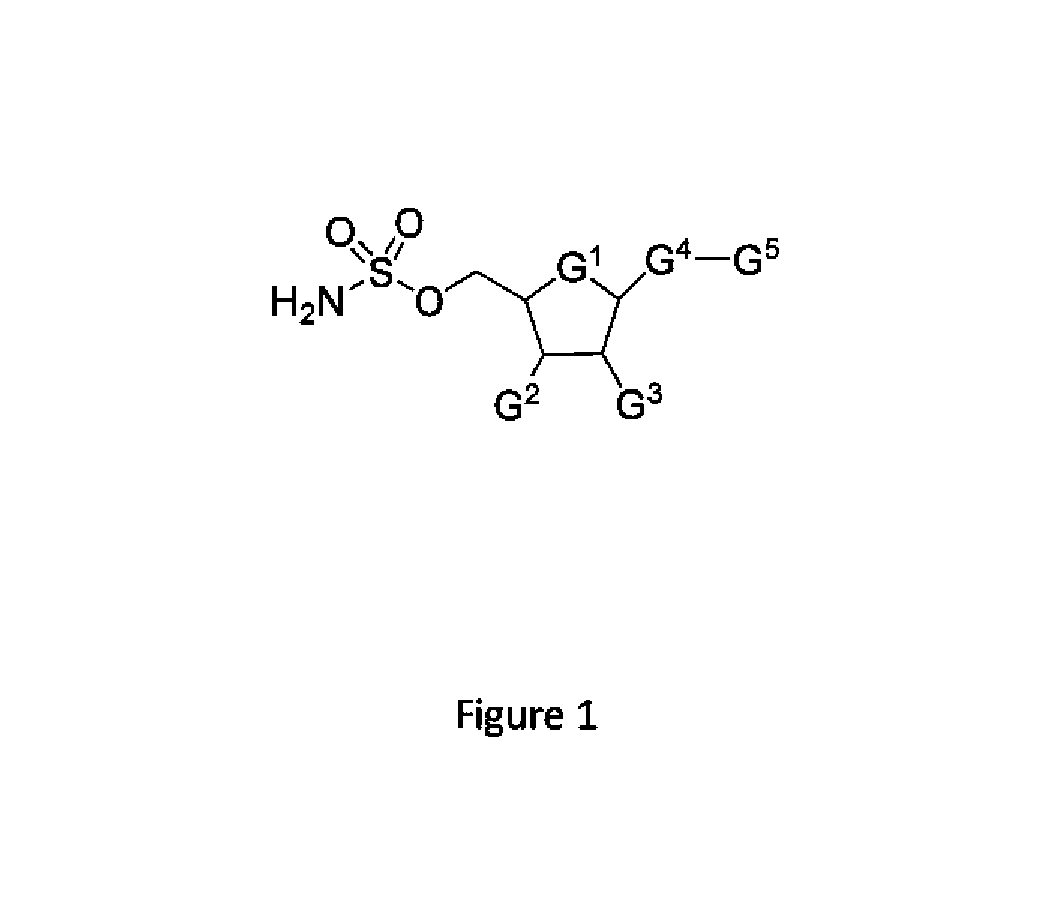
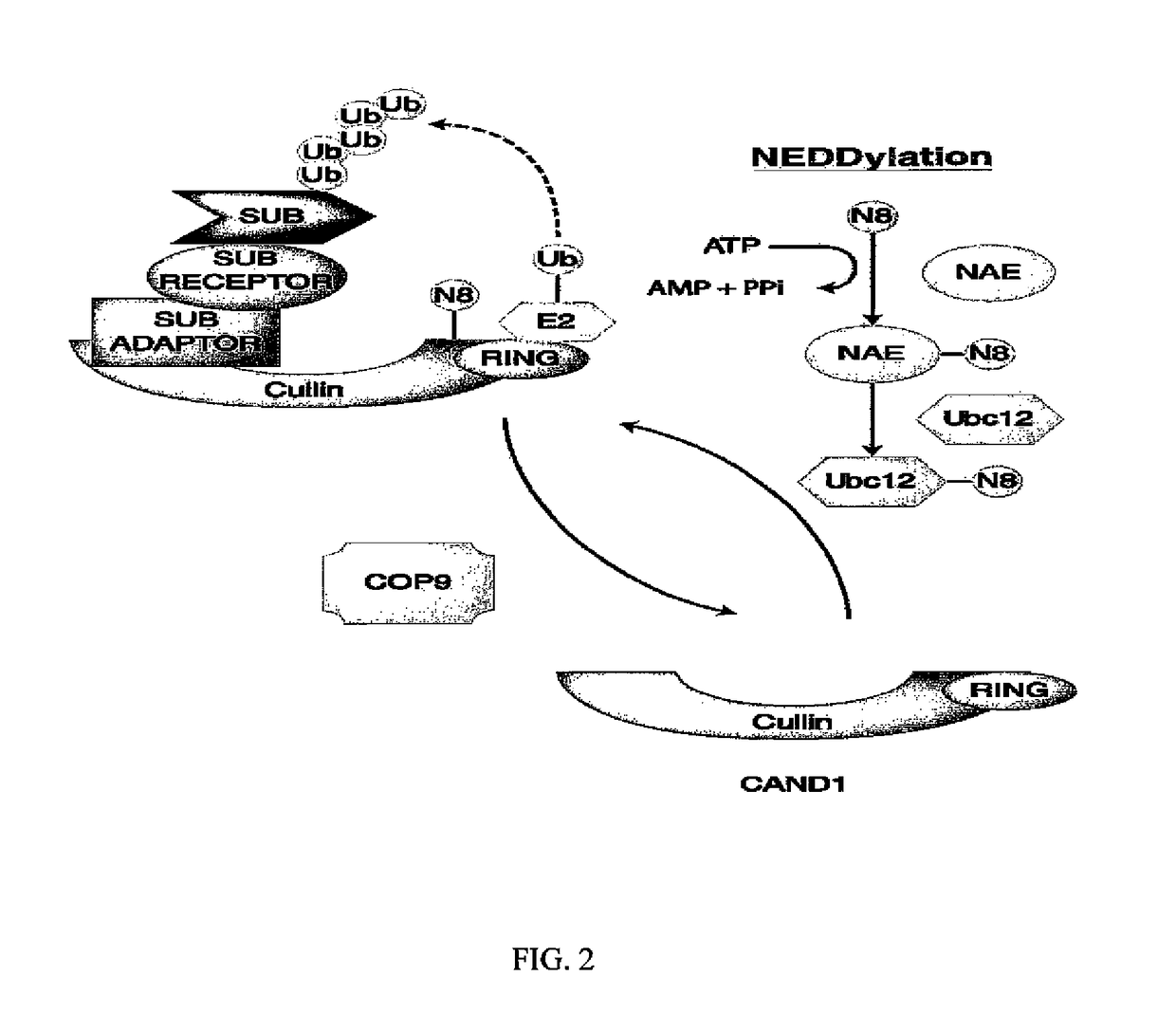
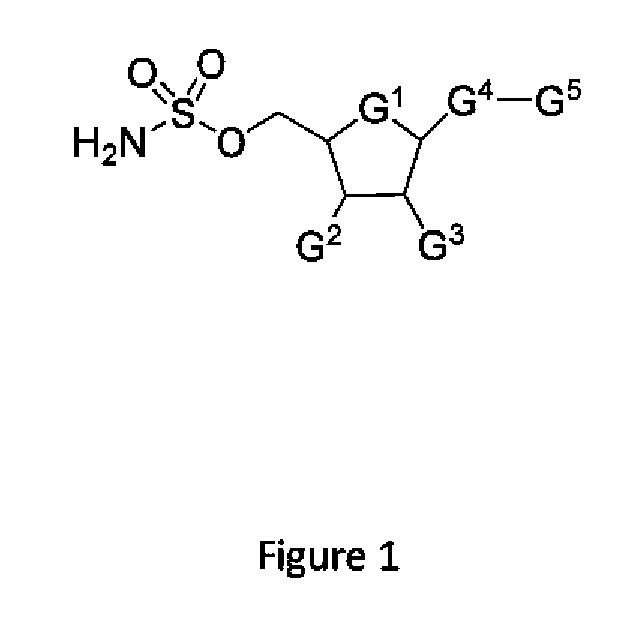
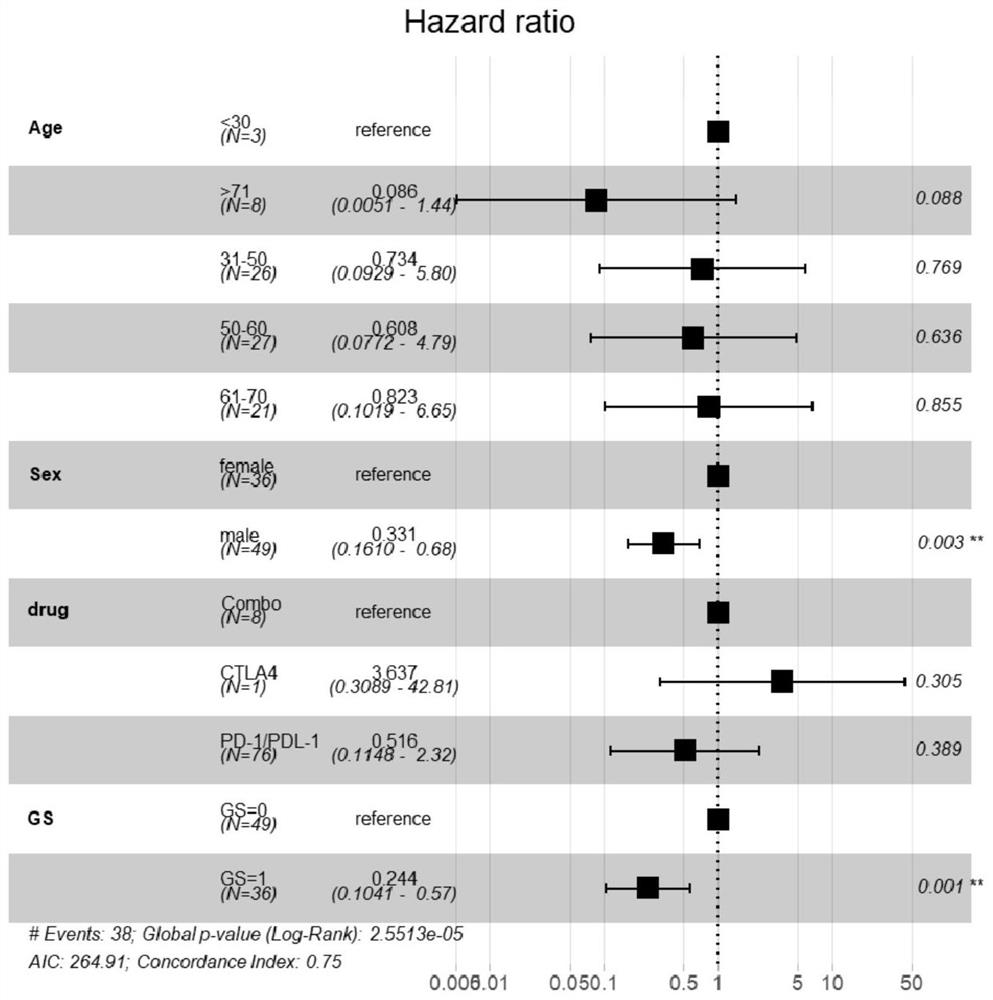
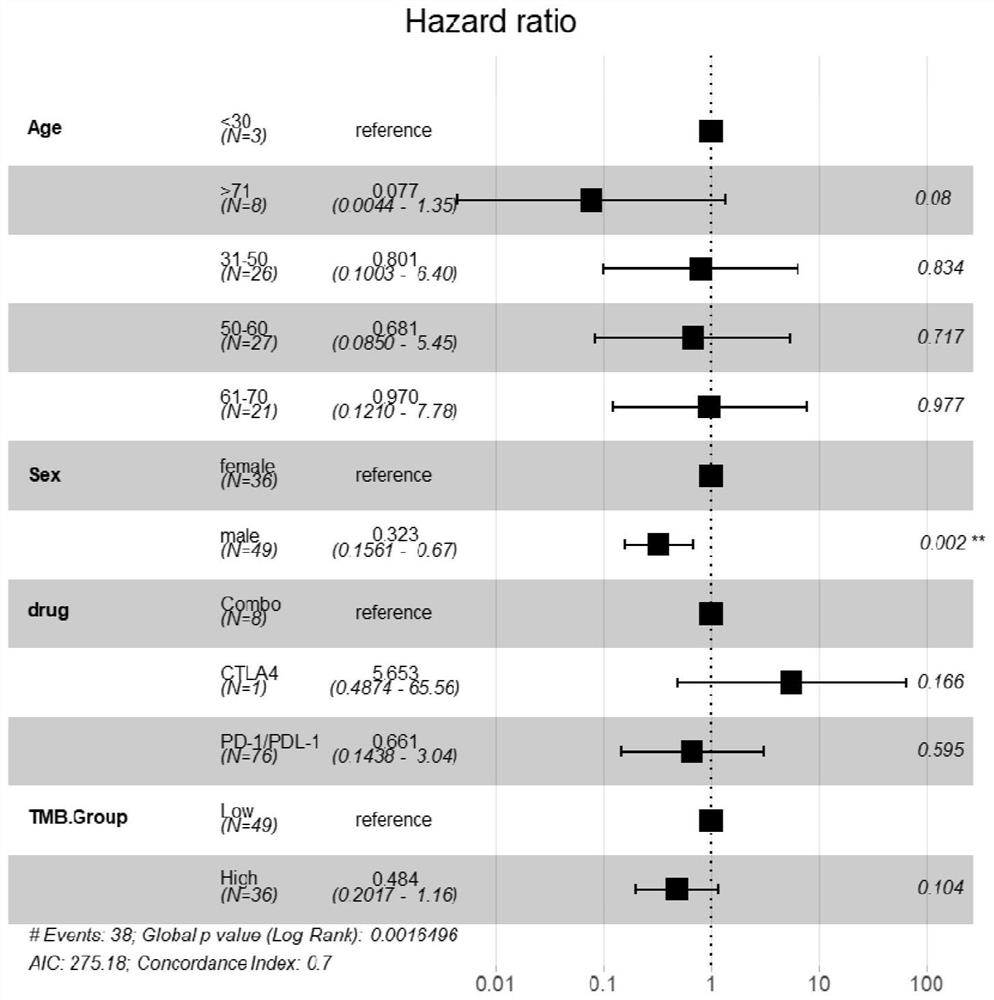
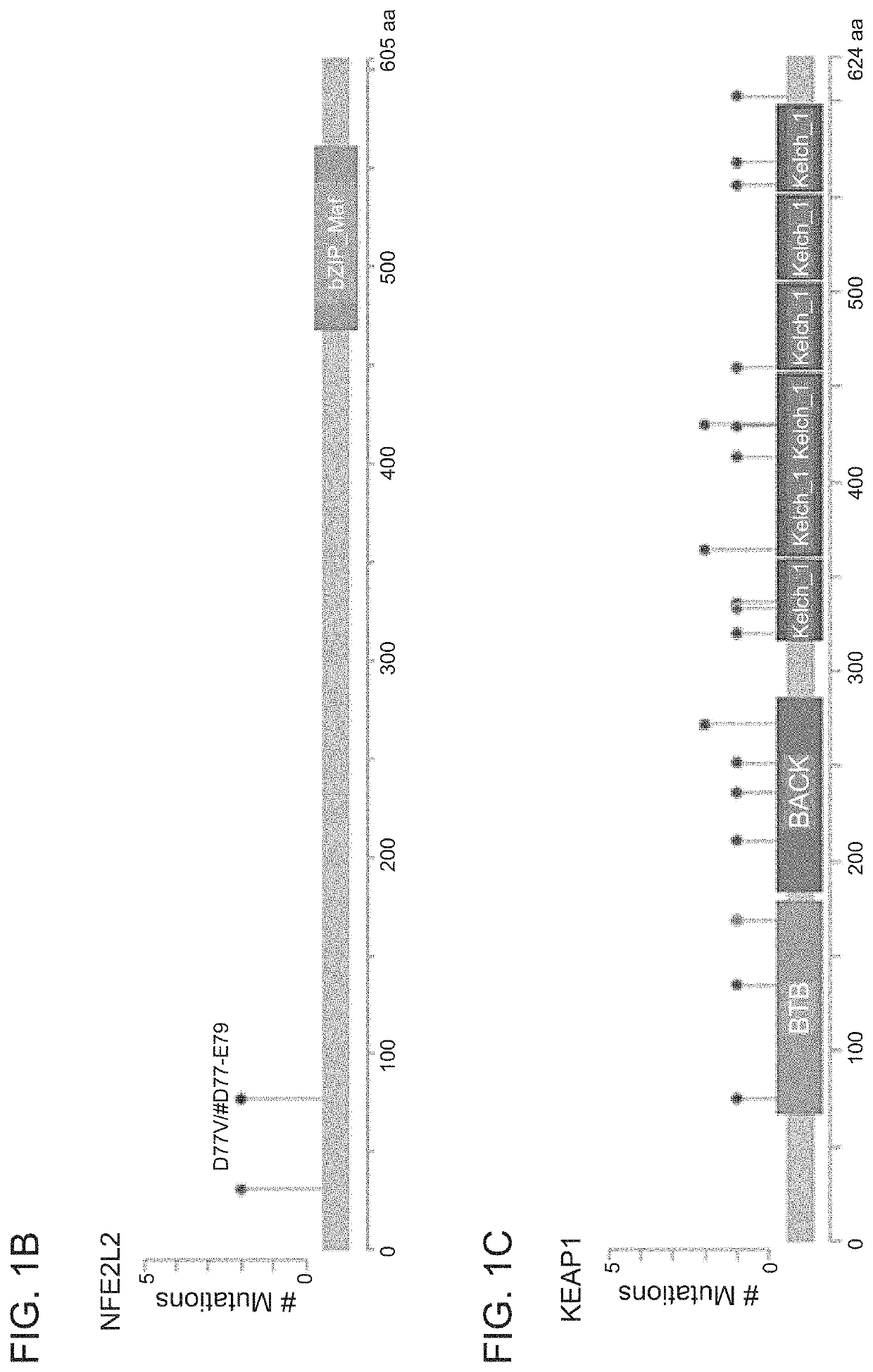
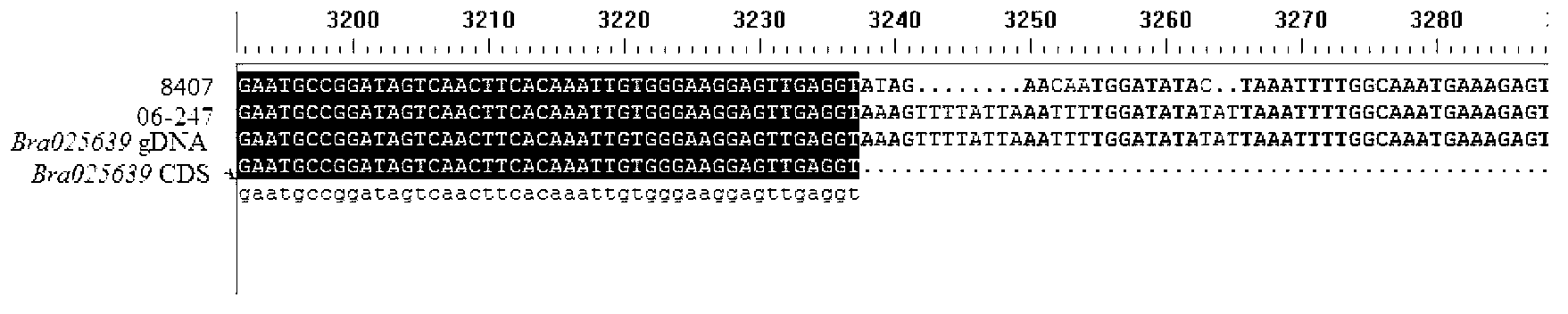Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
31 results about "Mutational status" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Compositions and methods for cancer diagnosis and therapy
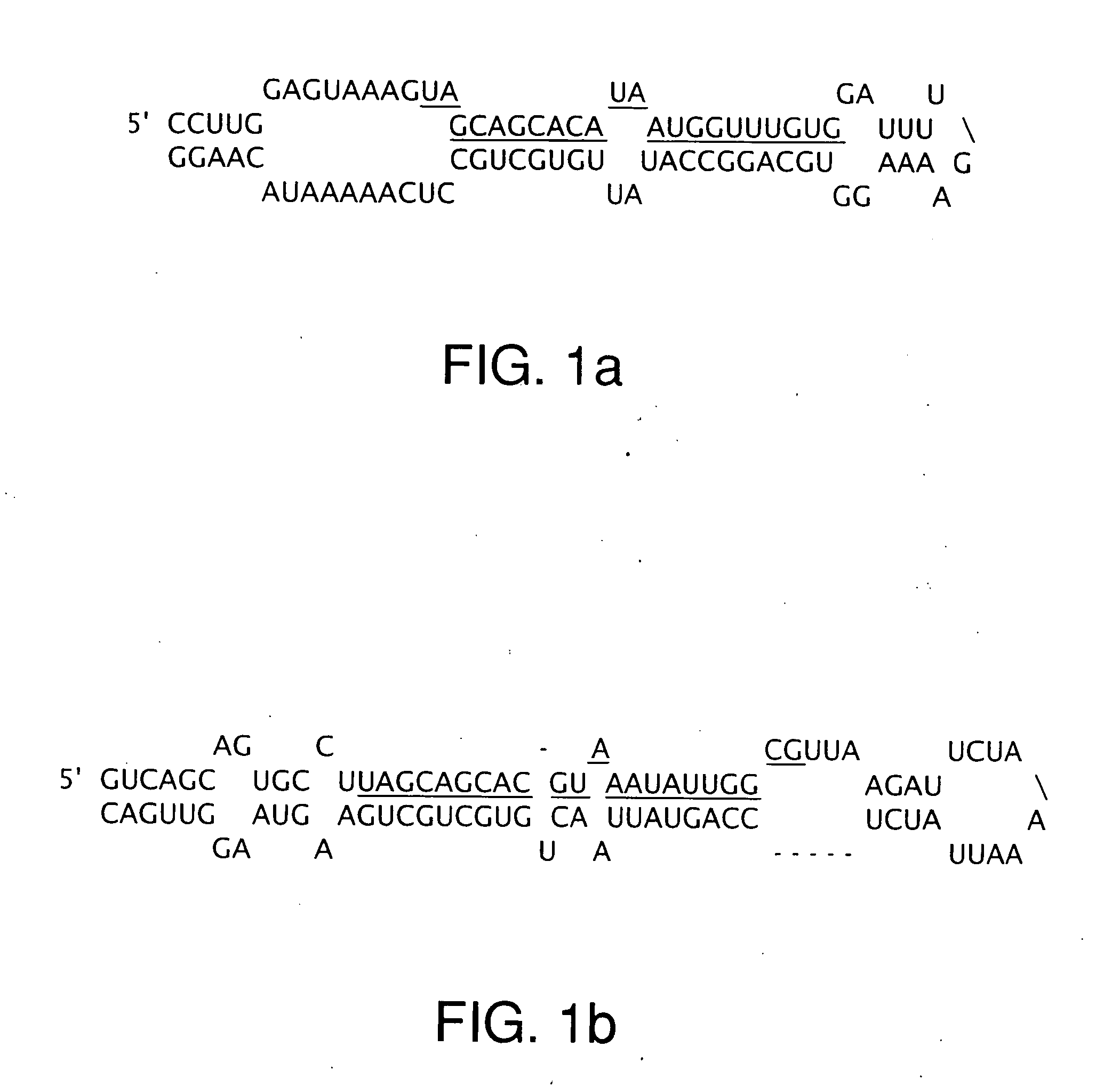
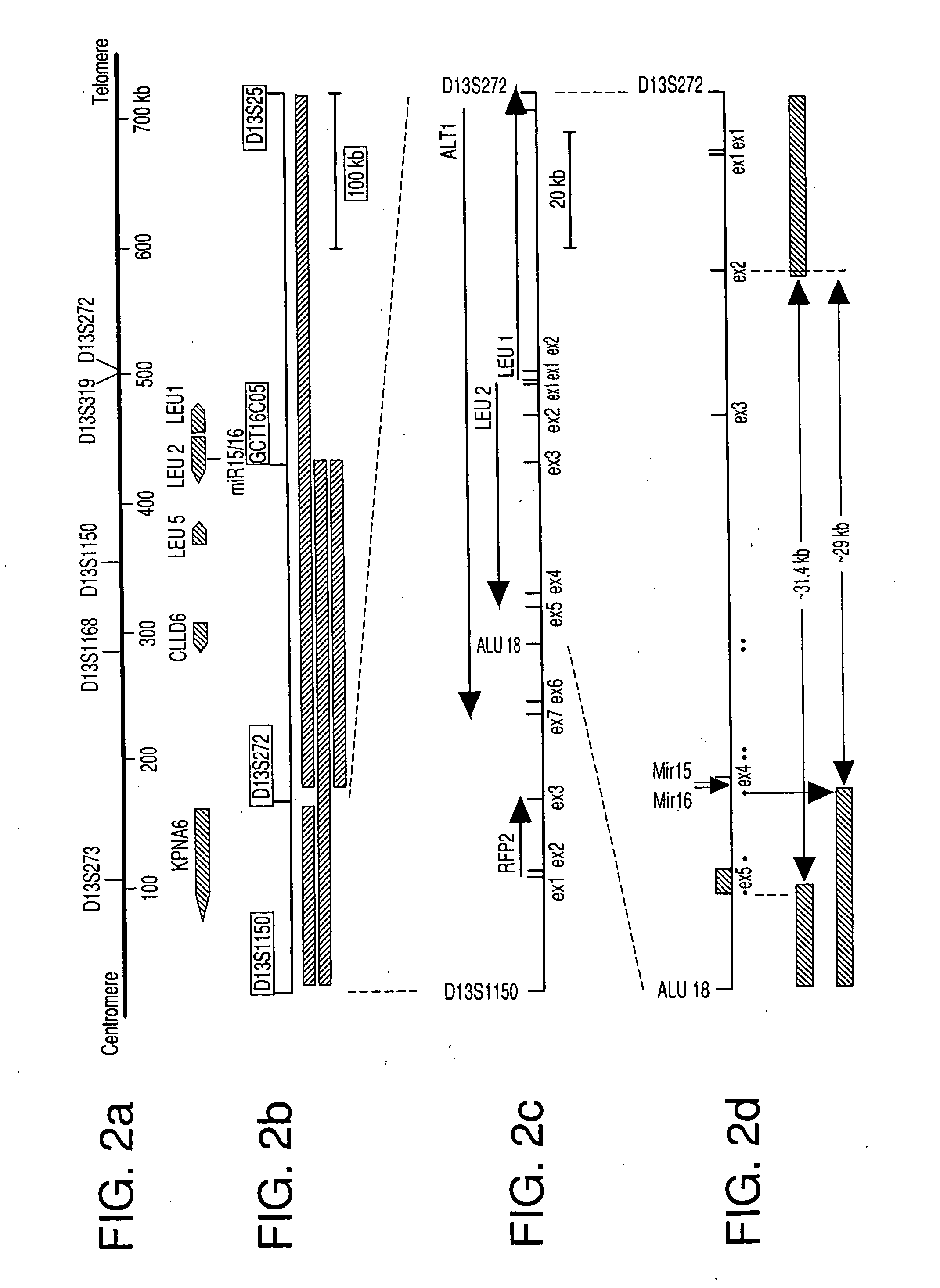
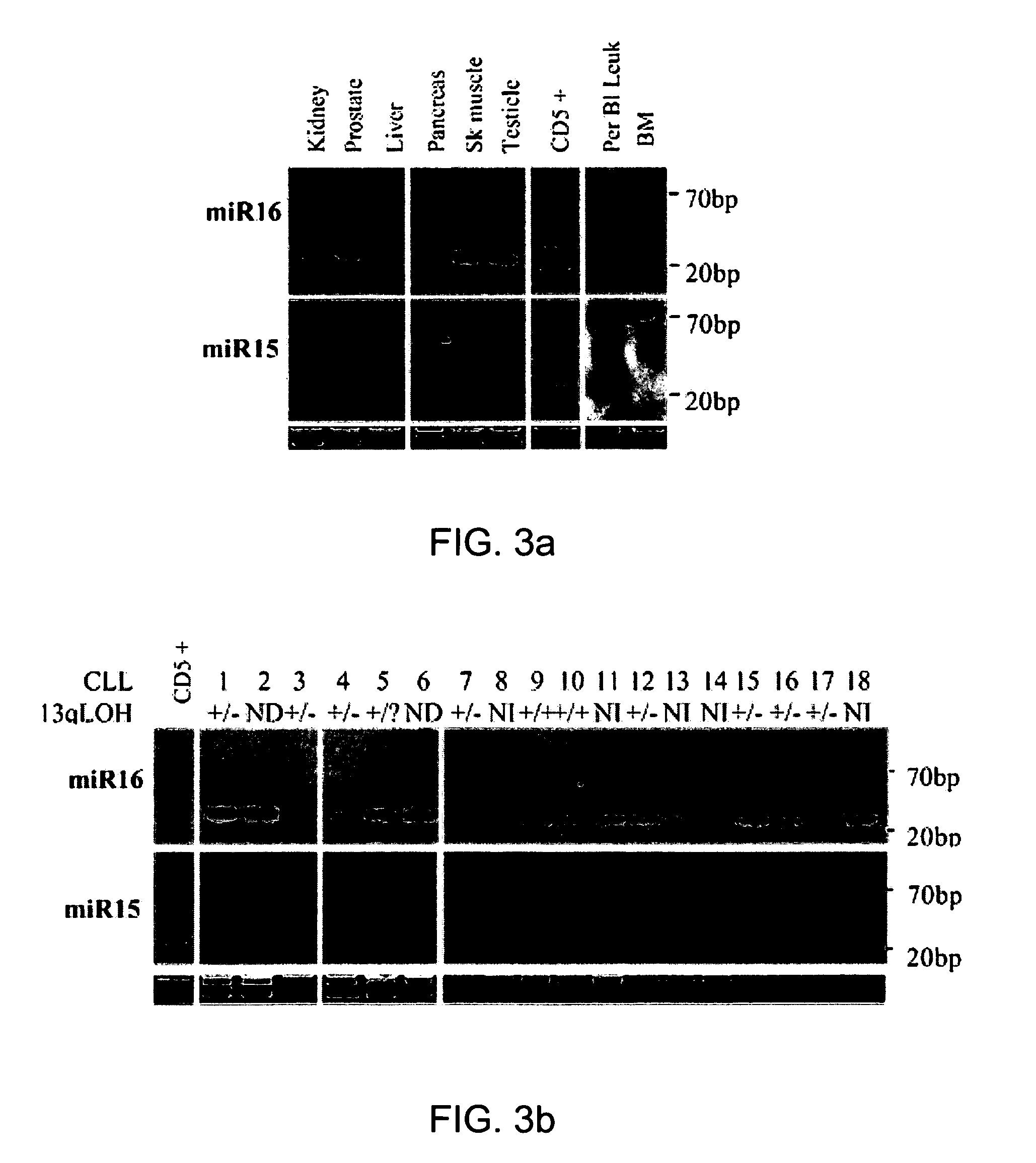
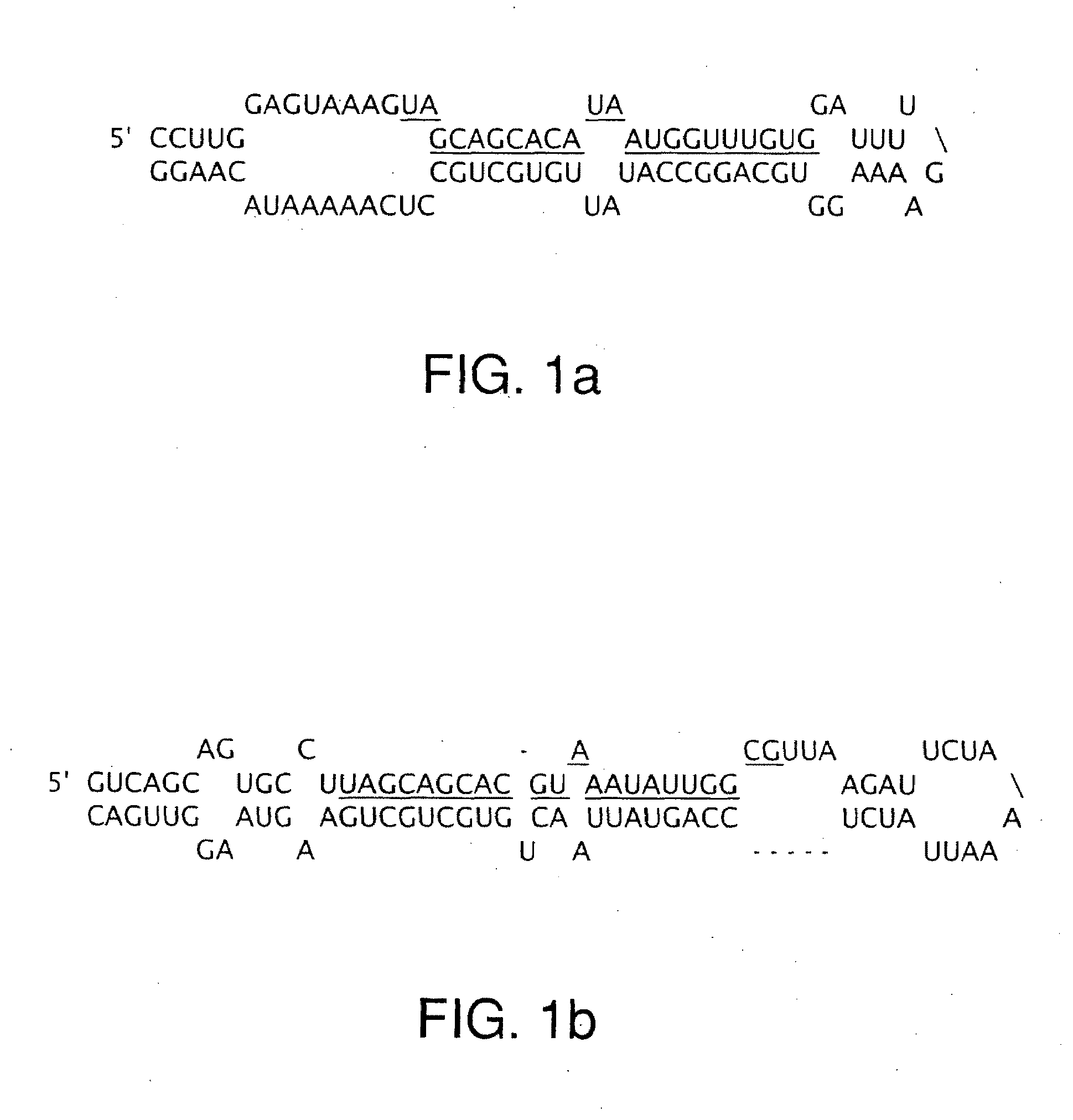
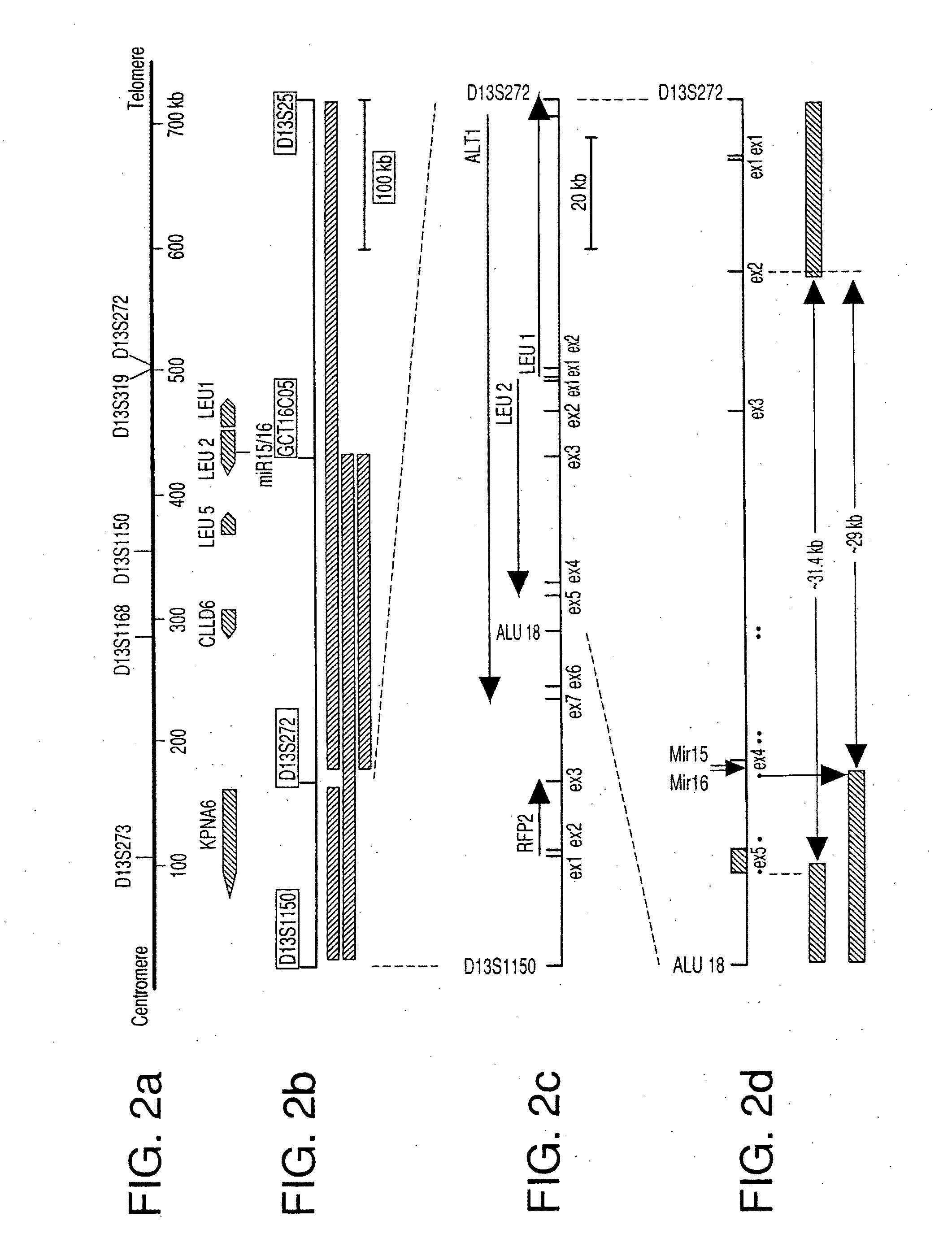
The miR15 and miR16 micro RNA genes are located at 13q14 within a 30 kb region of loss characteristic of cells from certain cancers, such as cells from chronic lymphocytic leukemia or prostate cancer. Chronic lymphocytic leukemia or prostate cancer can be diagnosed by detecting a reduction in miR15 or miR16 gene copy number, by determining miR15 or miR16 gene mutational status, or by detecting a reduction in the RNA transcribed from these genes. The miR15 or miR16 gene products can inhibit the neoplastic or tumorigenic growth of cancers such as chronic lymphocytic leukemia or prostate cancer cells when administered to subjects suffering from these diseases.
Owner:THOMAS JEFFERSON UNIV
Compositions and methods for cancer diagnosis and therapy
The miR15 and miR16 micro RNA genes are located at 13q14 within a 30 kb region of loss characteristic of cells from certain cancers, such as cells from chronic lymphocytic leukemia or prostate cancer. Chronic lymphocytic leukemia or prostate cancer can be diagnosed by detecting a reduction in miR15 or miR16 gene copy number, by determining miR15 or miR16 gene mutational status, or by detecting a reduction in the RNA transcribed from these genes. The miR15 or miR16 gene products can inhibit the neoplastic or tumorigenic growth of cancers such as chronic lymphocytic leukemia or prostate cancer cells when administered to subjects suffering from these diseases.
Owner:THOMAS JEFFERSON UNIV
Method for treating cancer based on mutation status of k-ras
The present invention provides methods and compositions for treating cancer by administering a) a composition comprising nanoparticles that comprise paclitaxel and an albumin and / or b) a therapeutic agent (e.g., gemcitabine) based upon K-ras mutation status.
Owner:ABRAXIS BIOSCI LLC
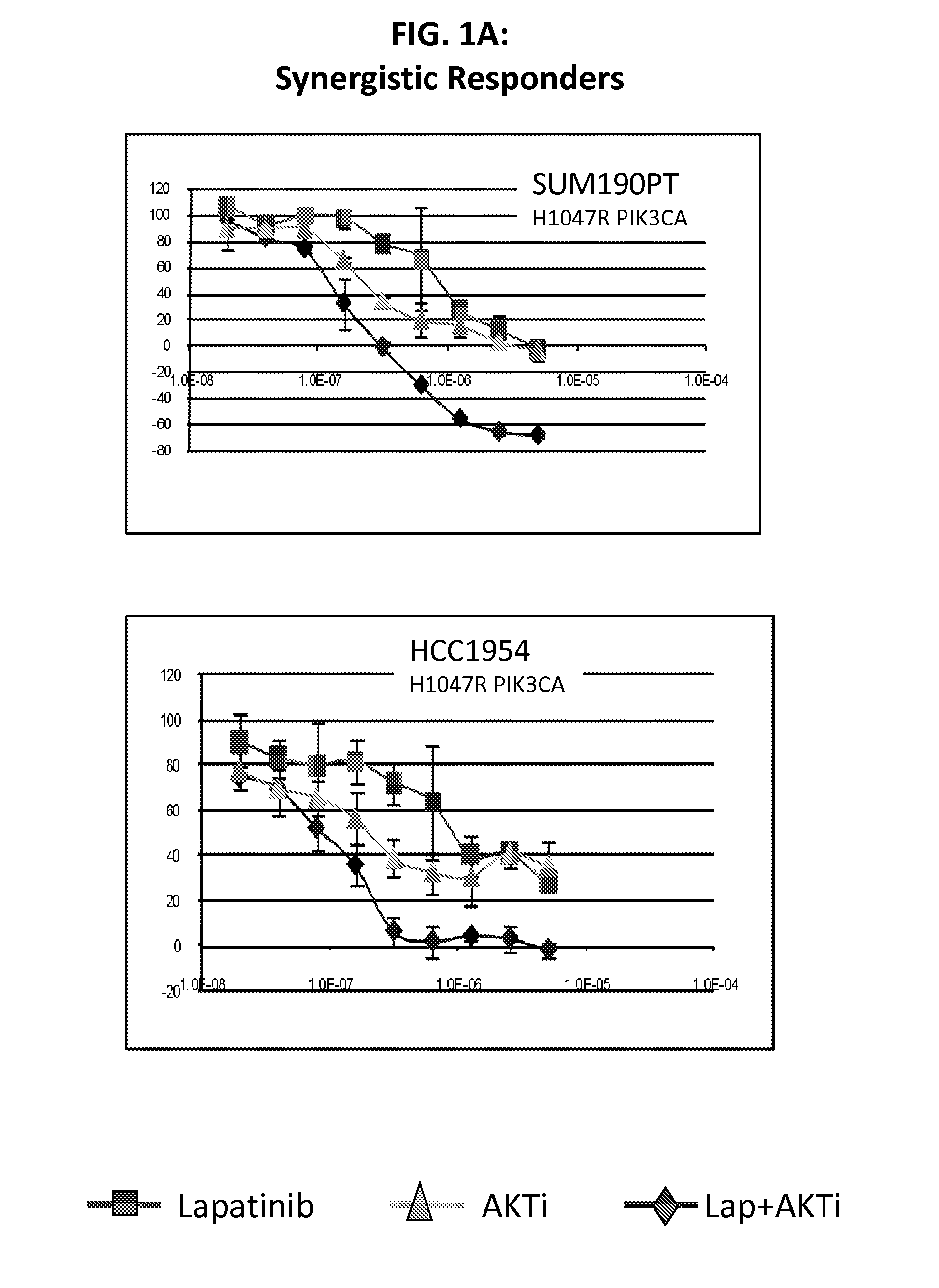
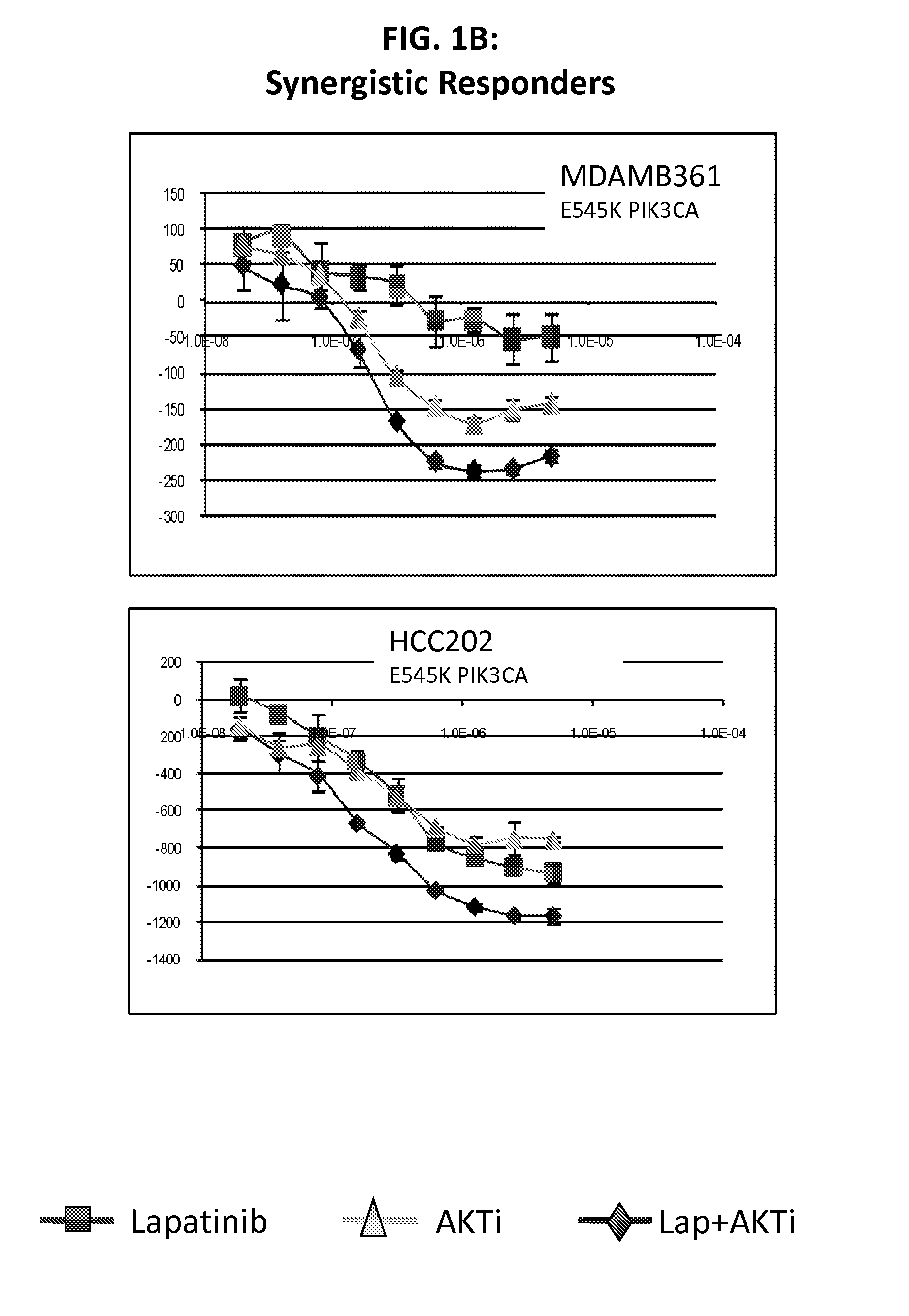
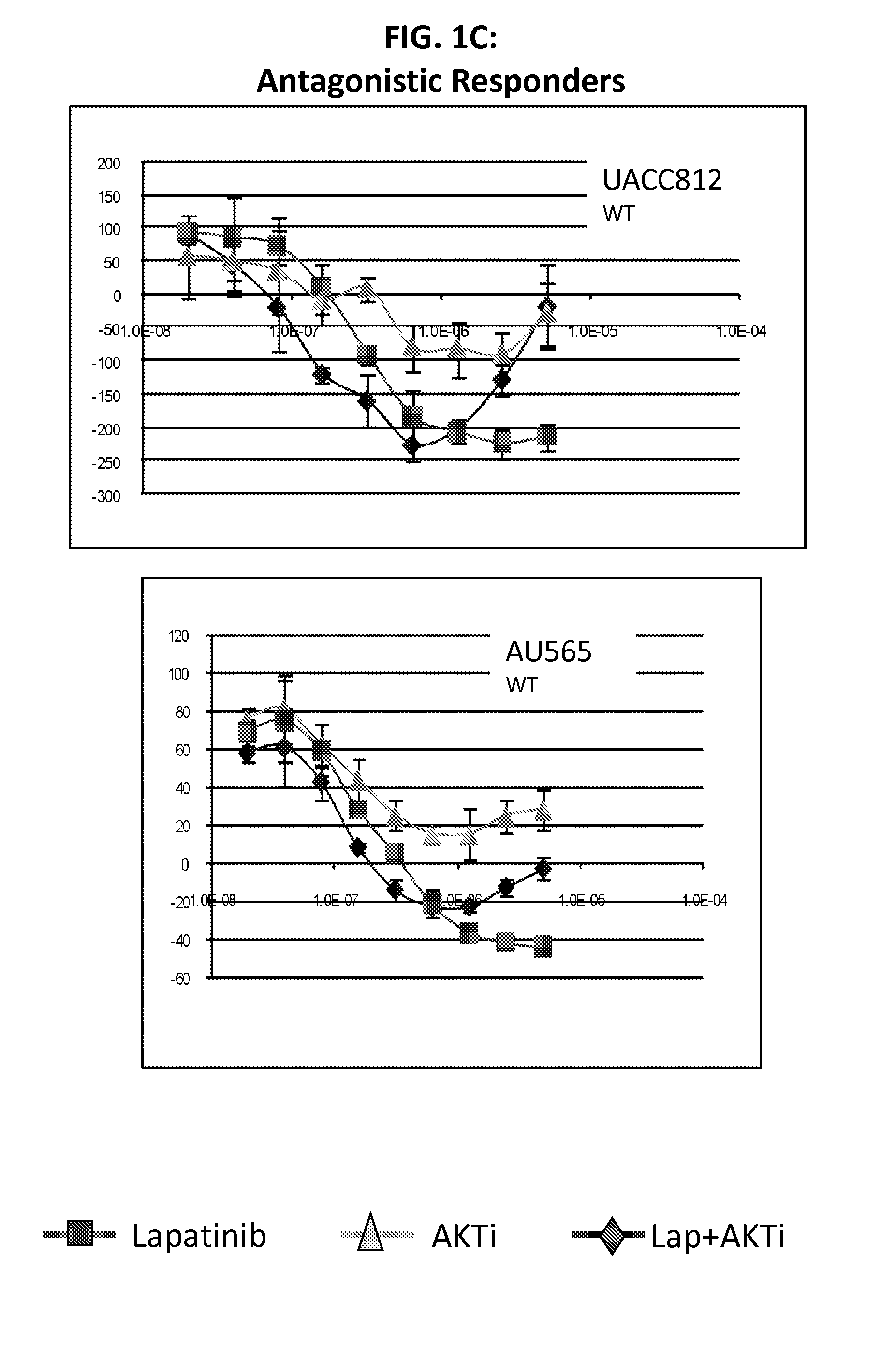
PIK3CA Mutation Status and SASH1 Expression Predicts Synergy Between Lapatinib and an AKT Inhibitor in HER2 Positive Breast Cancer
InactiveUS20130331405A1Improve expression levelOrganic active ingredientsBiocideProtein proteinHER2 Positive Breast Cancer
Methods for identifying a cancer patient, such as a breast cancer patient, suitable for treatment with a 4-anilinoquinazoline kinase inhibitor, such as lapatinib, and an AKT inhibitor, comprising detecting modulated expression of HER2 (ERBB2) and SASH1 or protein encoded thereof and detecting PIK3CA mutation status. High levels of expression in HER2 and high levels of SASH1 and / or positive PIK3CA mutation status indicate a patient that is suitable for treatment with a 4-anilinoquinazoline kinase inhibitor, such as lapatinib and an AKT inhibitor.
Owner:RGT UNIV OF CALIFORNIA
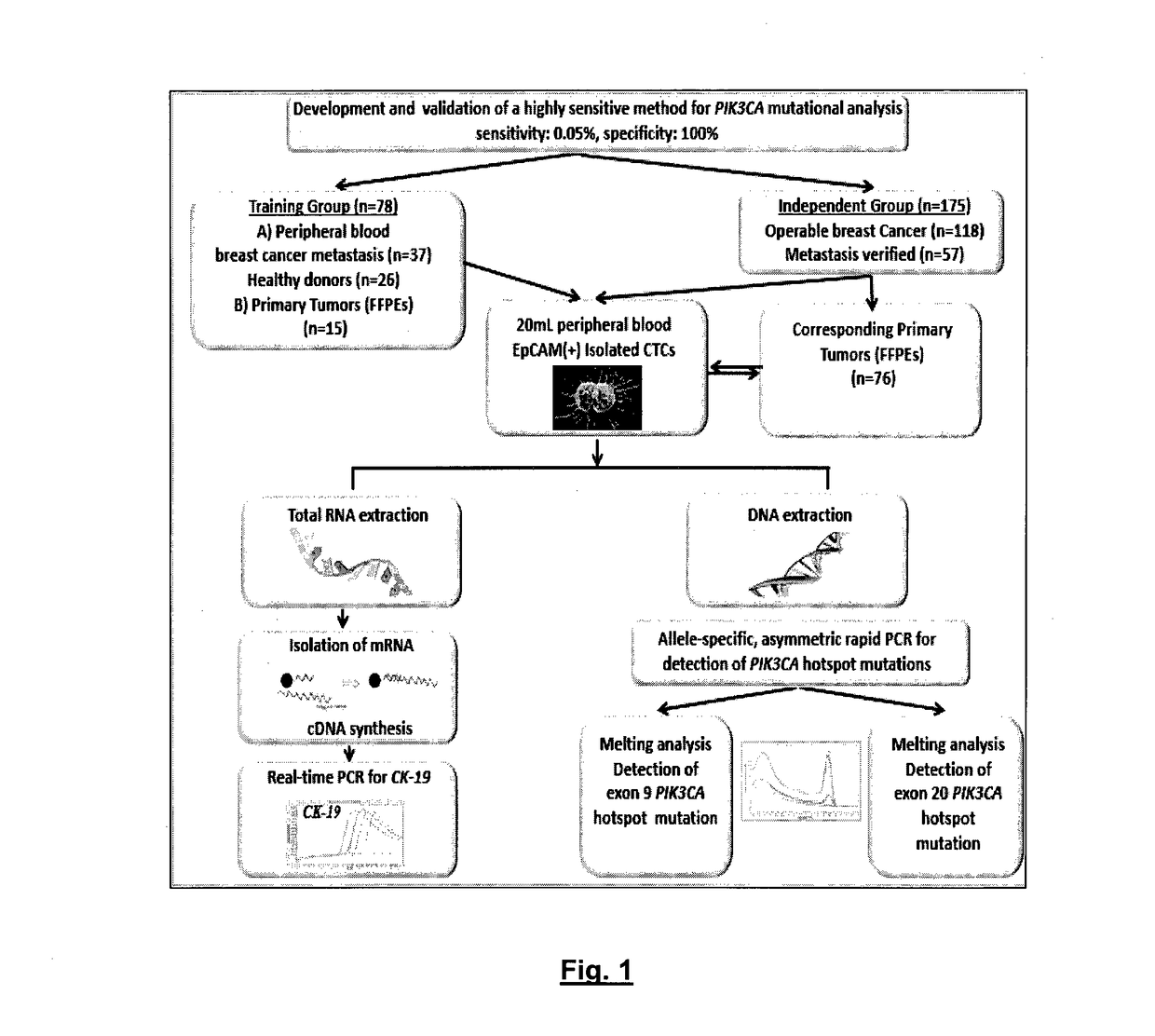
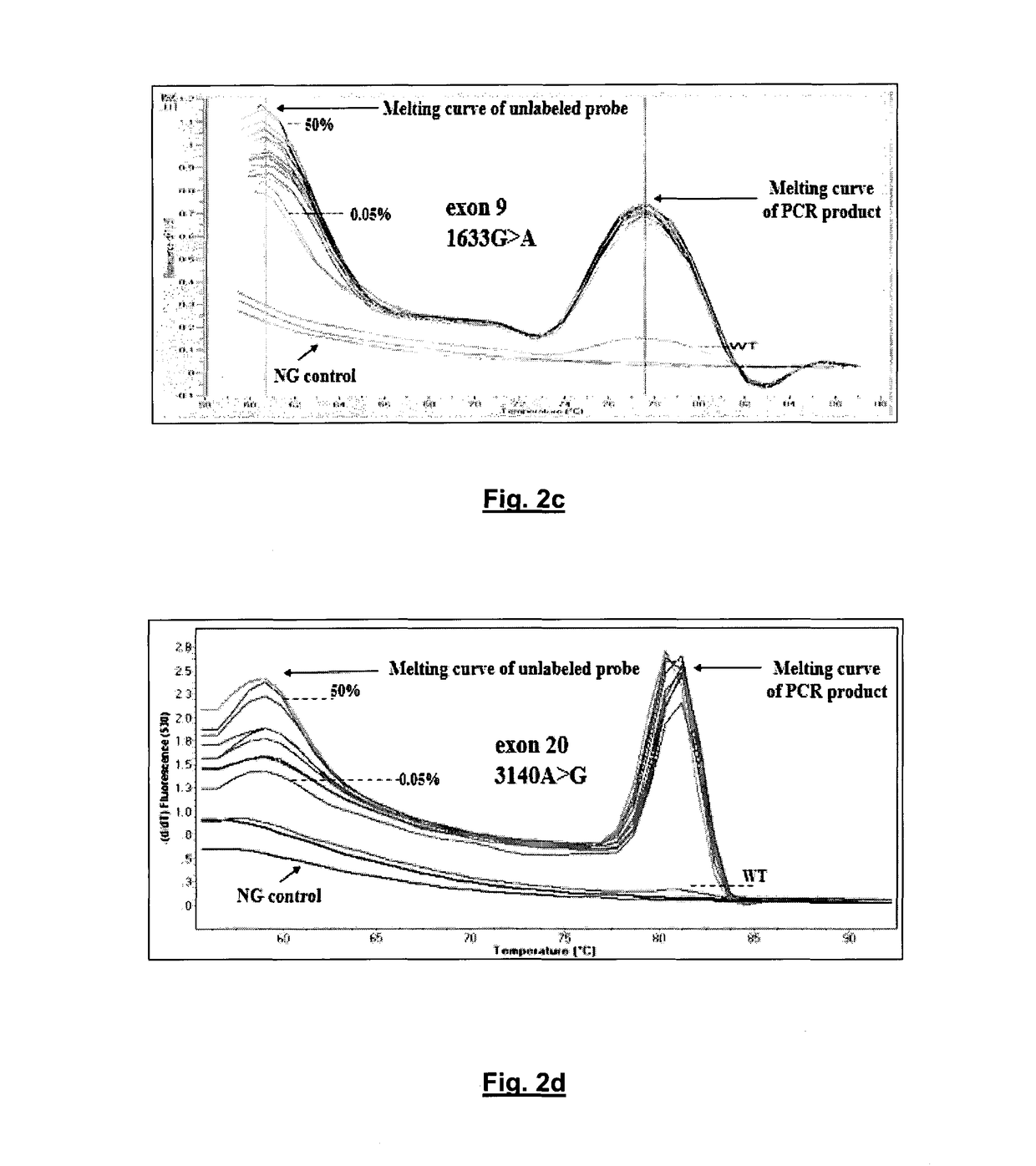
Method of determining pik3ca mutational status in a sample
ActiveUS20170233821A1High sensitivityEnhance rare allele detectionMicrobiological testing/measurementCell freeWild type
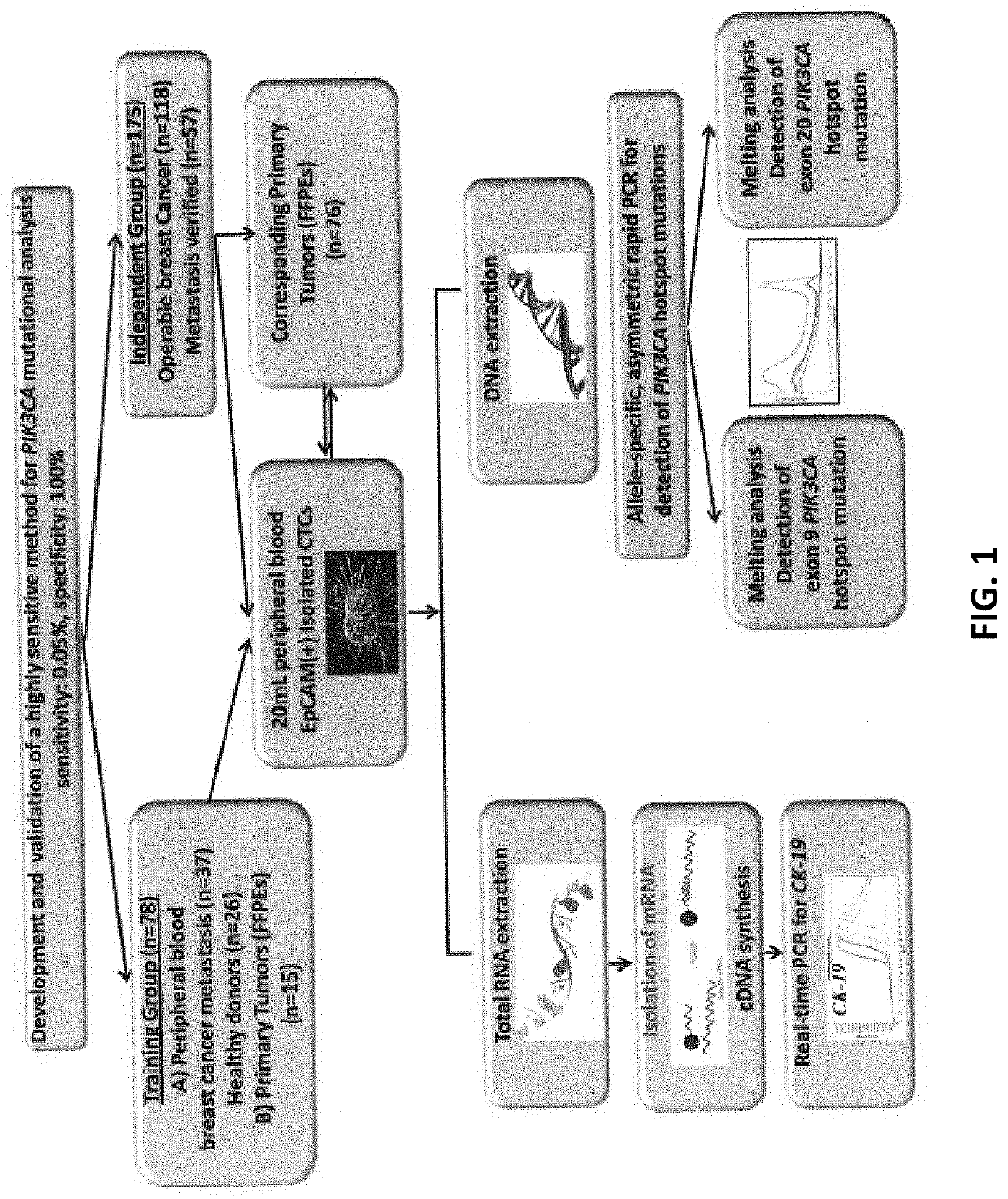
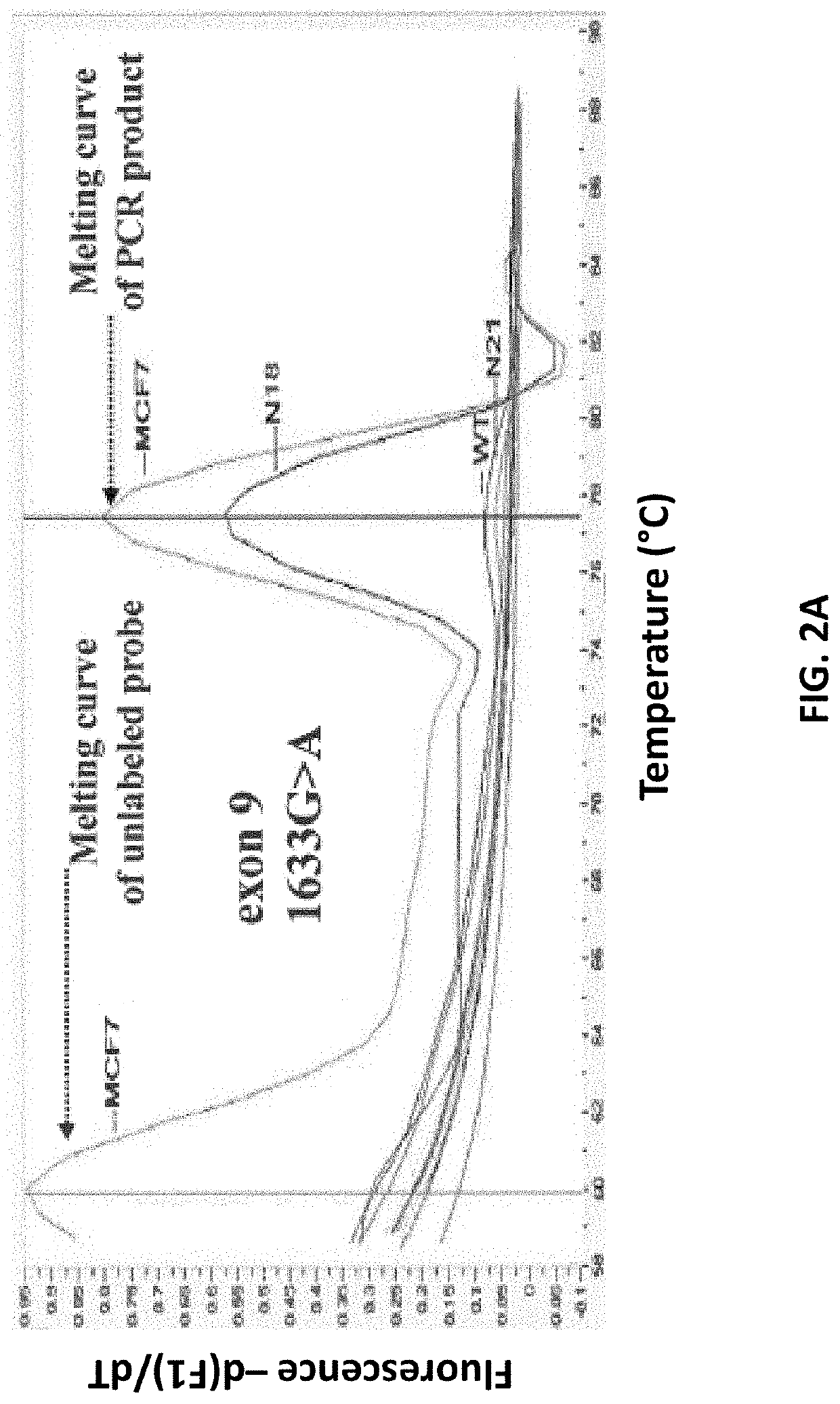
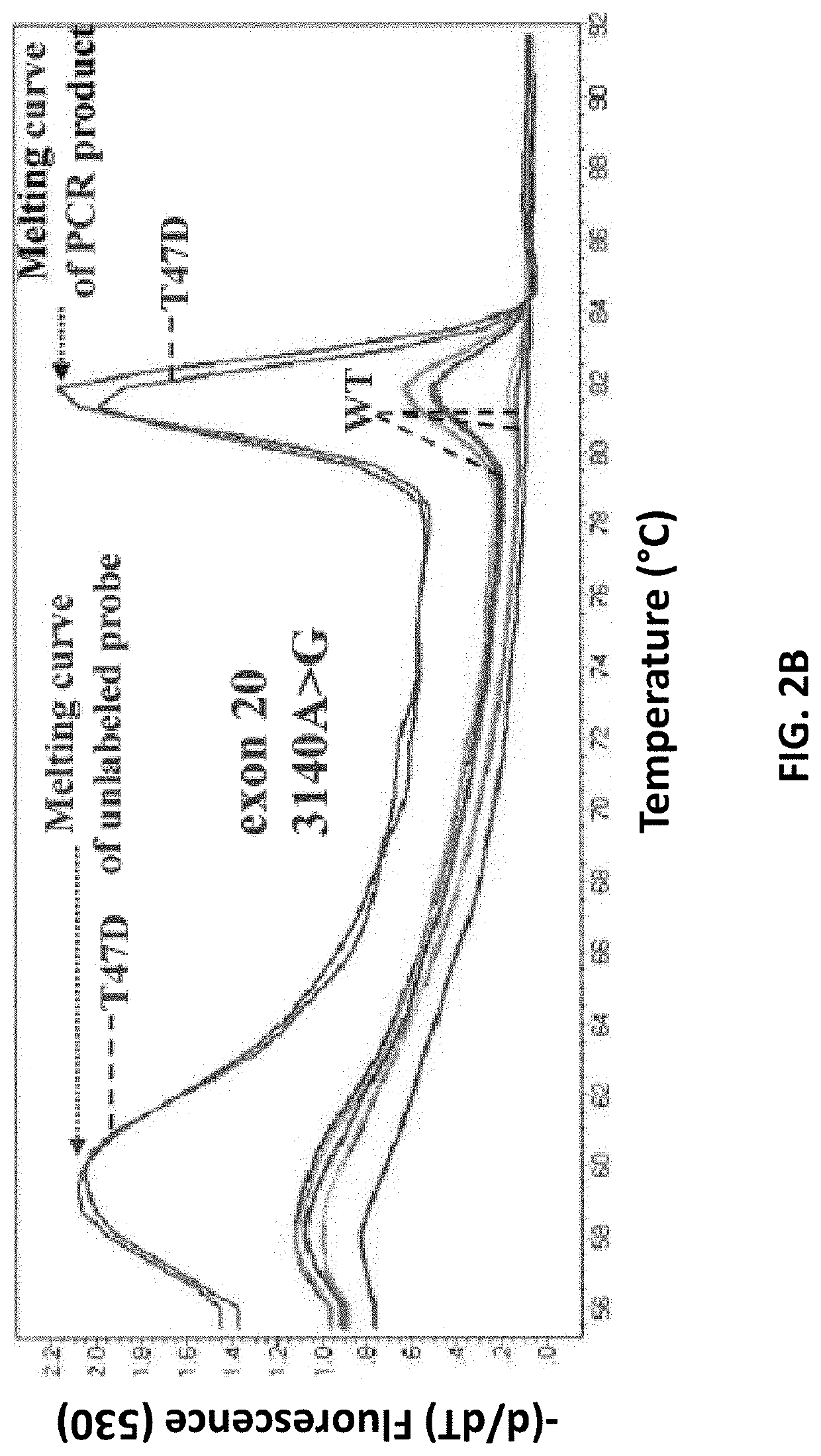
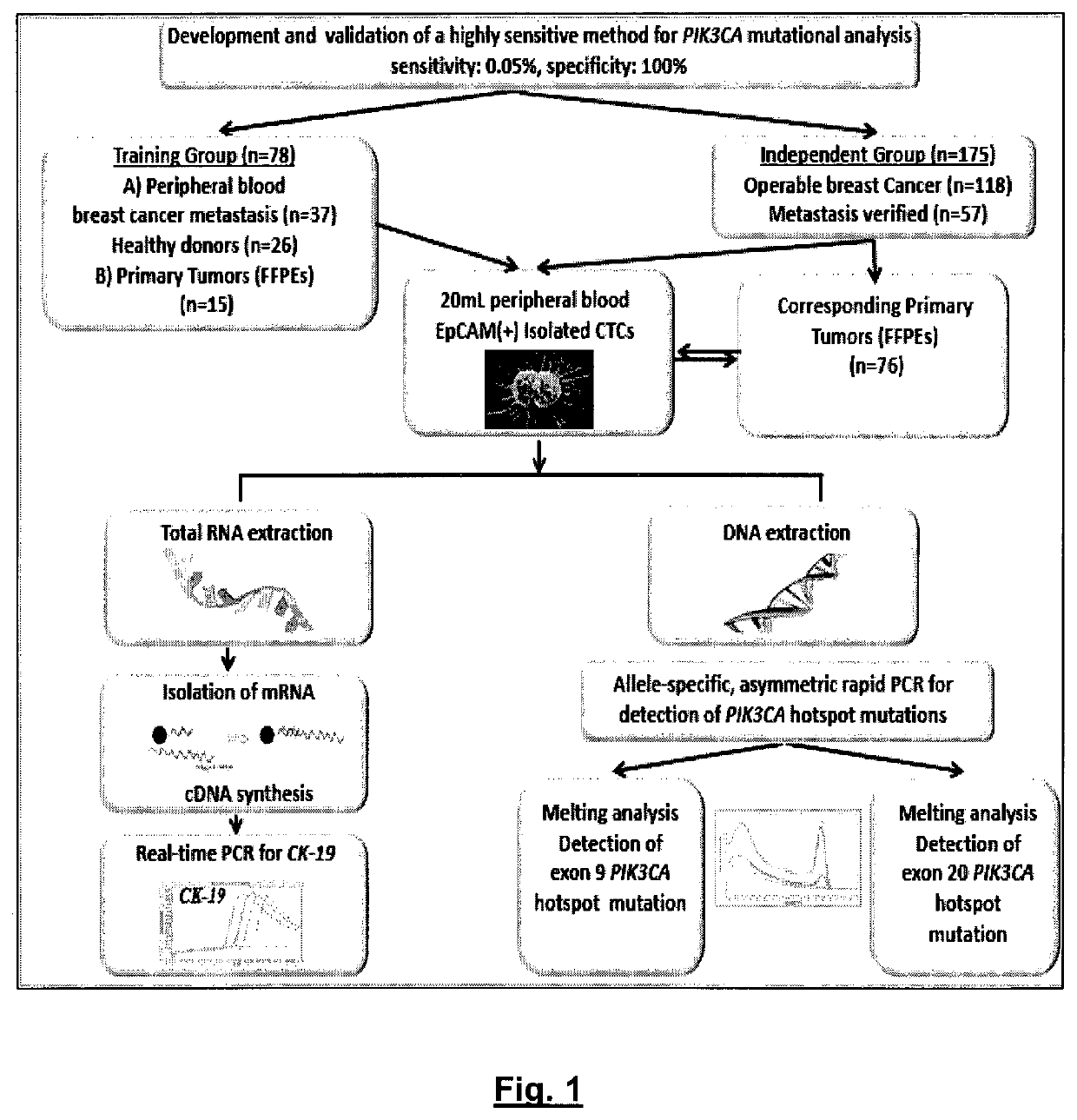
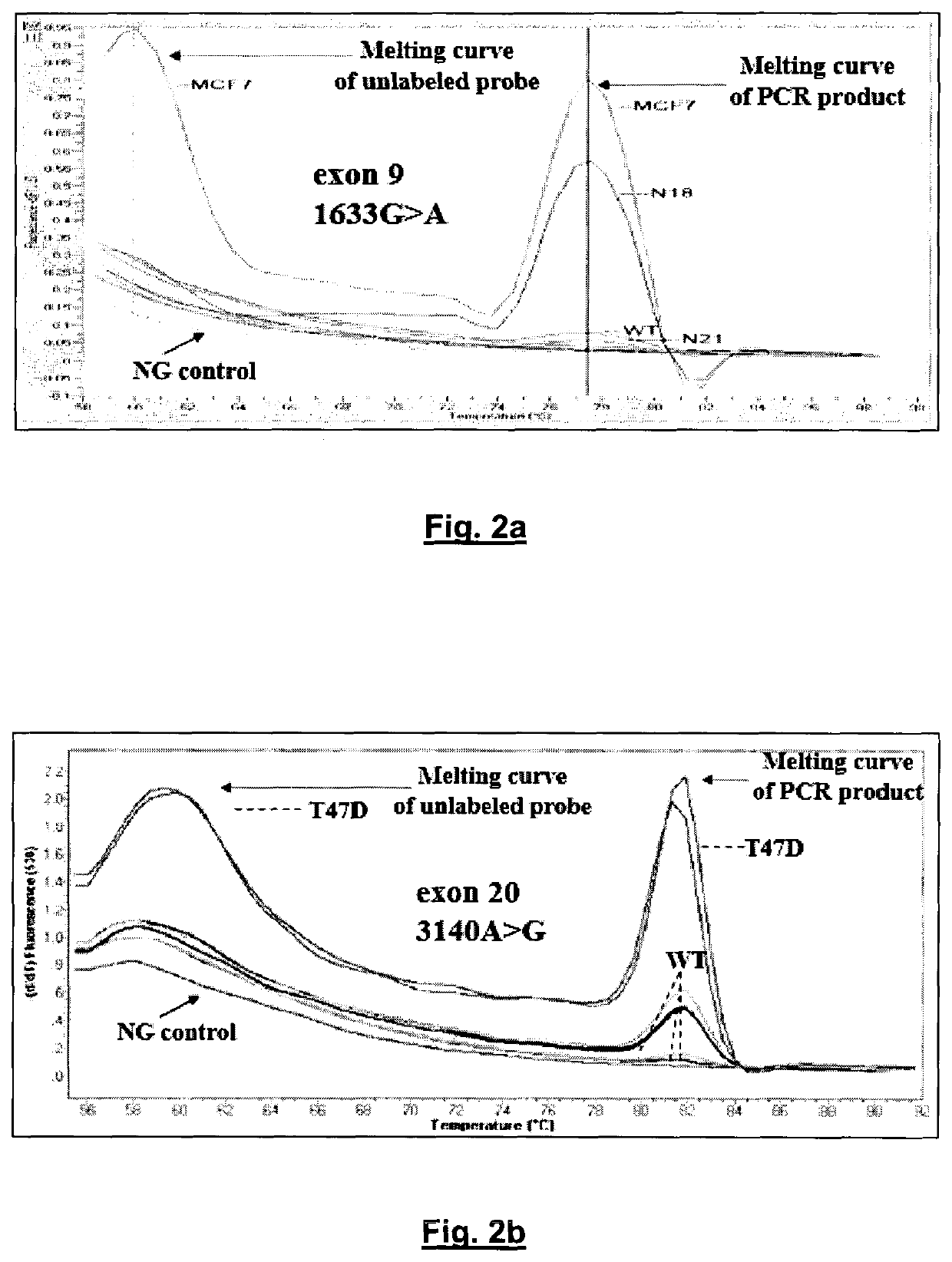
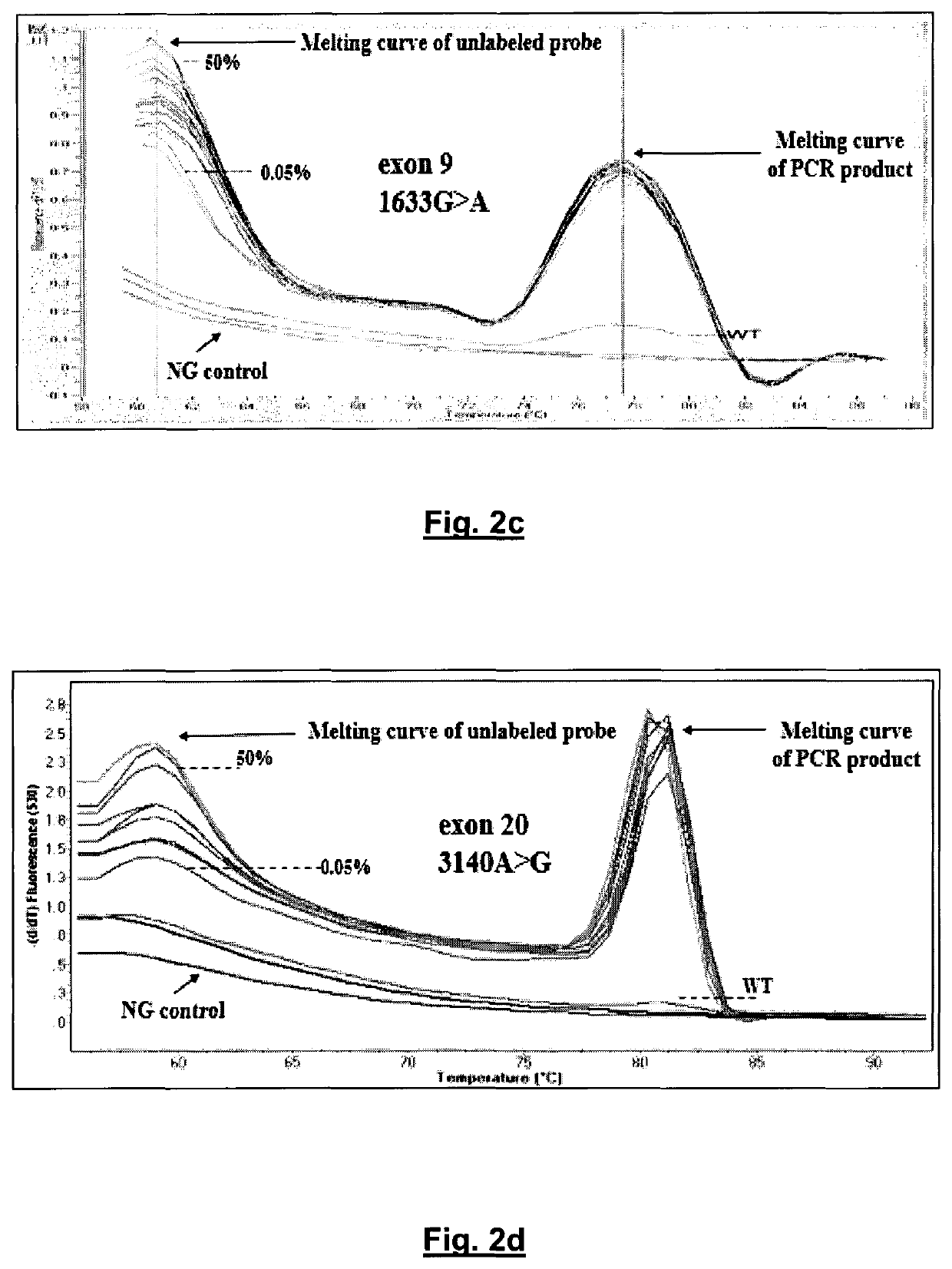
An ultra-sensitive, specific methodology for detecting PIK3CA mutations in biological samples of cancer patients, comprises a combination of allele-specific, asymmetric rapid PCR and melting analysis in a DNA sample from Circulating Tumor Cells, cell-free DNA in plasma / serum, or Formalin-Fixed Paraffin-Embedded tissues. Using the allele-specific primers for hotspot mutations in exons 9 and 20 (E545K and H1047R), detection can enhance amplification of mutant PIK3CA allele sequence, whereas presence of corresponding competitive blocking unlabeled probes for each exon can avoid non-specific amplification of wild-type PIK3CA sequence increasing the sensitivity and the specificity of method. The mutational detection is completed with melting curve analysis of the unlabeled probe and DNA template of the mutant PIK3CA sequence. Evaluation of PIK3CA mutational status on CTC in peripheral blood and cfDNA in plasma / serum of patients has potential for clinical applications and therapeutic interventions, since presence of PIK3CA mutations is associated with response to molecular targeted therapies.
Owner:PHARMASSIST
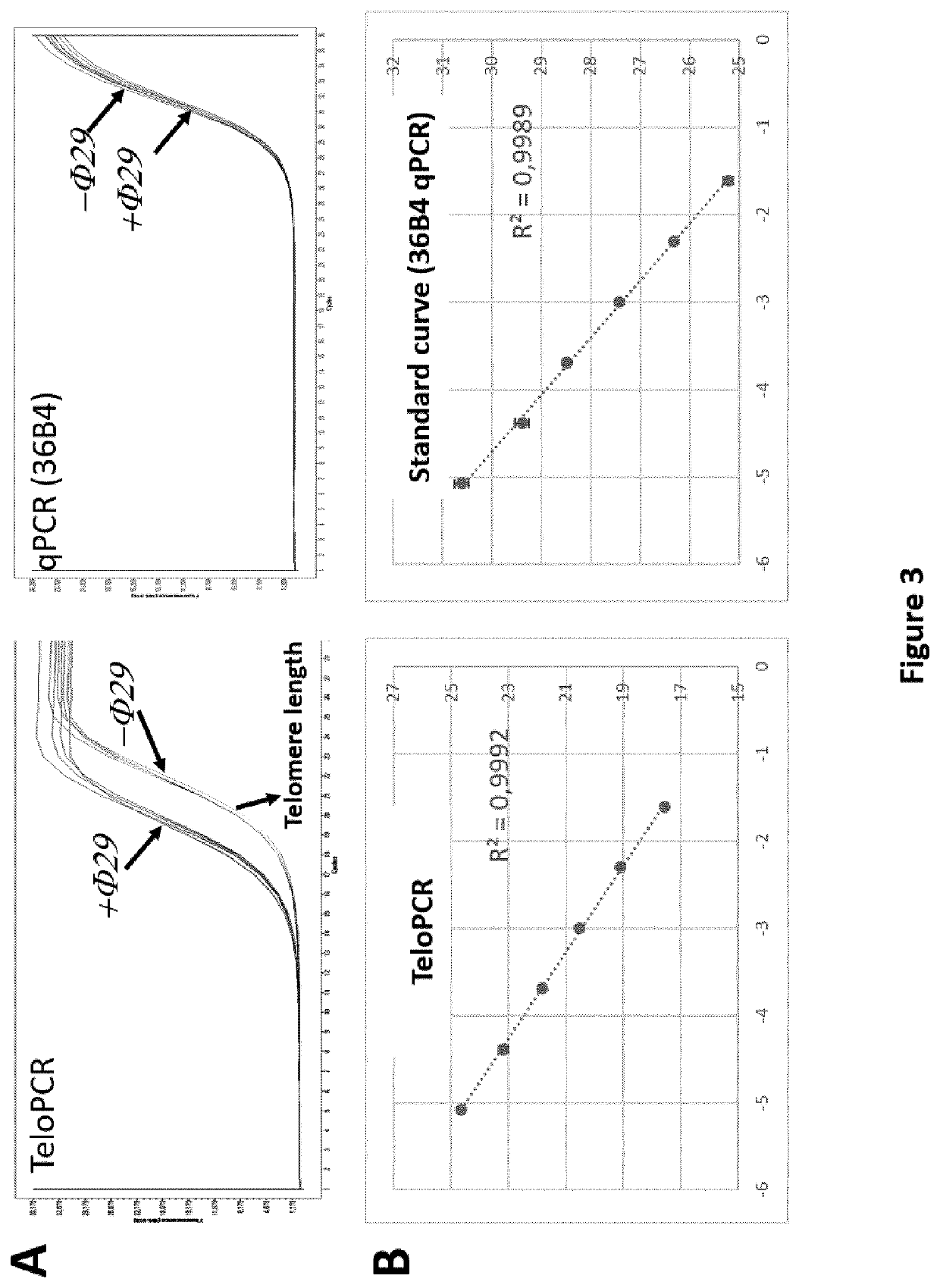
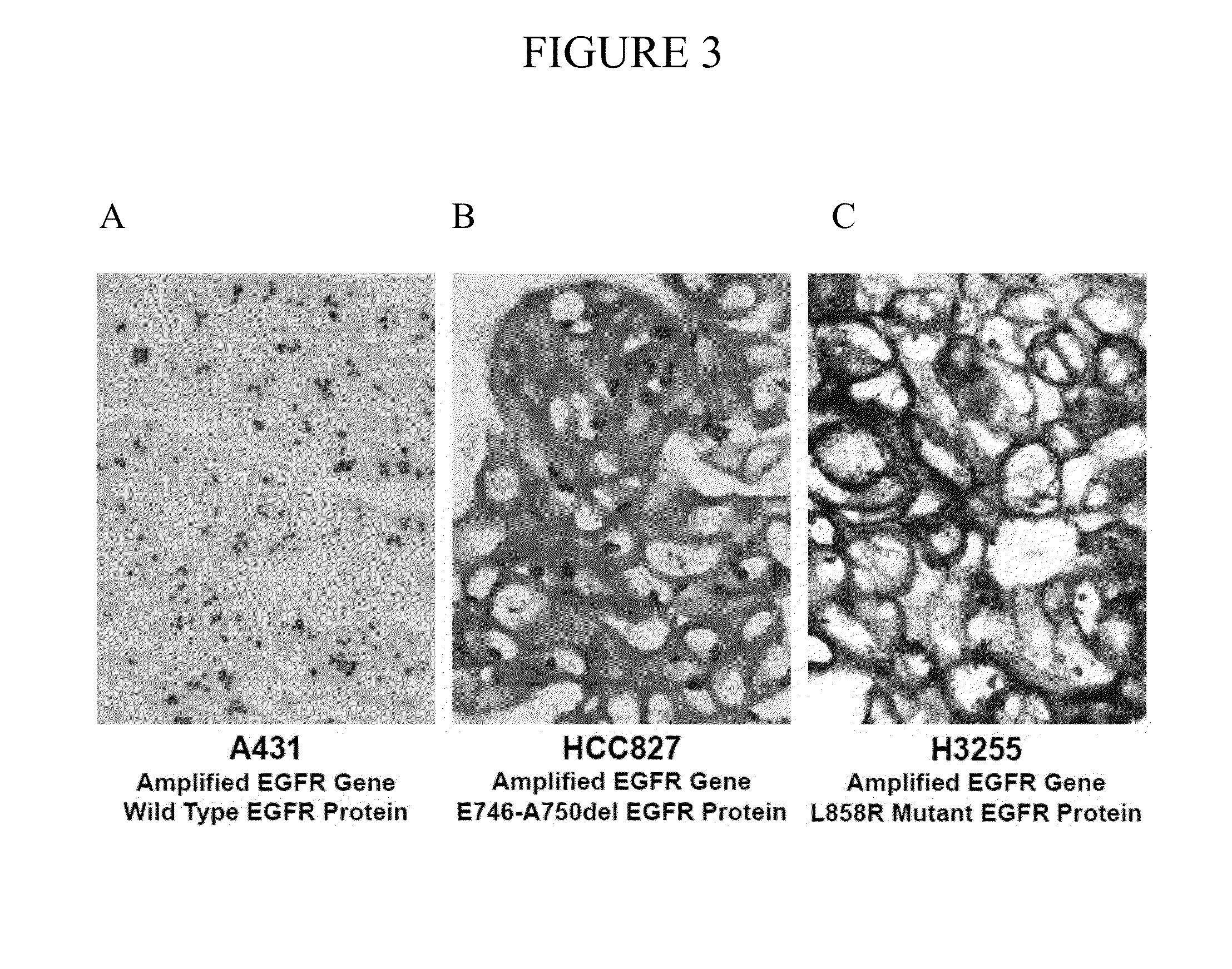
Simultaneous detection of mutational status and gene copy number
InactiveUS20120264127A1Material analysis by observing effect on chemical indicatorMicrobiological testing/measurementTissue sampleExon
The present invention provides compositions and methods for simultaneously detecting mutational status and gene copy number. In particular, the present invention provides simultaneous measurement of gene copy number and detection of the L858R and Exon 19 del mutations in a tissue sample.
Owner:VENTANA MEDICAL SYST INC
Methods for diagnosing and treating cancer by means of the expression status and mutational status of nrf2 and downstream target genes of said gene
ActiveUS20190218618A1Organic active ingredientsMicrobiological testing/measurementCancer researchAntagonist
The invention provides methods of identifying a subject having cancer, such as lung cancer, by analyzing expression levels of one or more NRF2 splice variants or NRF2 target genes. The invention also provides methods of treating cancer in a subject with a NRF2 pathway antagonist, wherein the subject expresses one or more NRF2 splice variants or overexpresses one or more NRF2 target genes.
Owner:GENENTECH INC +1
Methods for quantification of nucleosome modifications and mutations at genomic loci and clinical applications thereof
PendingCN111836907AMicrobiological testing/measurementOrganic compound librariesDiseaseEpigenetic Profile
The invention relates to clinical applications of quantitative chromatin mapping assays, such as chromatin immunoprecipitation assays and assays using tethered enzymes (e.g., chromatin immunocleavage(ChIC) and cleavage under targets & release using nuclease (CUT&RUN )). The methods may be used to detect and quantitate the presence of epigenetic modifications and mutations on nucleosomes (histonesand / or DNA) from biological samples, monitor changes in the status of modifications and mutations, monitor the effectiveness of epigenetic and mutation therapies, select suitable treatments for a disease, determine the prognosis of a subject, identify biomarkers of a disease, and screen for agents that modify epigenetic or mutation status. The invention further relates to kits for use in the methods of the invention.
Owner:埃皮赛佛尔有限公司
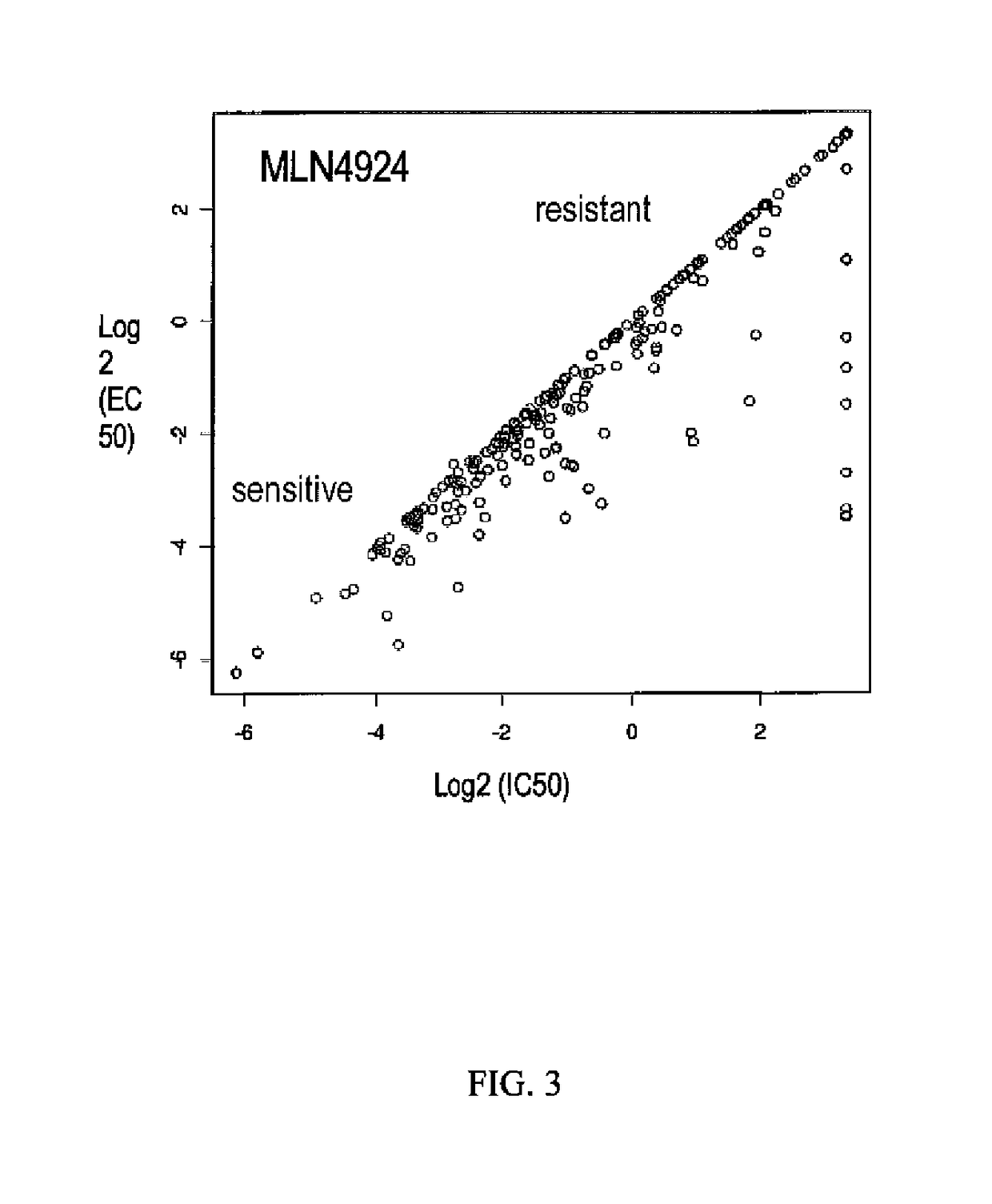
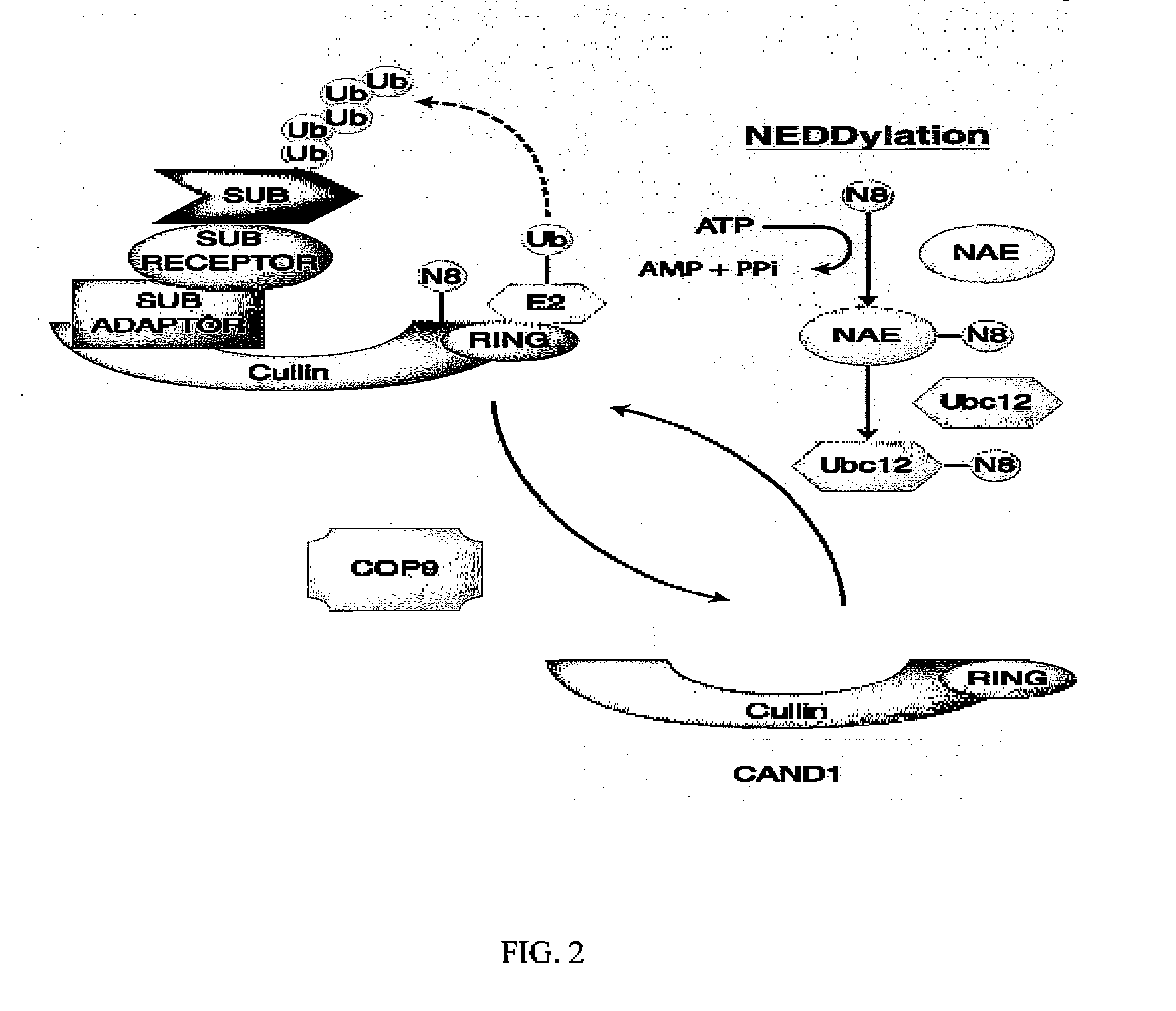
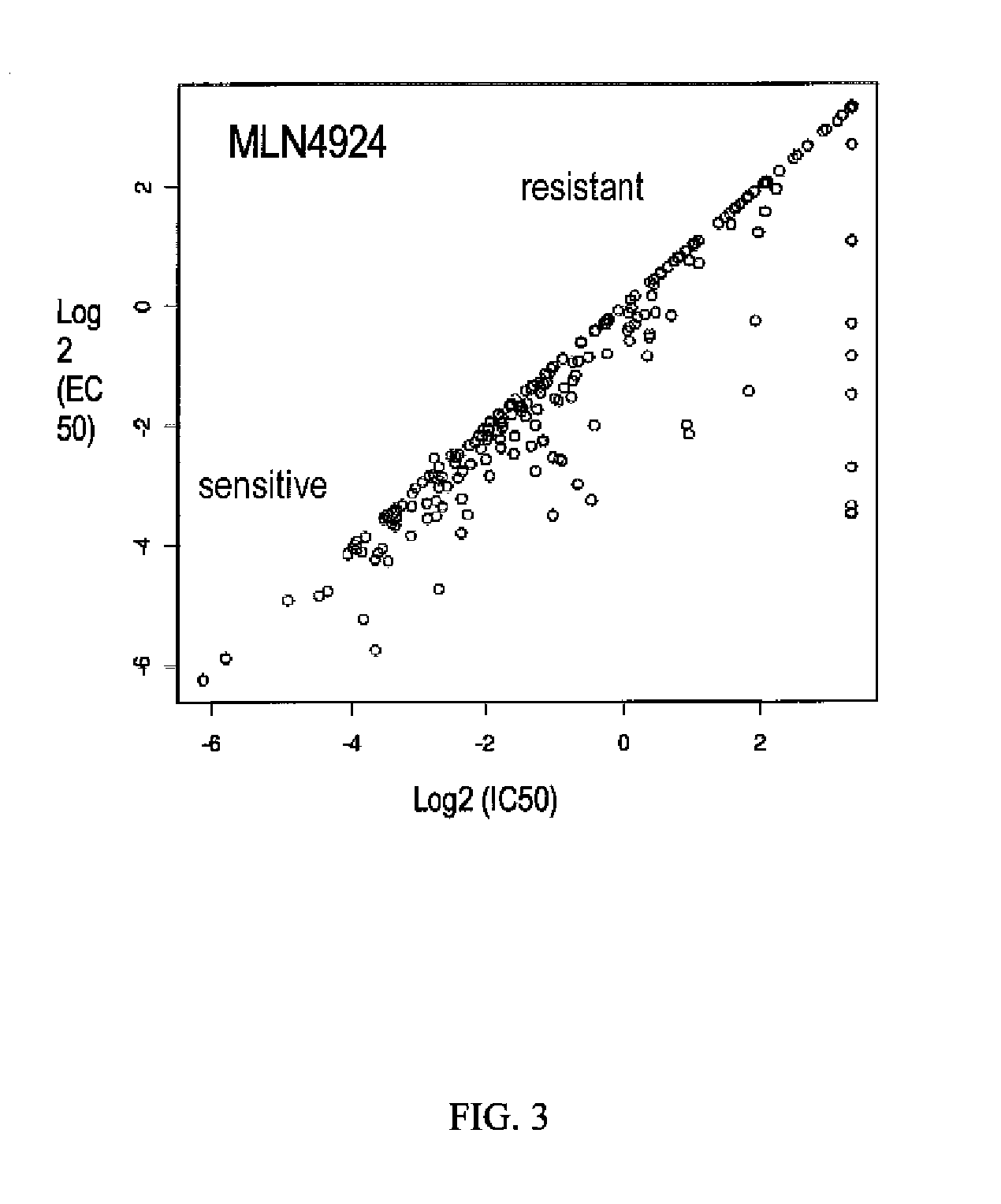
Biomarkers of response to NAE inhibitors
InactiveUS9827246B2Eliminate ineffective inappropriateEliminate ineffective and inappropriate useOrganic active ingredientsMechanical/radiation/invasive therapiesBiomarker (petroleum)Cancer research
Disclosed herein are markers whose mutational status is associated with sensitivity to treatment with NAE inhibitors. Mutational status is determined by measurement of characteristics of markers associated with the marker genes. Compositions and methods are provided to assess markers of marker genes to predict response to NAE inhibition treatment.
Owner:MILLENNIUM PHARMA INC
PCR (Polymerase Chain Reaction) detection method for mutation of human BRAF (v-raf Murine Sacoma Viral Oncogene Homolog) gene V600E, primer, probe and kit
InactiveCN107400716AAchieving Specific DetectionEasy to detectMicrobiological testing/measurementDNA/RNA fragmentationViral OncogeneBraf genes
Owner:SHANGHAI BIOCHIP
Molecular marker for identifying wild state/mutation state of Chinese cabbage TNL-E gene and applications of marker
InactiveCN103290125ARich genetic backgroundConducive to screeningMicrobiological testing/measurementDNA/RNA fragmentationForward primerGerm plasm
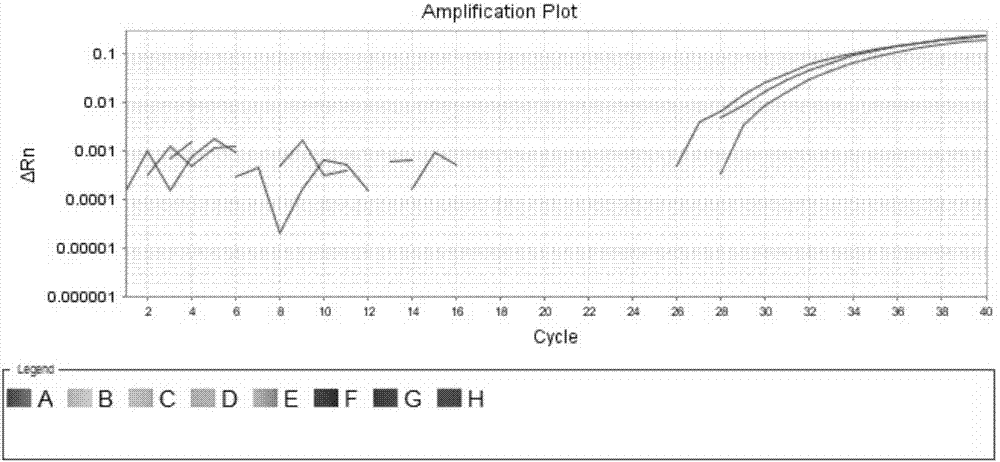
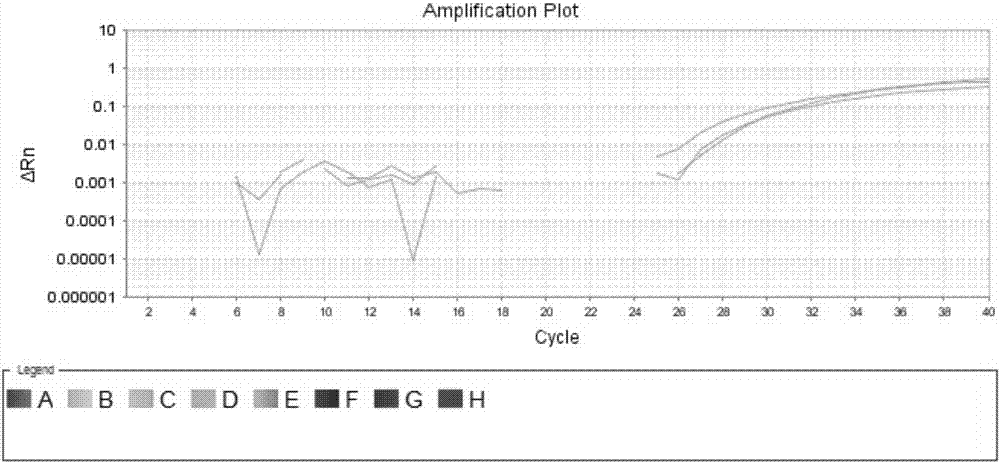
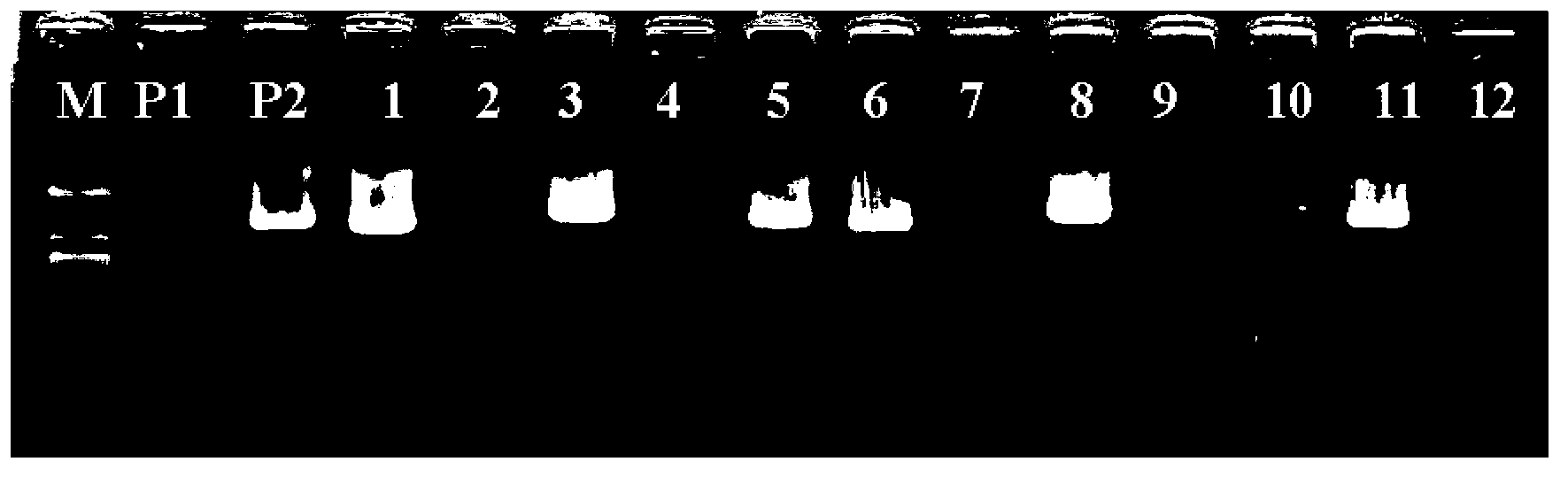
The invention discloses a dominant marker ASM-TNL-E-01 for identifying a wild state / mutation state of a Chinese cabbage TNL-E family gene, wherein a marker segment is 1367bp in size and a nucleotide sequence of the marker segment is as shown in SEQIDNo.1. Primers for identifying the specific molecular marker include a forward primer TNL1F:5'-TGAAACTTTCATTCAGCTTCAAGT-3' and a reverse primer TNL1R:5'-TTGGGTTAGTTTTCAACTTTCACA-3'; the sequences of the two primers are as shown in SEQIDNO.2 and SEQIDNO.3. The molecular marker can be used for identifying Chinese cabbage germ-plasm resources and assisting selective breeding, and a wild-type germ-plasm material or transformed offspring containing the TNL-E in 8407 is selected. A detailed method comprises the steps of: implementing PCR (Polymerase Chain Reaction) amplification on a genomic DNA (Deoxyribose Nucleic Acid) of an individual to be detected by utilizing the primers; detecting whether an amplified segment is generated; if a band is amplified, determining that a genome contains wild-type genes; otherwise, determining that the gene is mutated.
Owner:VEGETABLE RES INST OF SHANDONG ACADEMY OF AGRI SCI
Biomarkers of response to nae inhibitors
InactiveUS20150105411A1Eliminate ineffective and inappropriate useAggressive administrationOrganic active ingredientsMechanical/radiation/invasive therapiesMutational statusMolecular biology
Disclosed herein are markers whose mutational status is associated with sensitivity to treatment with NAE inhibitors. Mutational status is determined by measurement of characteristics of markers associated with the marker genes. Compositions and methods are provided to assess markers of marker genes to predict response to NAE inhibition treatment.
Owner:MILLENNIUM PHARMA INC
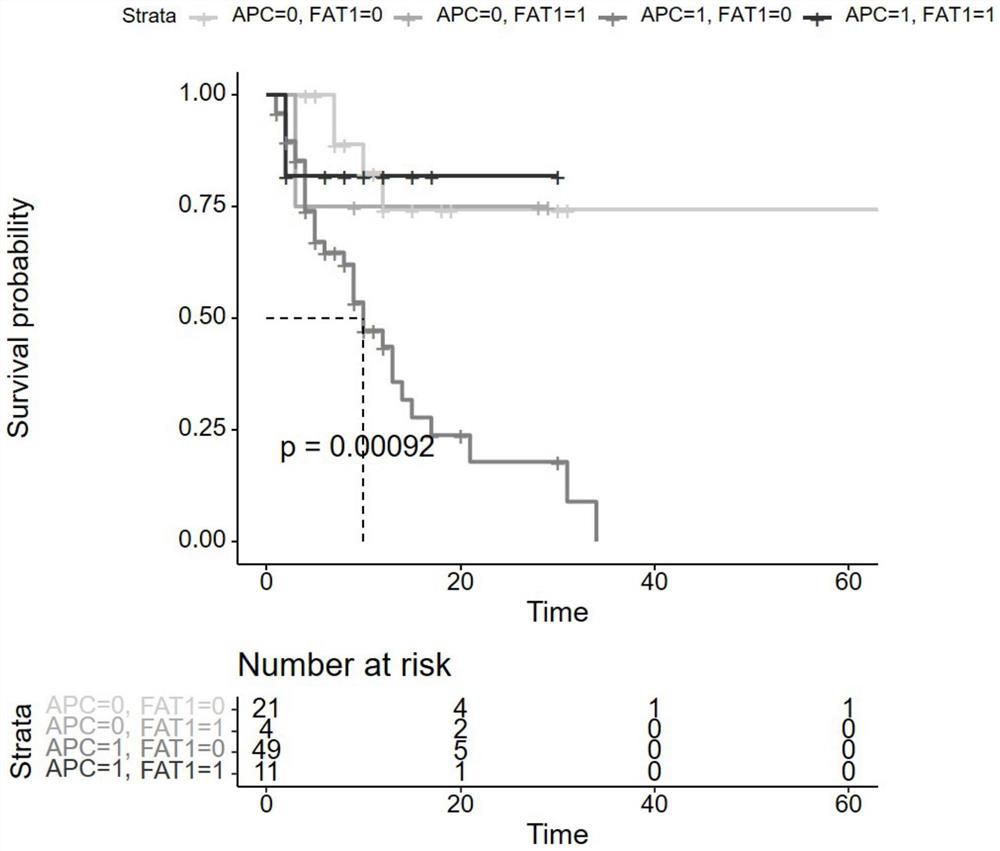
Marker, kit and device for predicting sensitivity of colorectal cancer patients to immunotherapy
PendingCN113957148APredictive validityReduce testing costsMicrobiological testing/measurementProteomicsBiologic markerImmunotherapy
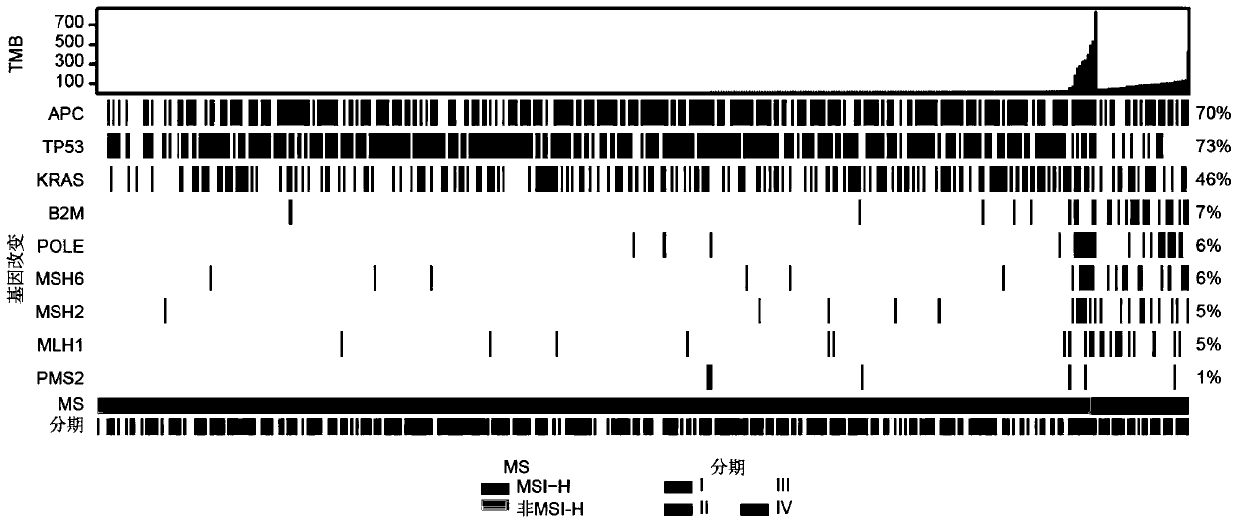
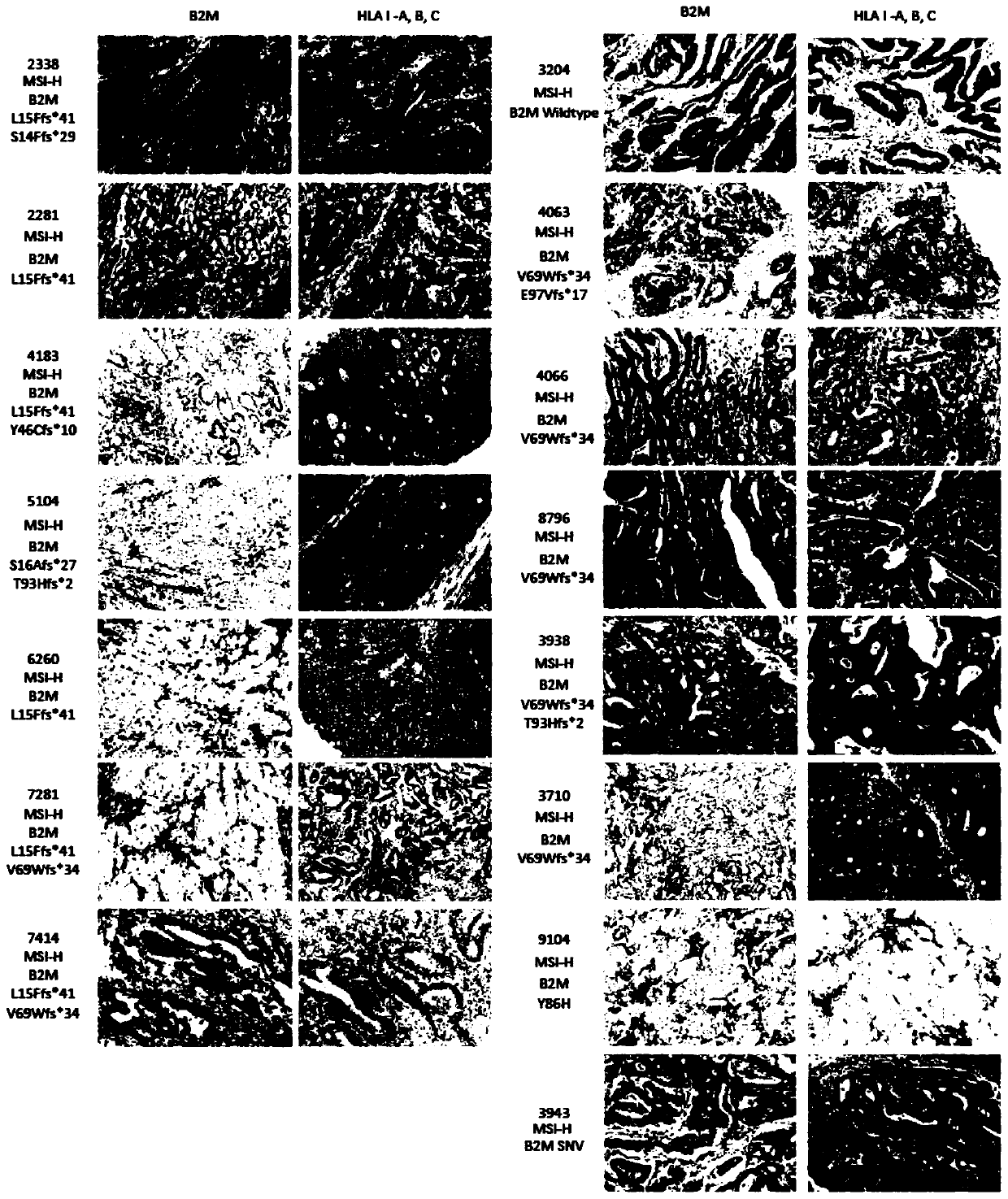
The invention relates to a gene marker combination for predicting sensitivity of colorectal cancer patients to immunotherapy. The gene marker combination comprises an APC gene and an FAT1 gene. Wherein mutation states of the APC gene and the FAT1 gene have significant correlation with microsatellite instability (MSI) and tumor mutational burden (TMB) states, and the APC gene and the FAT1 gene can be used as new biomarkers to predict the sensitivity of the colorectal cancer patients to immunotherapy and predict the effectiveness of the immunotherapy in colorectal cancer patients. Compared with TMB which needs to detect at least hundreds of genes for calculation, the gene marker combination disclosed by the invention, as a biomarker, can predict the immunotherapy effect only by detecting mutations of a small number of key genes, so that the detection cost is greatly reduced; compared with MSI, the gene marker combination can cover more people according to gene detection and expand potential beneficiaries of immunotherapy.
Owner:SHANGHAI ORIGIMED CO LTD +1
Methods for diagnosing and treating cancer by means of the expression status and mutational status of NRF2 and downstream target genes of said gene
The invention provides methods of identifying a subject having cancer, such as lung cancer, by analyzing expression levels of one or more NRF2 splice variants or NRF2 target genes. The invention also provides methods of treating cancer in a subject with a NRF2 pathway antagonist, wherein the subject expresses one or more NRF2 splice variants or overexpresses one or more NRF2 target genes.
Owner:GENENTECH INC +1
Immune sensitization predicted biomarker component, use and kit equipment storage medium
PendingCN110656179ASave treatment timeSave moneyBioreactor/fermenter combinationsBiological substance pretreatmentsMedicineMicrosatellite
Owner:SHANGHAI ORIGIMED CO LTD
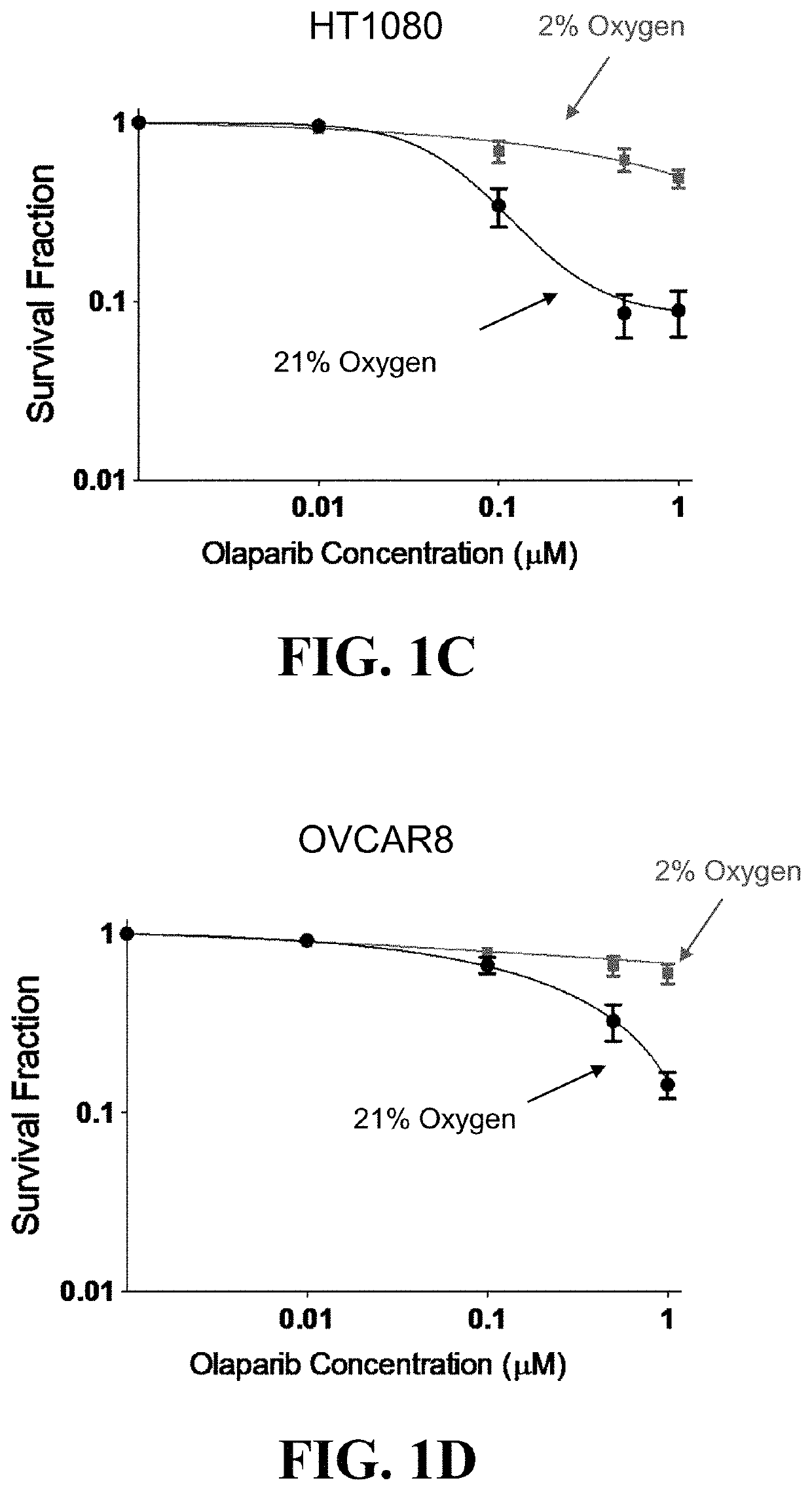
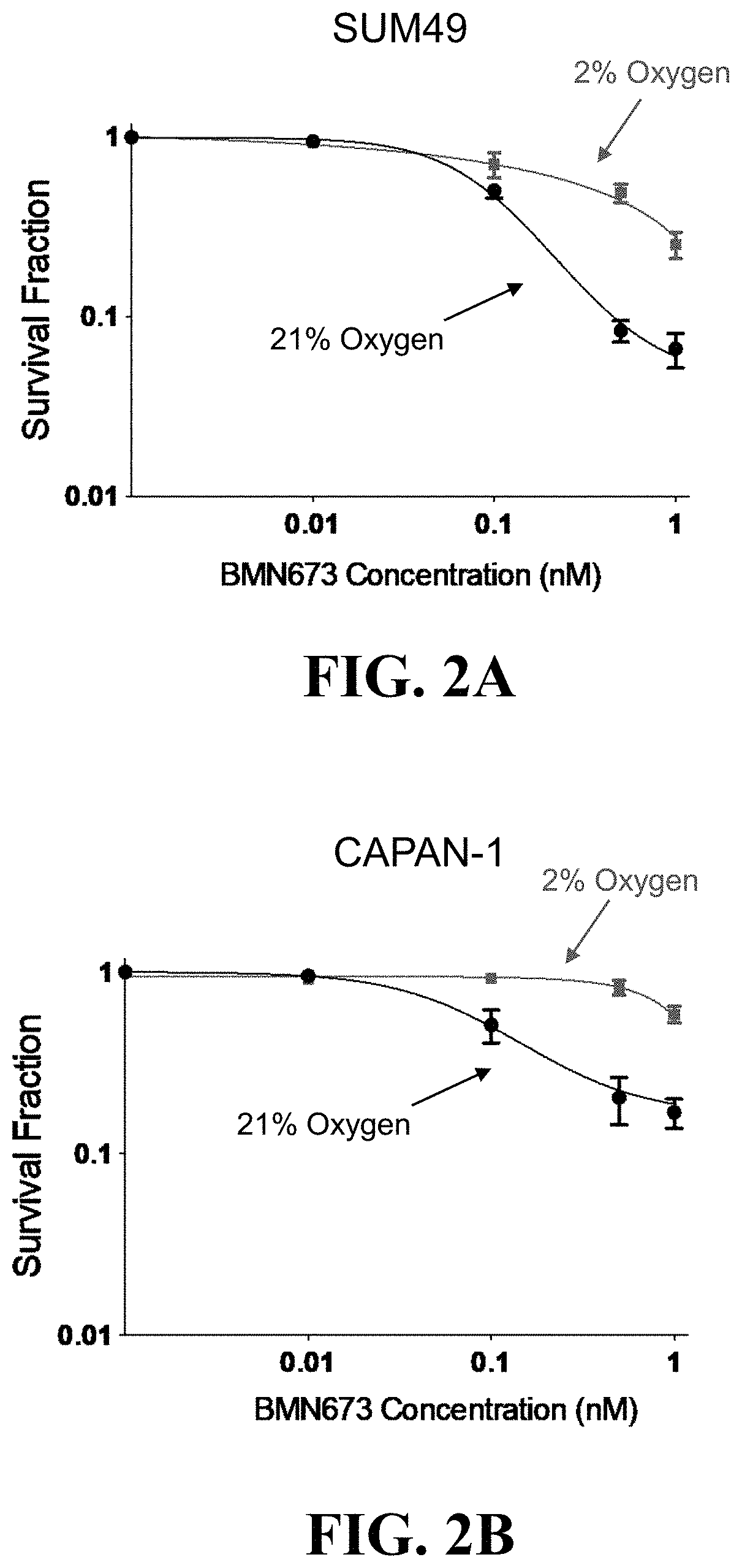
Hypoxia Targeting Compositions and Combinations Thereof with a PARP Inhibitor and Methods of Use Thereof
PendingUS20220249522A1Antineoplastic agentsHeterocyclic compound active ingredientsPARP inhibitorPharmaceutical formulation
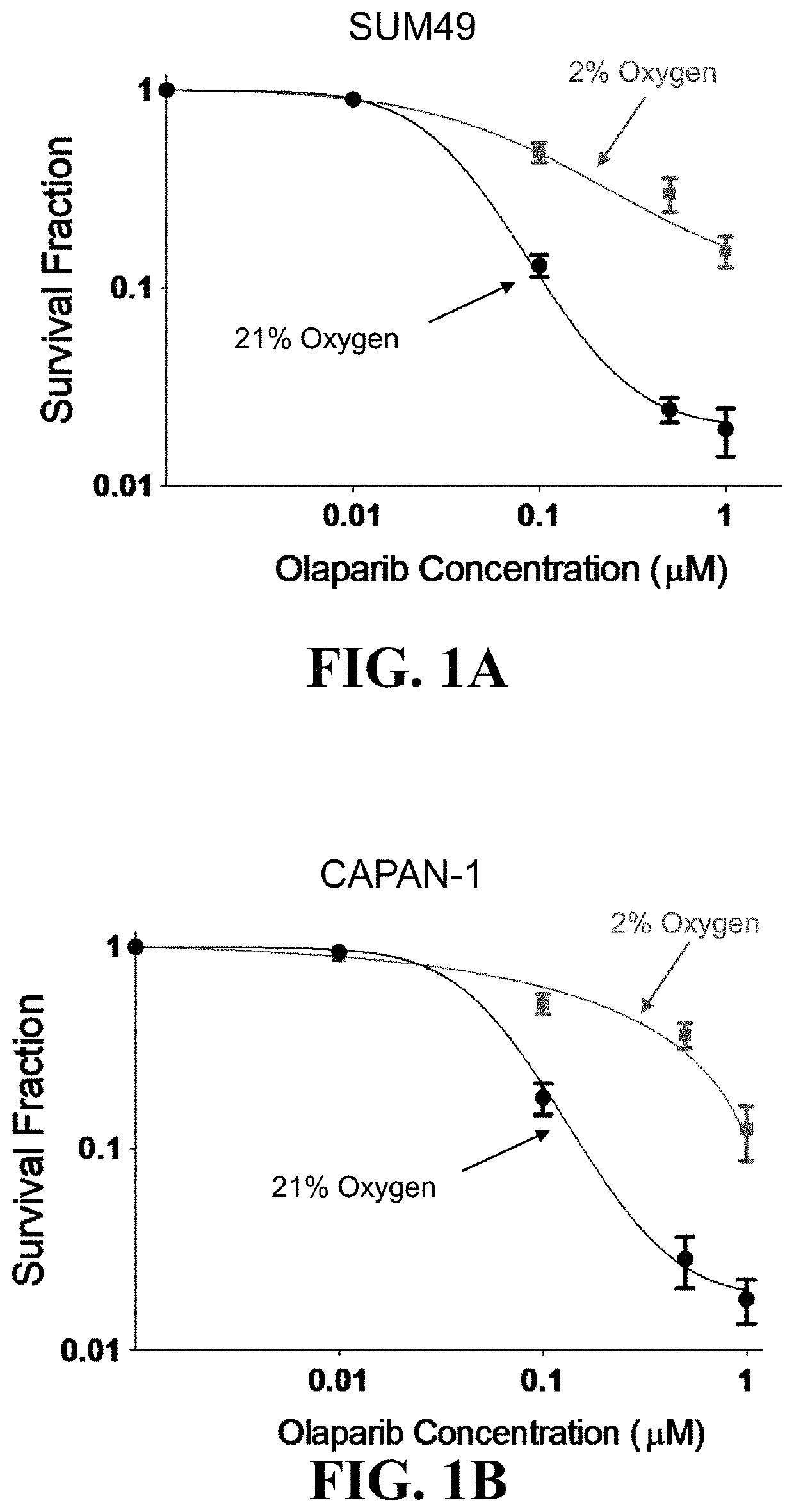
Methods for treating a cancer are provided, the methods comprising administering to an individual an effective amount of a hypoxia targeting composition, such as a hypoxia-activated drug or a prodrug thereof, and combinations thereof with an effective amount of a poly(ADP-ribose) polymerase (PARP) inhibitor. In some instances, one or more of a homology recombination (HR) efficiency status, an IDH mutation status, and a hypoxia status of a cancer is used as a basis for selecting an individual for a treatment disclosed herein. Also provided are compositions (such as pharmaceutical formulations), medicine, kits, and unit dosages useful for the methods described herein.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
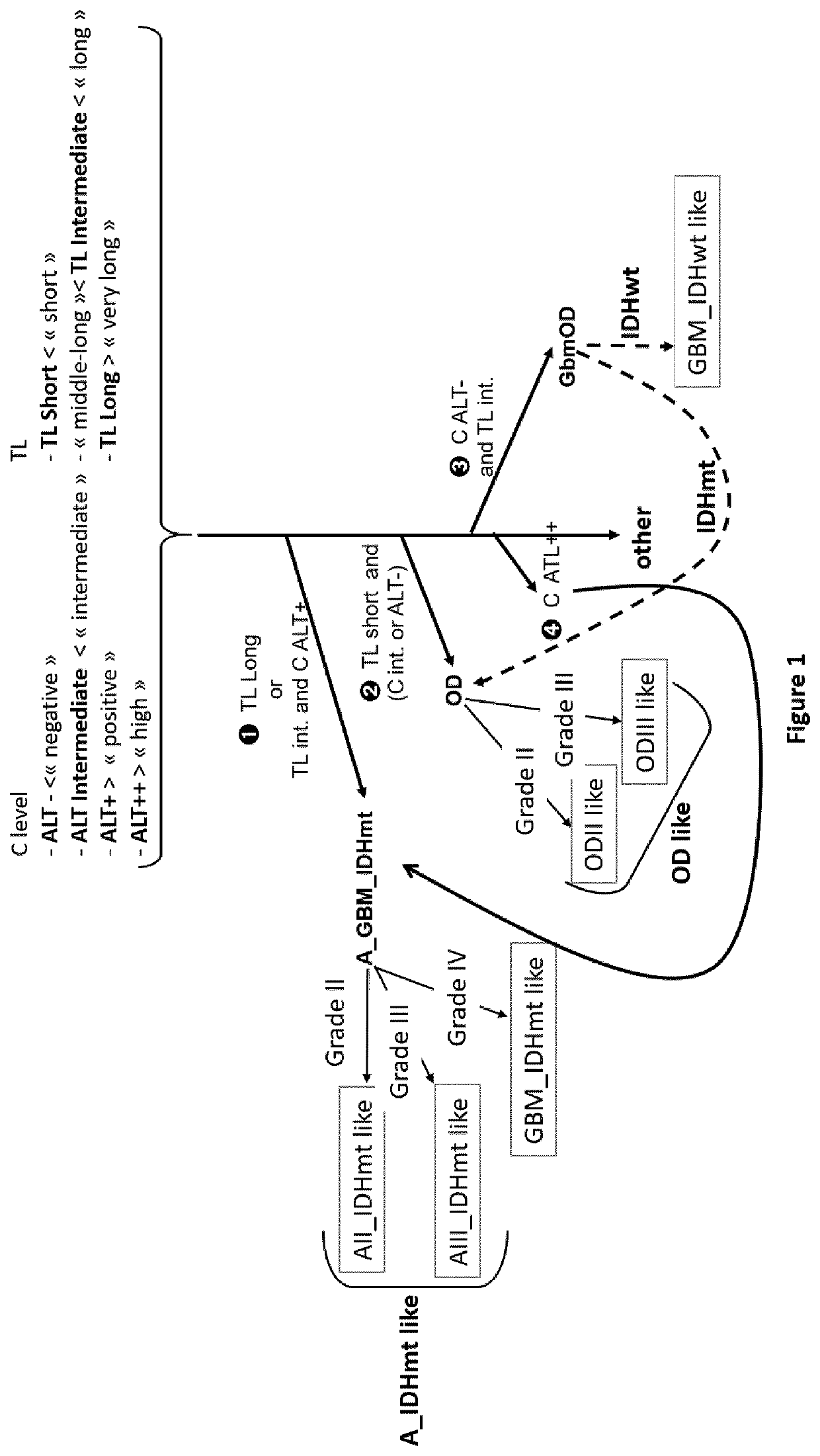
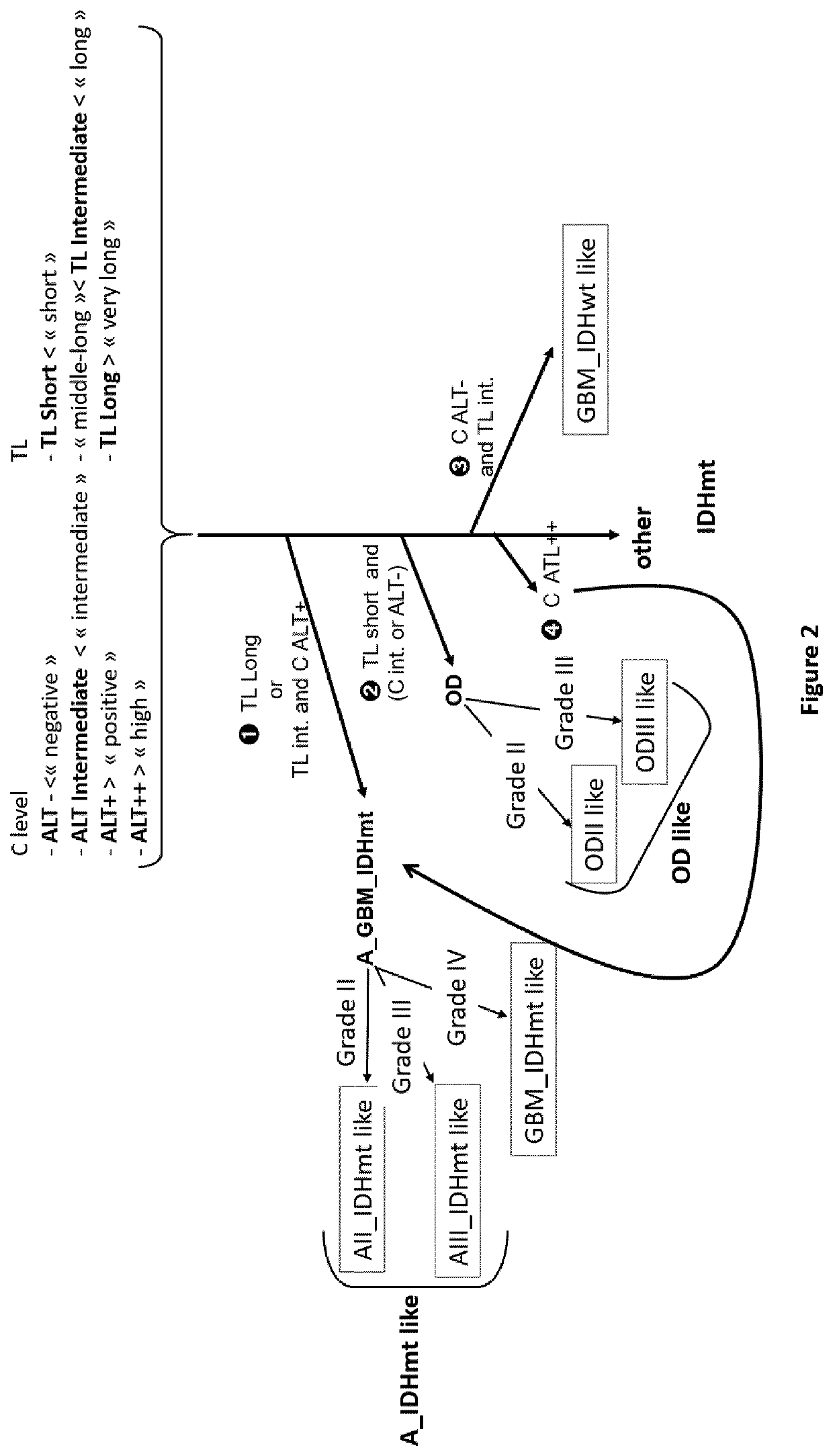
Process for classification of glioma
PendingUS20220119893A1Reduce in quantitySame prognosisMicrobiological testing/measurementGenes mutationBlastoma
The invention relates to an in vitro process for classifying a glioma, comprising the following steps: a. Measuring at least, from a glioma patient biological sample, the Alternative Lengthening of Telomeres (ALT) status of said glioma; b. Optionally, determining the isocitrate dehydrogenase genes mutation status (IDH status) of said glioma; c. Based on the data obtained in steps (a) and optionally (b) and, if available, on the histological grade of said glioma, classifying said glioma in one of the five following classes: oligodendroglioma-like, glioblastoma IDHwt-like, glioblastoma IDHmt-like, low-grade astrocytoma-like, and other gliomas.
Owner:HOSPICES CIVILS DE LYON +1
Simultaneous detection of mutational status and gene copy number
The present invention provides compositions and methods for simultaneously detecting mutational status and gene copy number. In particular, the present invention provides simultaneous measurement of gene copy number and detection of the L858R and Exon 19 del mutations in a tissue sample.
Owner:VENTANA MEDICAL SYST INC
Invasive glioma classification device based on DNA methylation
PendingCN113308545ARapid determinationEconomic measurementMicrobiological testing/measurementDNA methylationMedicine
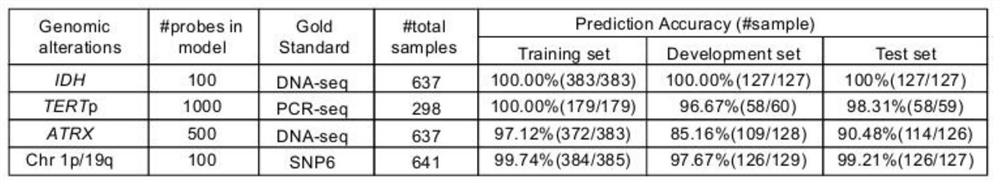
The invention discloses an invasive glioma classification device based on DNA methylation. The device comprises the following parts: a detection module which is used for carrying out methylation chip detection or methylation sequencing on extracted tumor sample DNA; and a classification module which is used for preprocessing the original data and then detecting the mutation states of IDH, TERTp and ATRX, the chr1p19qcodel state and the gene expression subtype of the invasive glioma through a classification system. According to the invention, different methylation probe combinations are utilized to rapidly, economically and efficiently determine the IDH, TERTp and ATRX mutation states, the chr1p19qcodel state and the gene expression subtype of the invasive glioma; and in addition, the requirement for a sample to be detected is very low, and the subtype can be accurately recognized by using a small amount of FFPE tissue.
Owner:NANJING MEDICAL UNIV
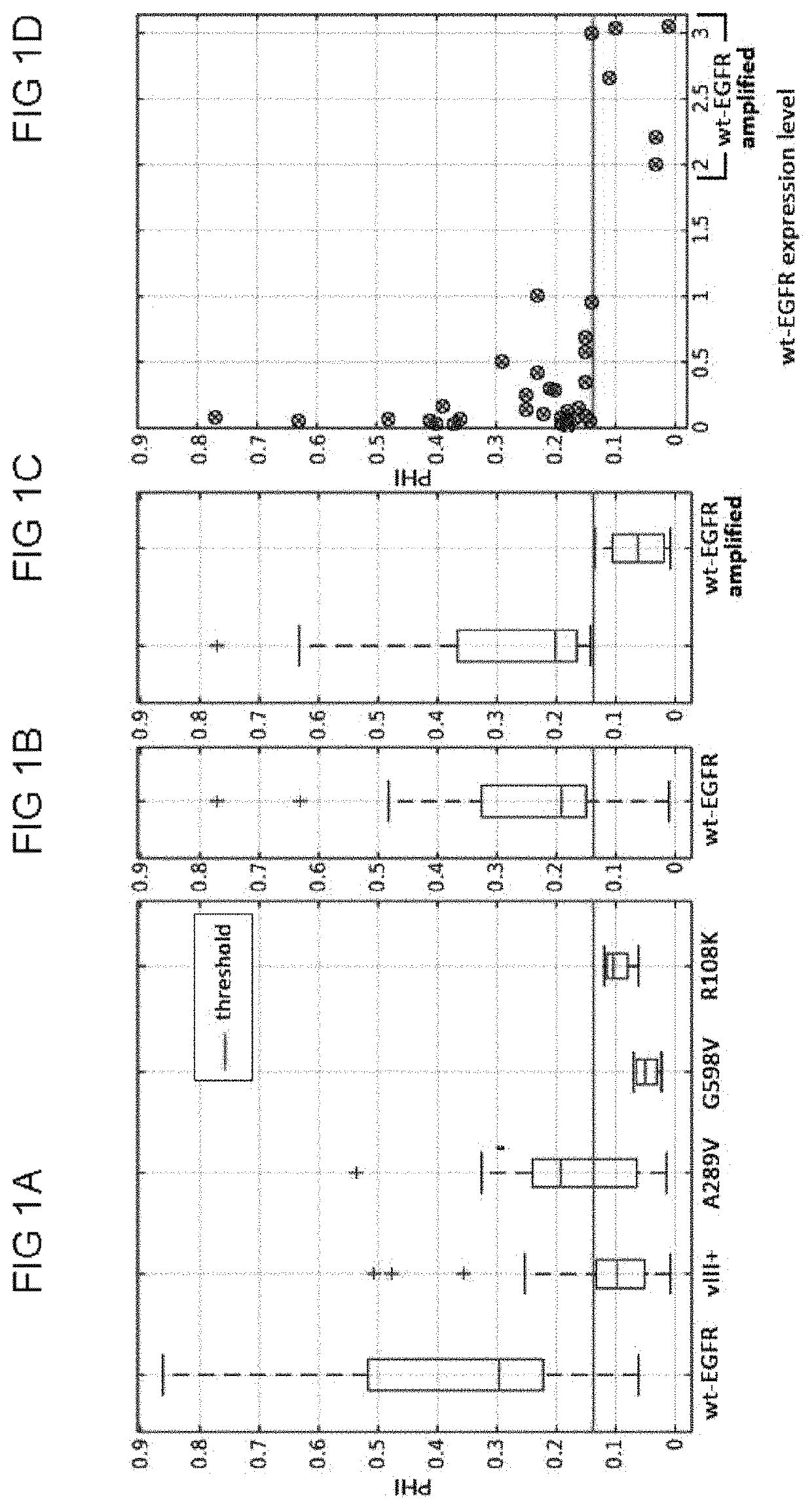
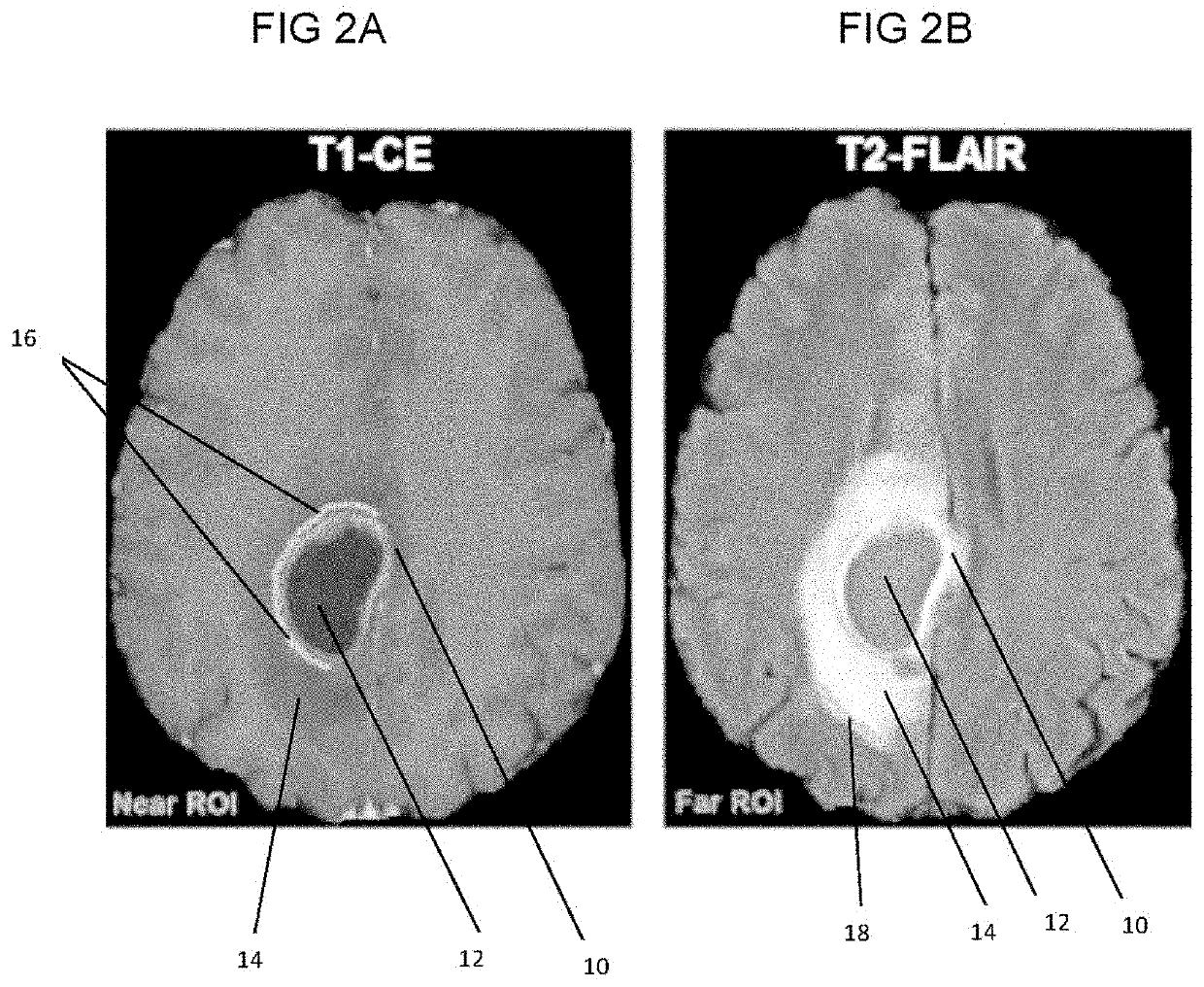
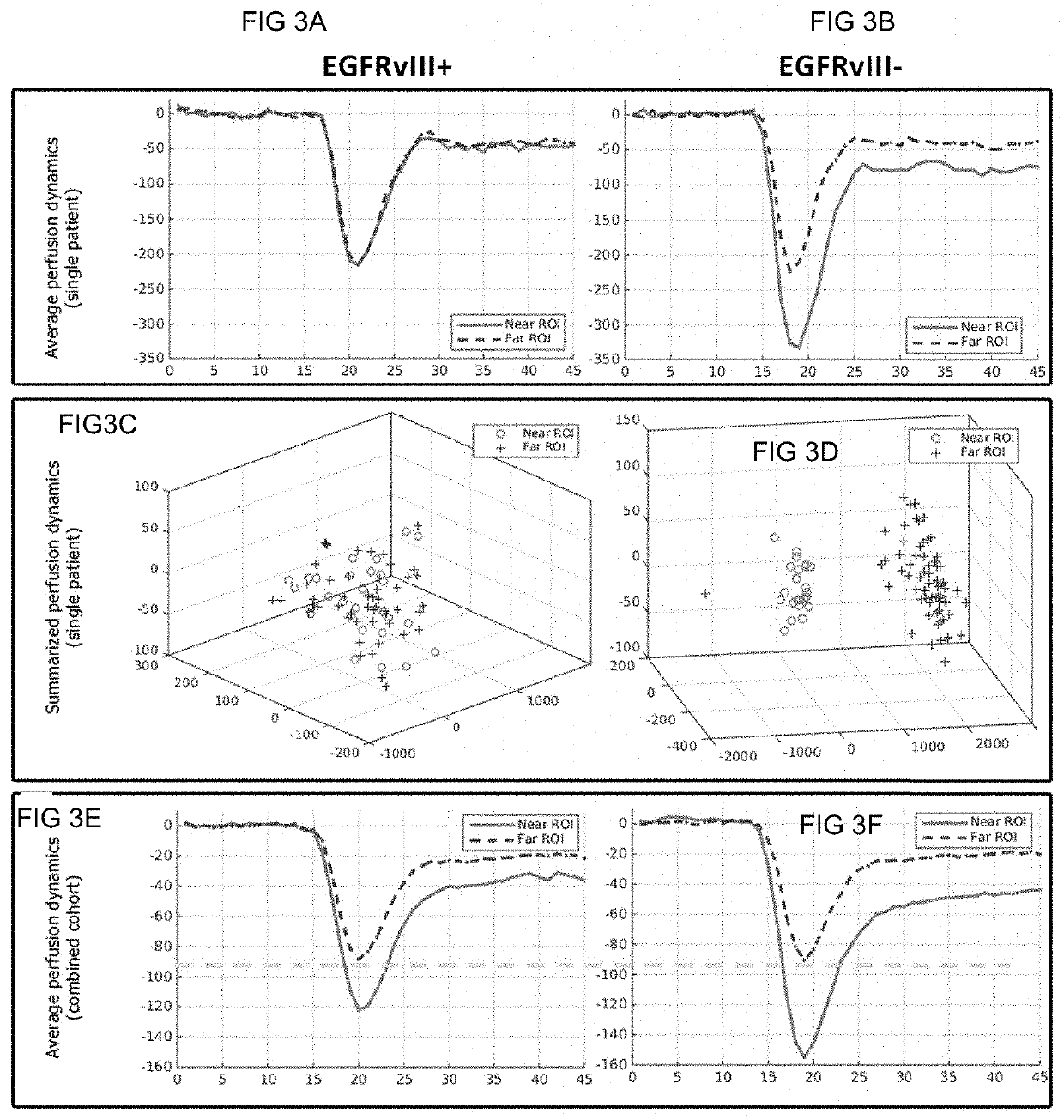
In vivo detection of EGFR mutation in glioblastoma via MRI signature consistent with deep peritumoral infiltration
A method, including a computer-implemented method, is provided for in vivo detection of epidermal growth factor receptor (EGFR) mutation status within peritumoral edematous tissue of a patient. The method includes performing quantitative pattern analysis of magnetic resonance imaging (MRI) data corresponding to MRI of in vivo peritumoral edematous tissue to determine a level of spatial heterogeneity or similarity within the in vivo peritumoral edematous tissue. EGFR mutation status is assigned as one of negative or positive based on the level of spatial heterogeneity or similarity determined. A non-transitory computer-readable storage medium and a system are also provided.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
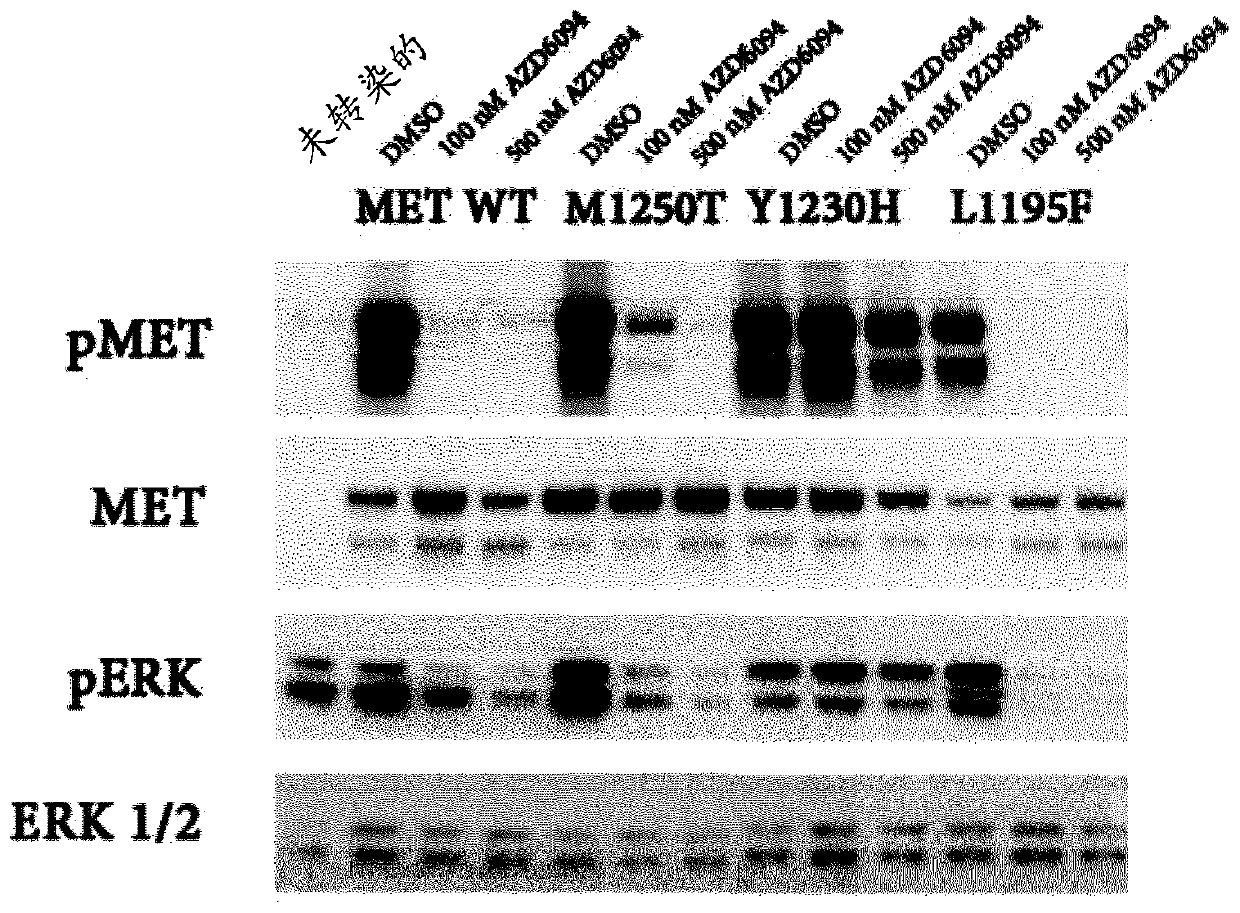
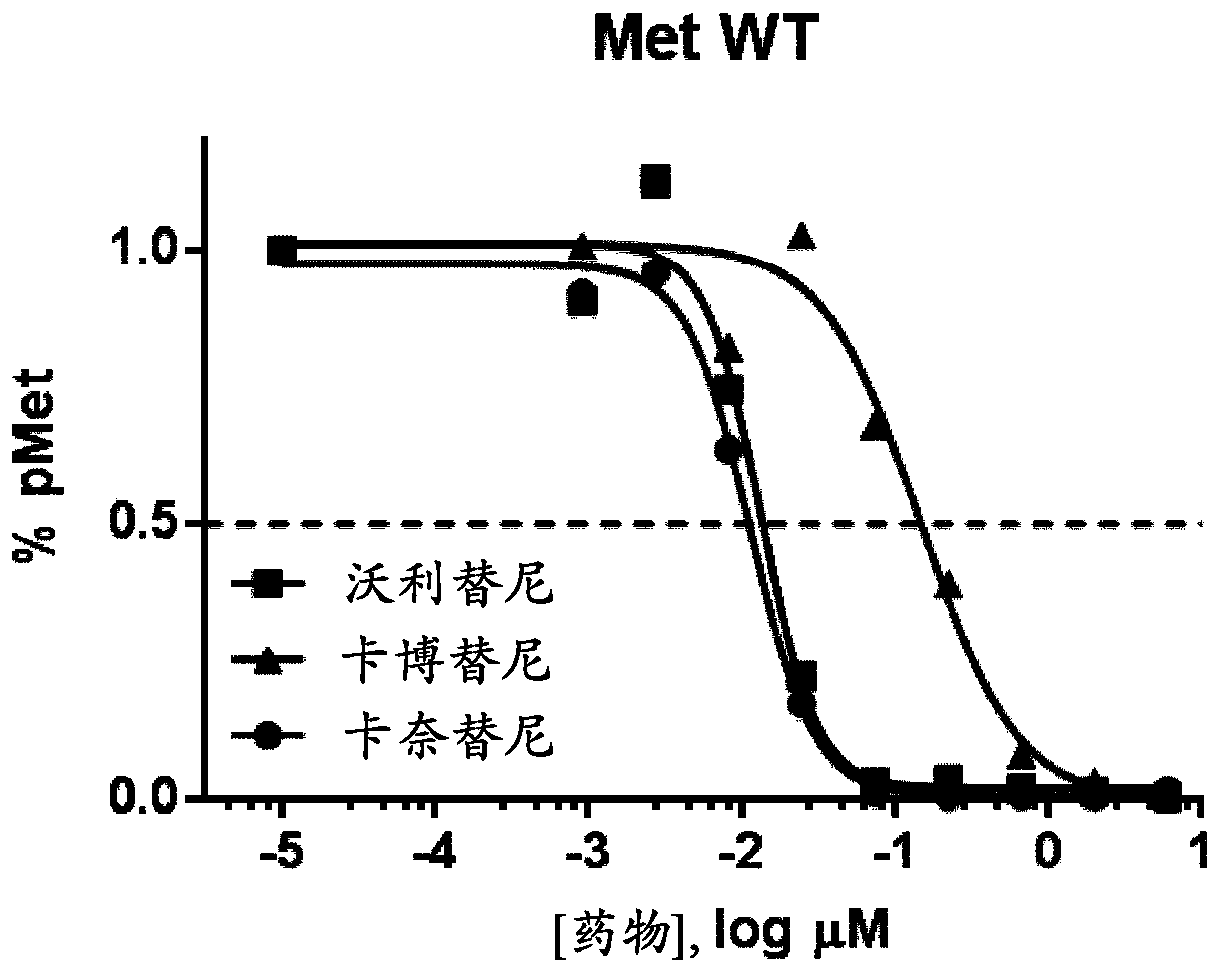
Use of c-met inhibitors to treat cancers harbouring met mutations
PendingCN109789108AMicrobiological testing/measurementPharmaceutical delivery mechanismTyrosine-kinase inhibitorTyrosine
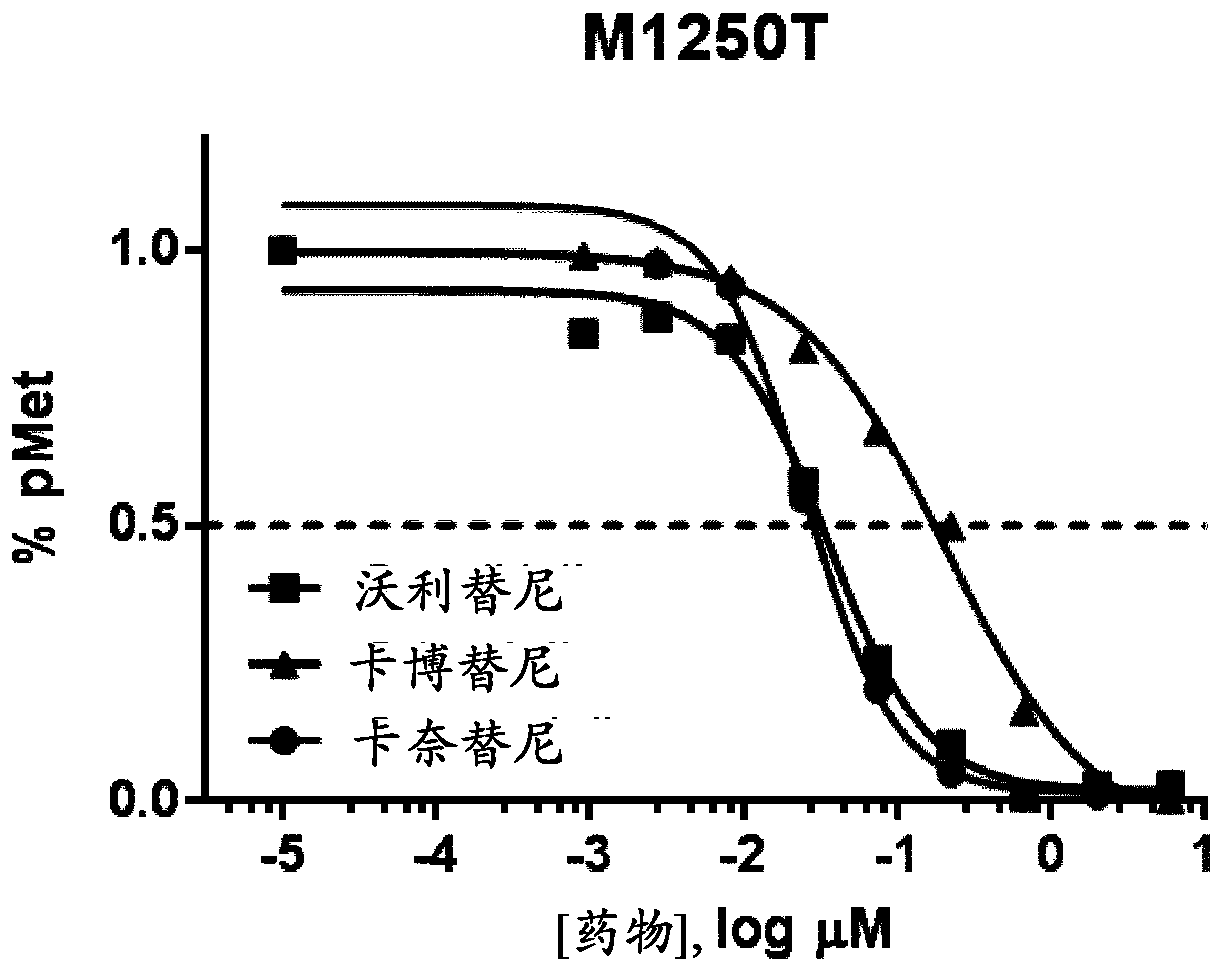
This specification relates to c-Met receptor tyrosine kinase ("c-Met") inhibitors and their use in the treatment of cancers characterised by certain MET mutations (e.g. cancers comprising cells that express a MET protein having a MET V1092I, MET H1094L or MET L1195F mutation); the use of MET mutation status to select patients suitable for treatment with a c-Met inhibitor; and to methods of treating cancers comprising cells that are characterised by certain MET mutations with a c-Met inhibitor.
Owner:ASTRAZENECA AB
Analyzing device, analyzing method and storage medium storing program
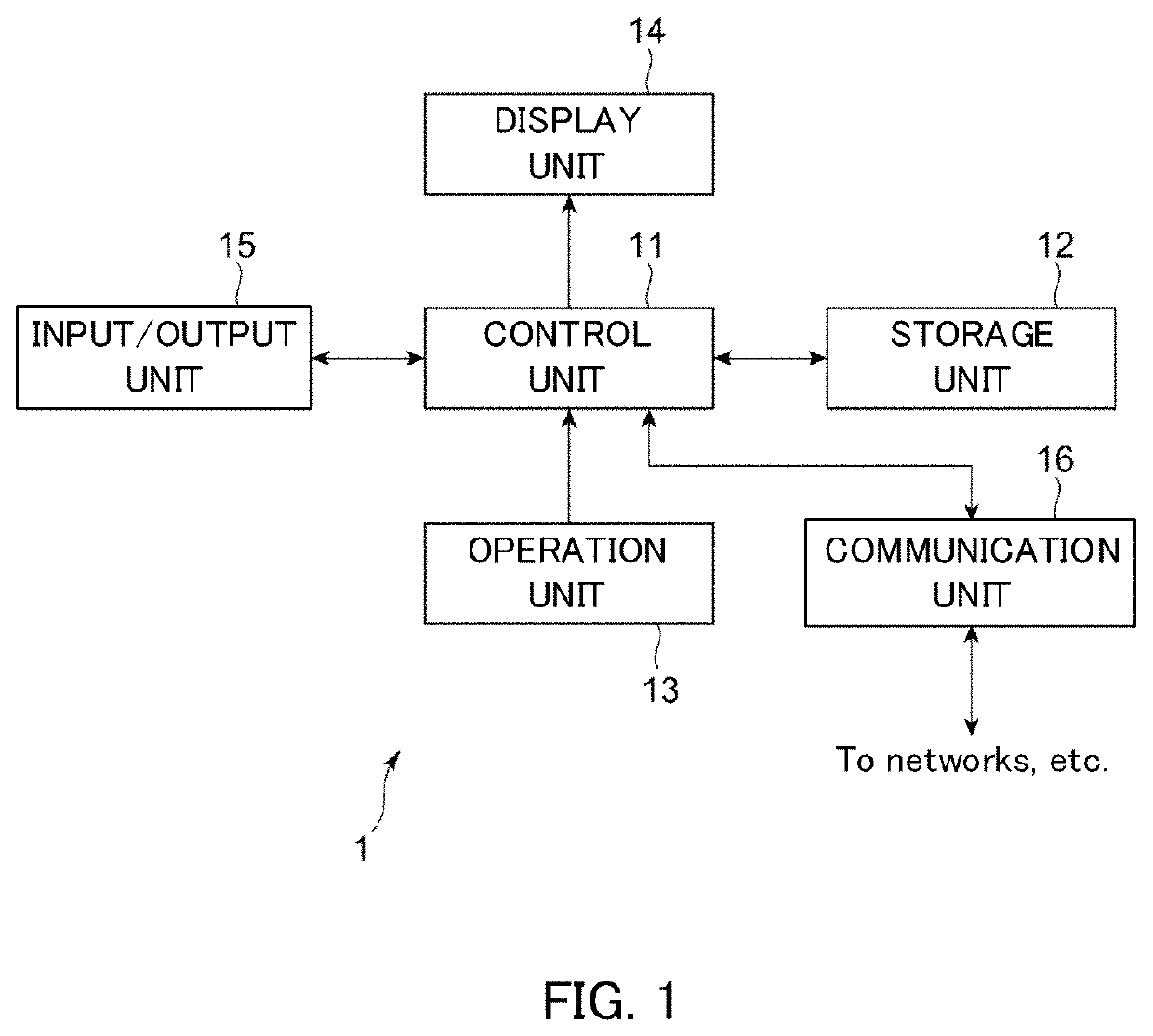
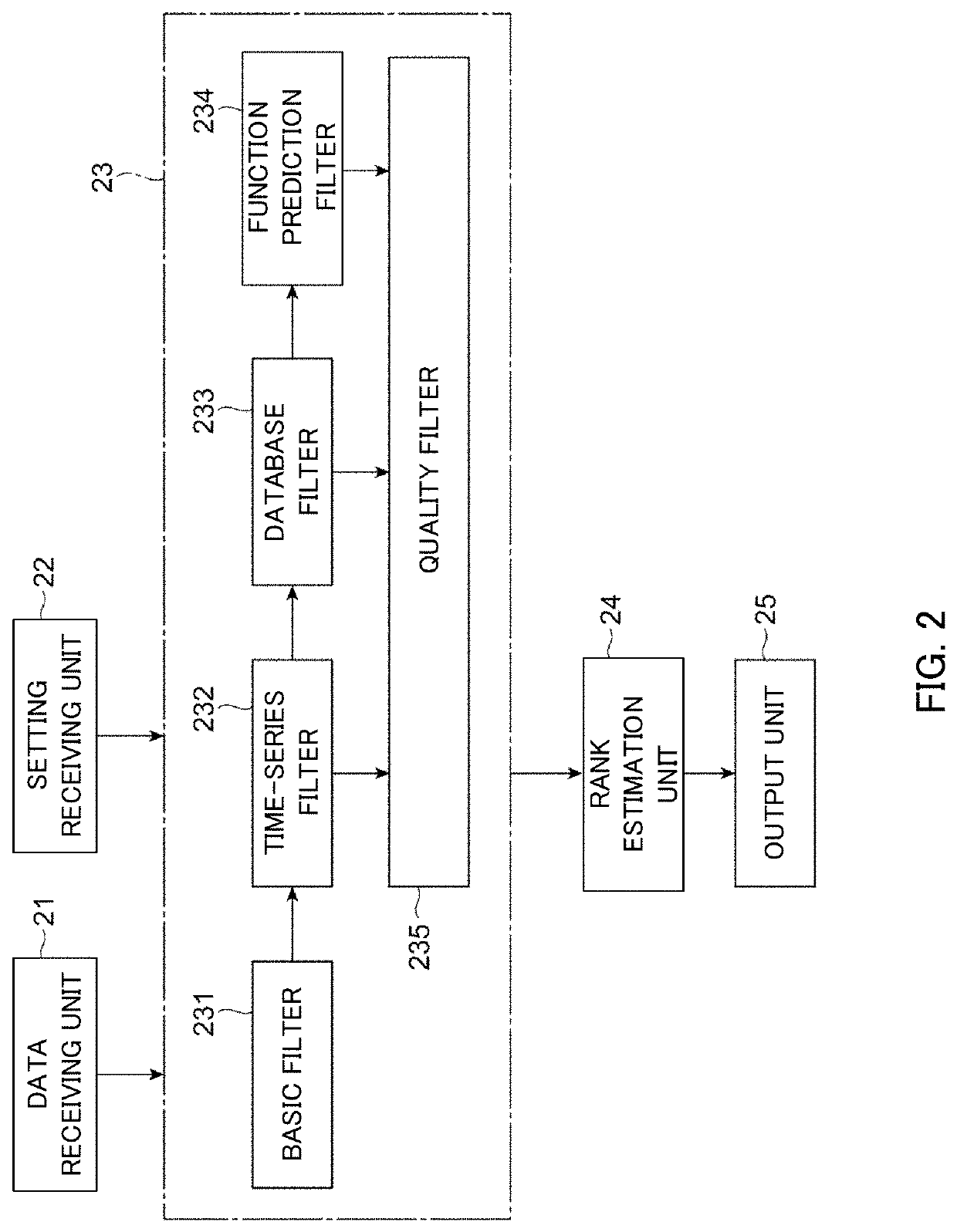
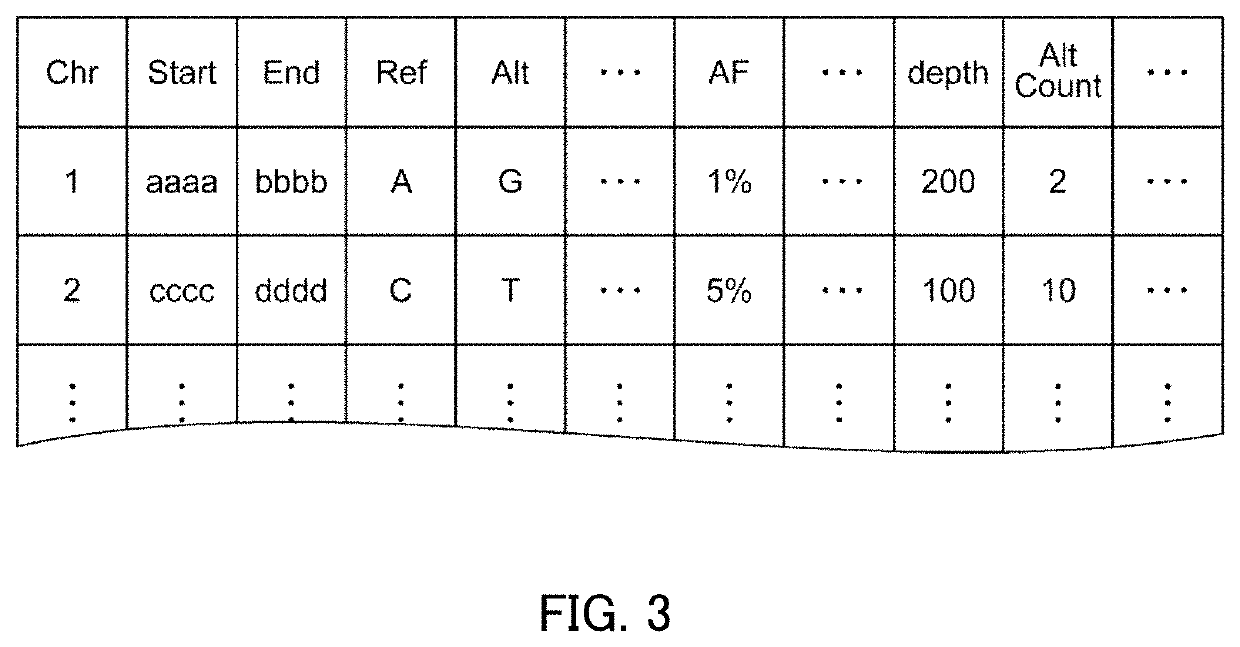
PendingUS20220223229A1Health-index calculationMicrobiological testing/measurementAlgorithmEngineering
An analysis apparatus is provided for analyzing a mutation state, including a mutation location in a base sequence and the content of the mutation, extracted from genetic information of a sample to be analyzed by sequence alignment. The apparatus includes: a receiving unit that receives mutant base sequence information representing the mutation state; a filter processing unit that that outputs a score determined depending on whether or not the mutation state represented by the mutant base sequence information received by the receiving unit is a structural mutation across multiple genes; and an output unit that creates rank information representing the degree of possibility that the mutation state is pathologic based on the score output by the filter processing unit and outputs the created rank information.
Owner:THE UNIV OF TOKYO +1
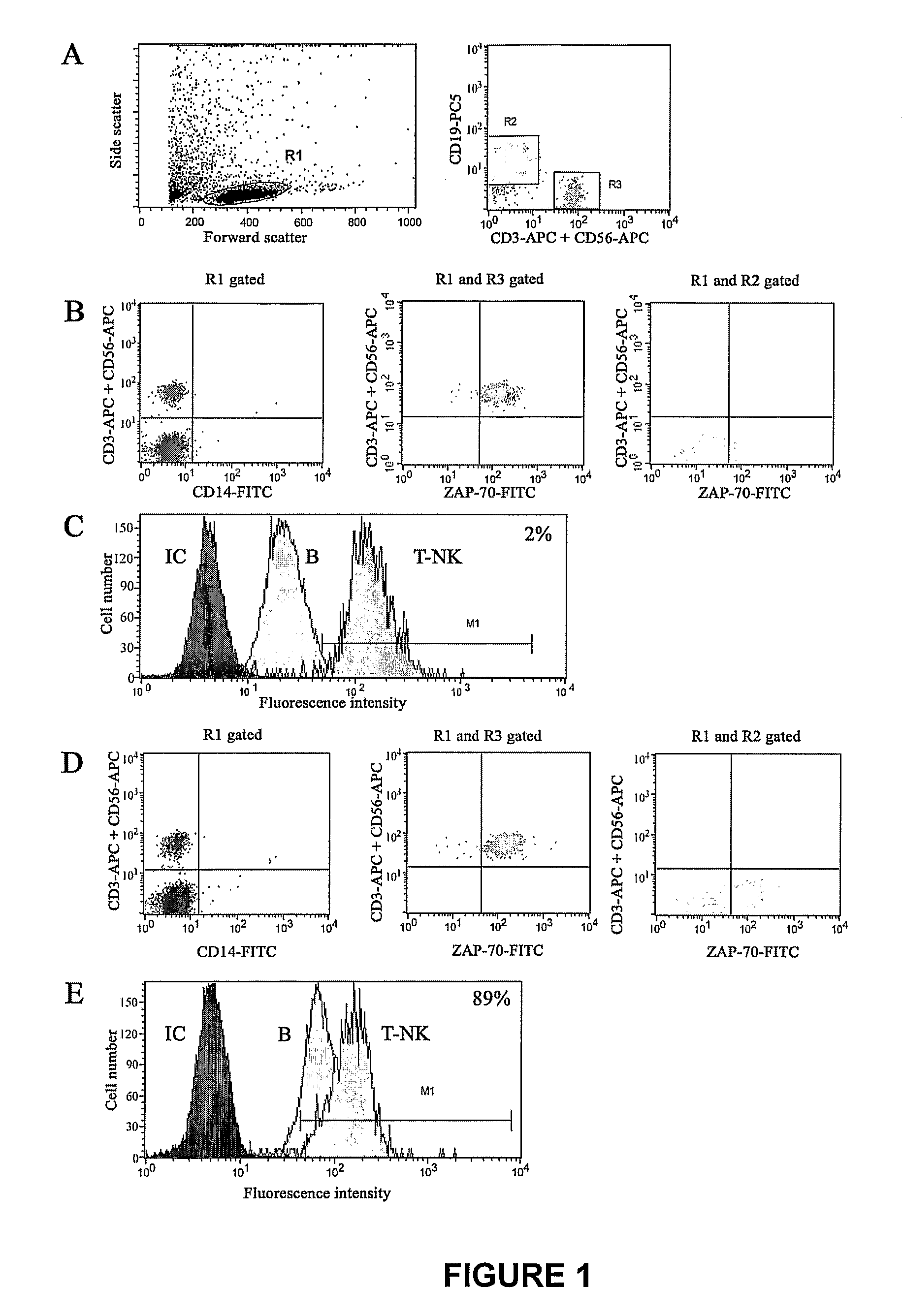
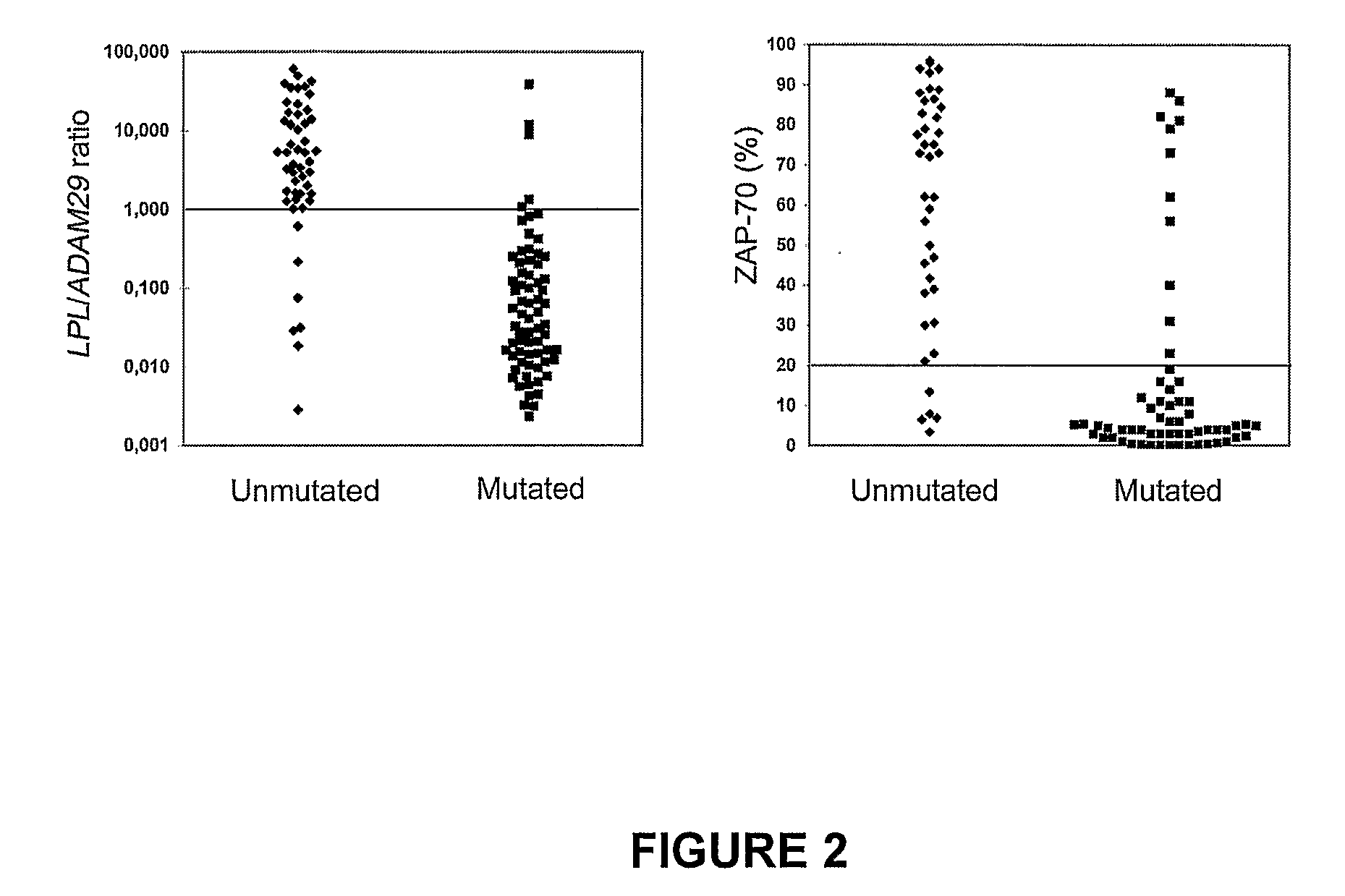
Method of Diagnosis/Prognosis of Human Chronic Lymphocytic Leukemia Comprising the Profiling of Lpl/Adam Genes
InactiveUS20080261212A1Eliminate needAccurate estimateMicrobiological testing/measurementChronic lymphocytic leukemiaIgVH Mutation
The present invention provides methods of diagnosis and prognosis of human chronic lymphocytic leukemia (CLL) in a patient in need thereof and methods to determine IgVH mutational status. The methods of the present invention involve measuring the expression profile of two known genes: LPL and ADAM29; and comparing the ratio of their expression to diagnose the presence of CLL or to prognose the likelihood of developing CLL or the symptoms consistent with CLL.
Owner:INST PASTEUR +2
A preparation and recurrence risk model for judging the prognosis of early endometrial cancer
ActiveCN112961921BImprove predictive performanceLow diagnostic agreementMicrobiological testing/measurementBiological material analysisIntermediate riskIndividualized treatment
Owner:PEKING UNION MEDICAL COLLEGE HOSPITAL CHINESE ACAD OF MEDICAL SCI
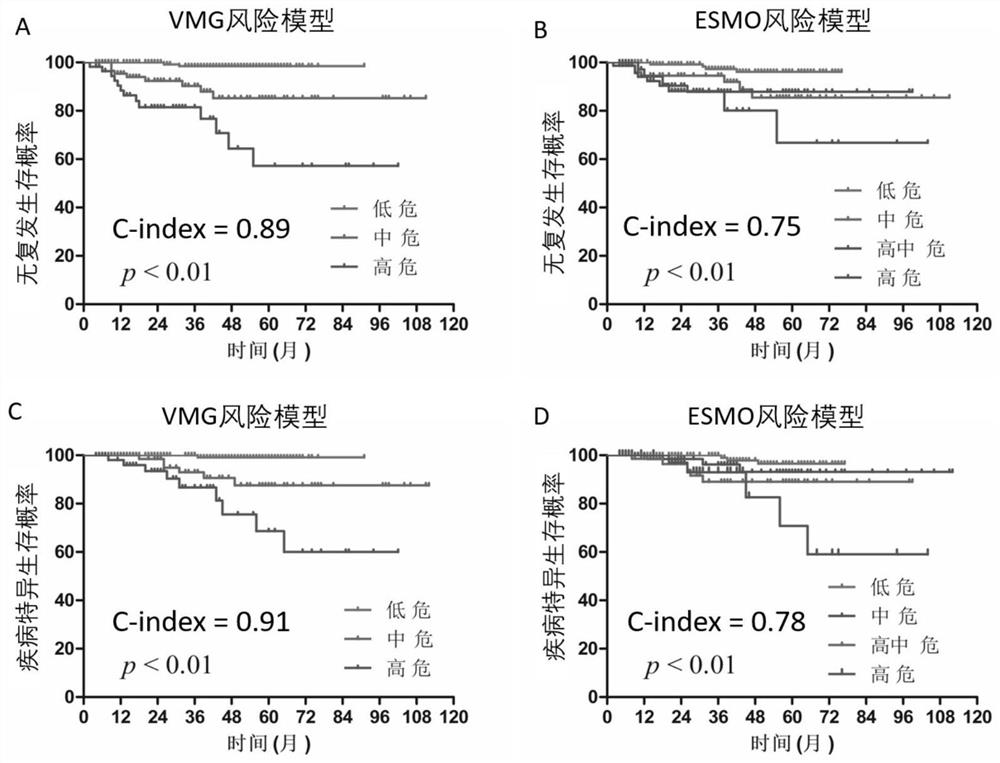
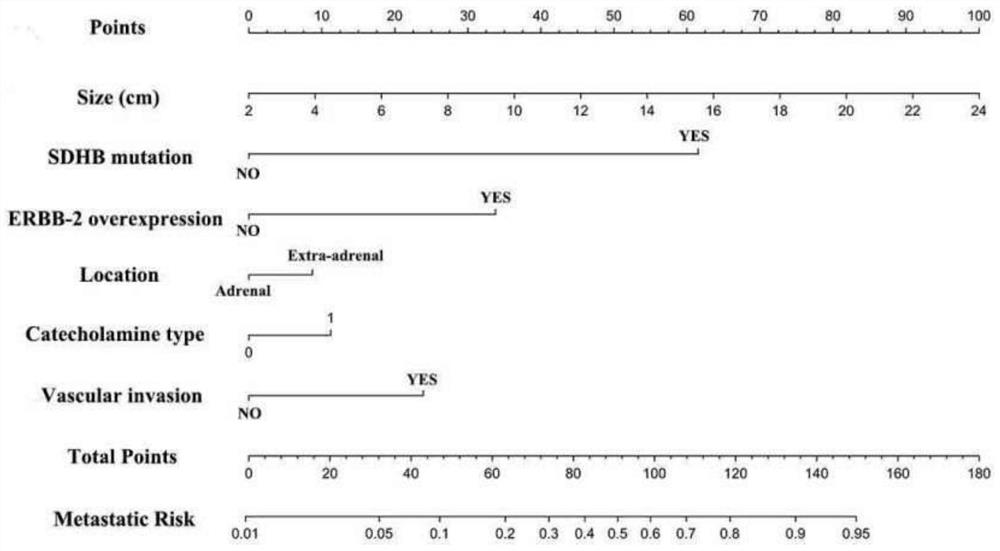
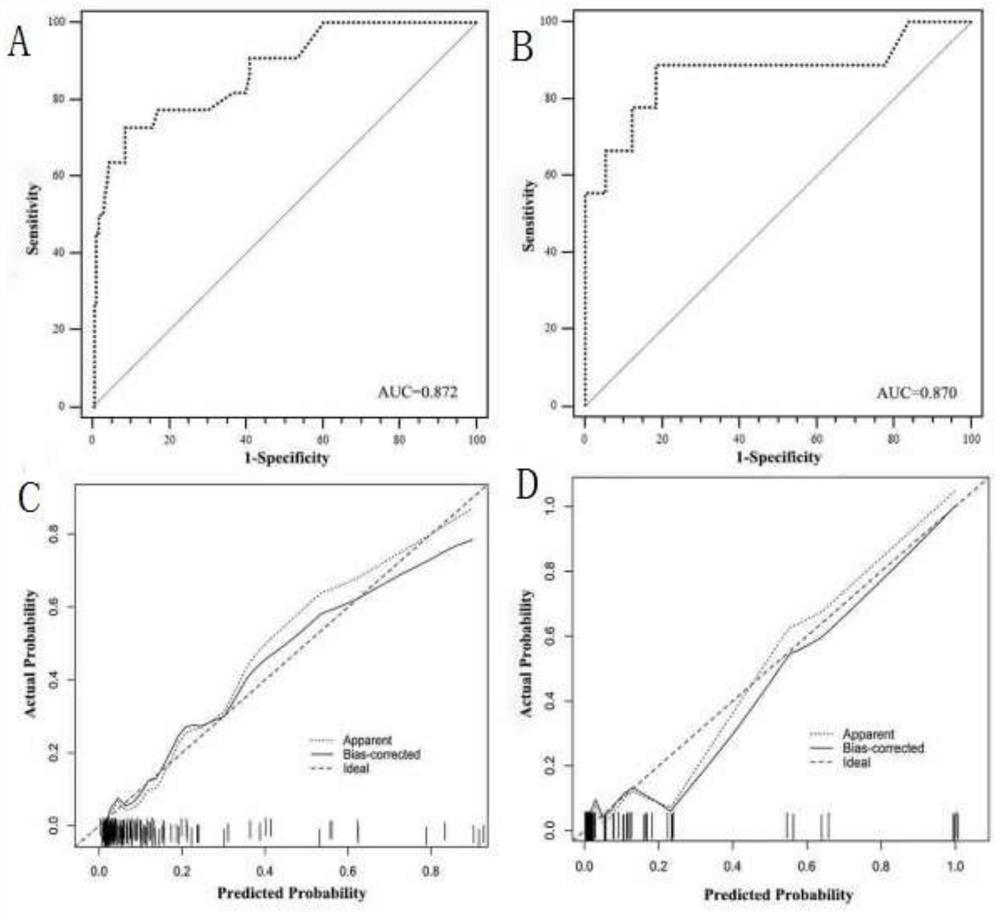
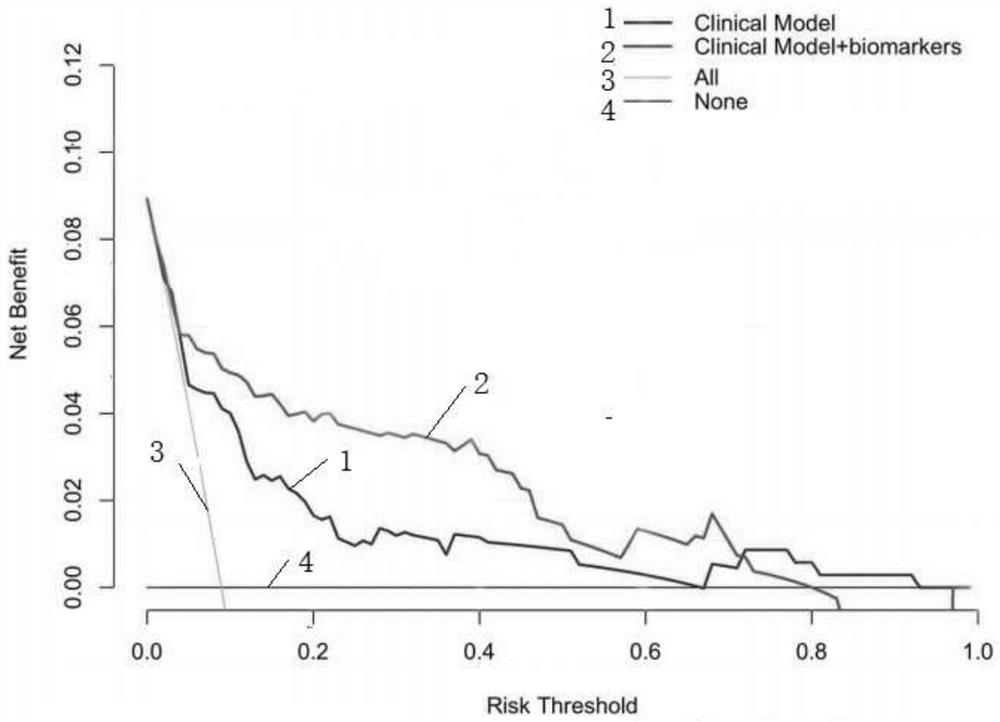
Pheochromocytoma Metastasis Prediction System Based on Molecular Markers
ActiveCN107545144BEasy to distinguishEasy CalibrationHealth-index calculationPheochromocytomaGenes mutation
Owner:SHANGHAI INST FOR ENDOCRINE & METABOLIC DISEASES +1
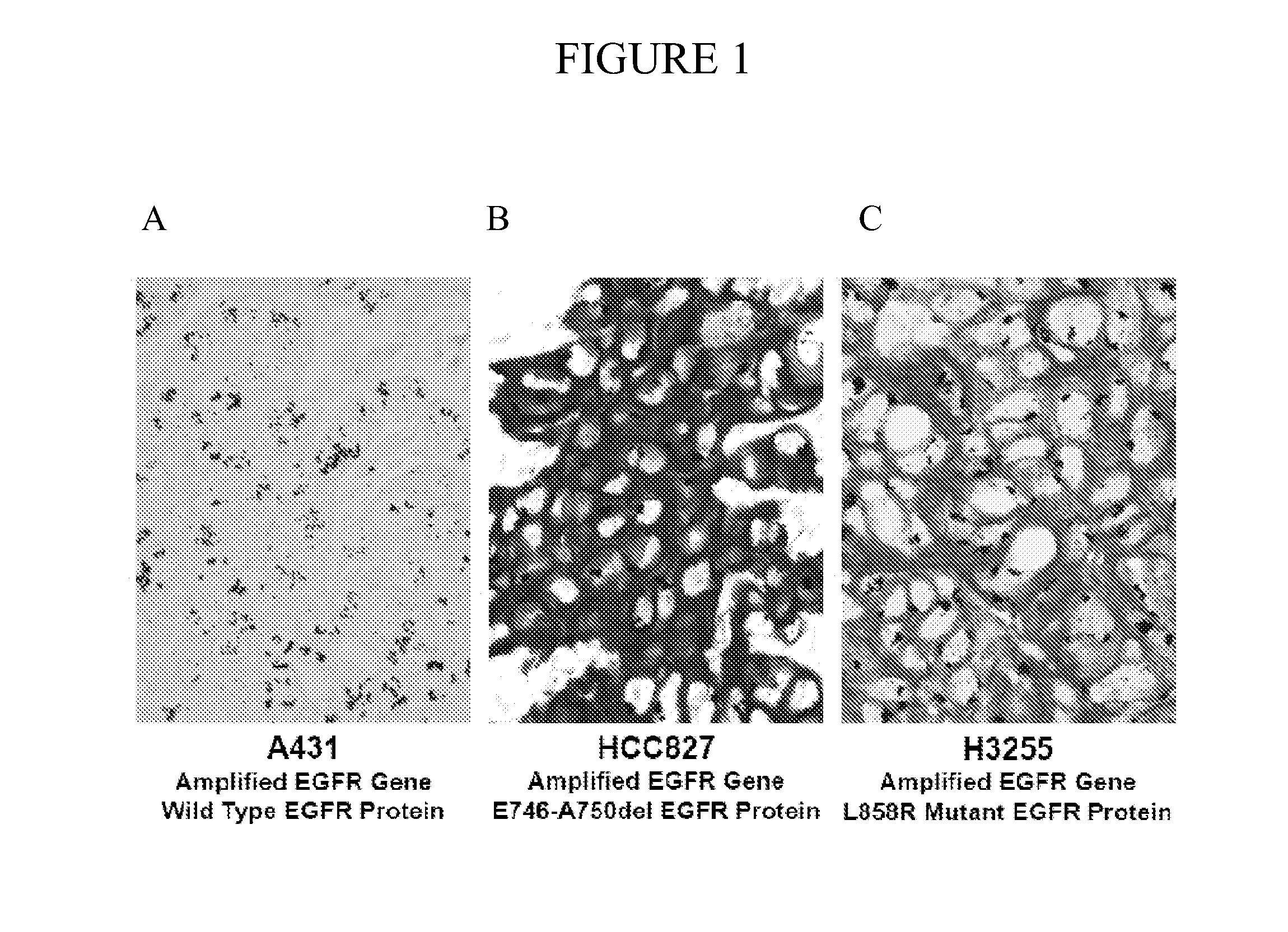
Radiolabeled macrocyclic EGFR inhibitor
ActiveUS10716869B2Isotope introduction to heterocyclic compoundsRadioactive preparation carriersPatient stratificationTherapeutic treatment
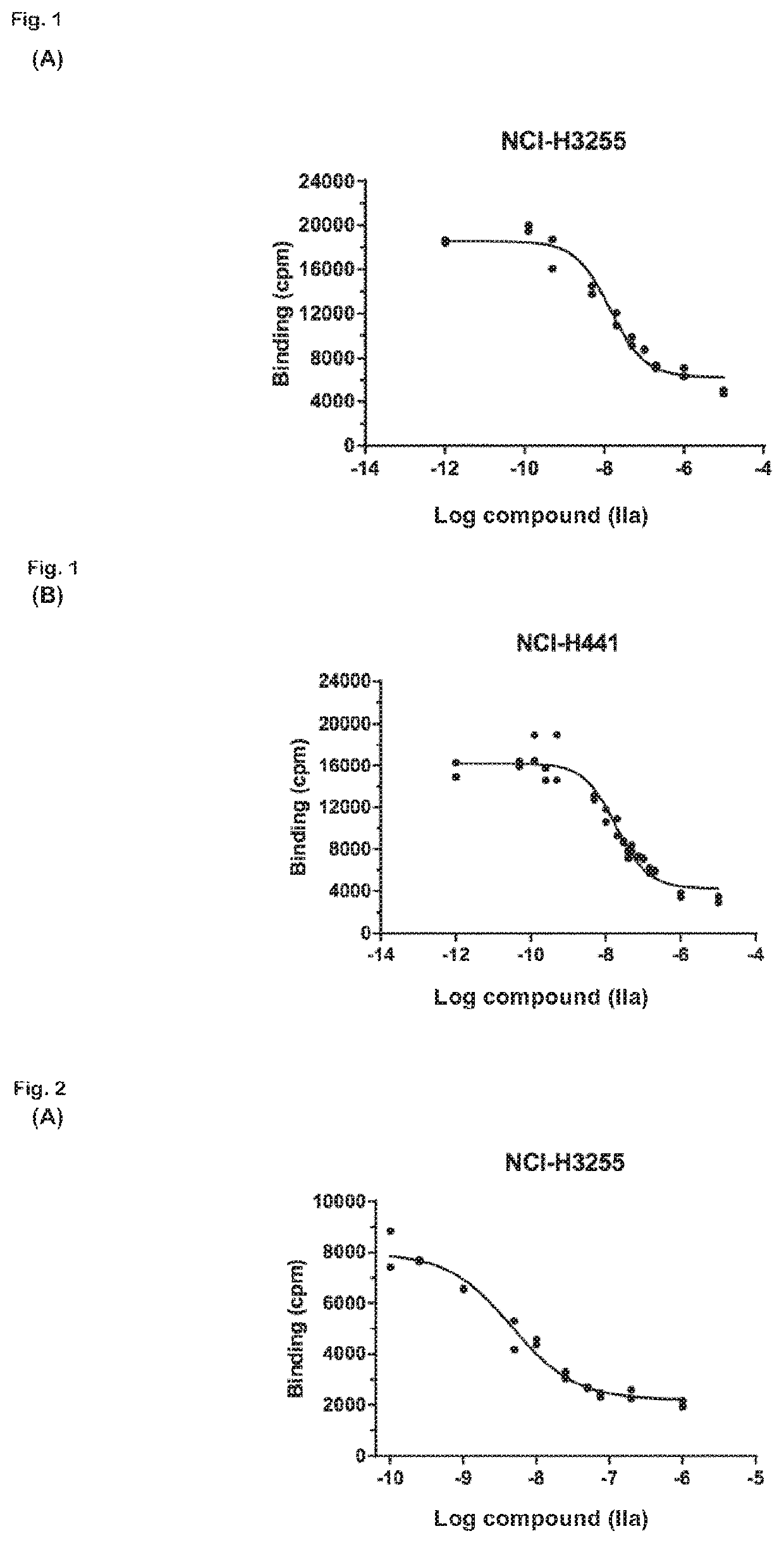
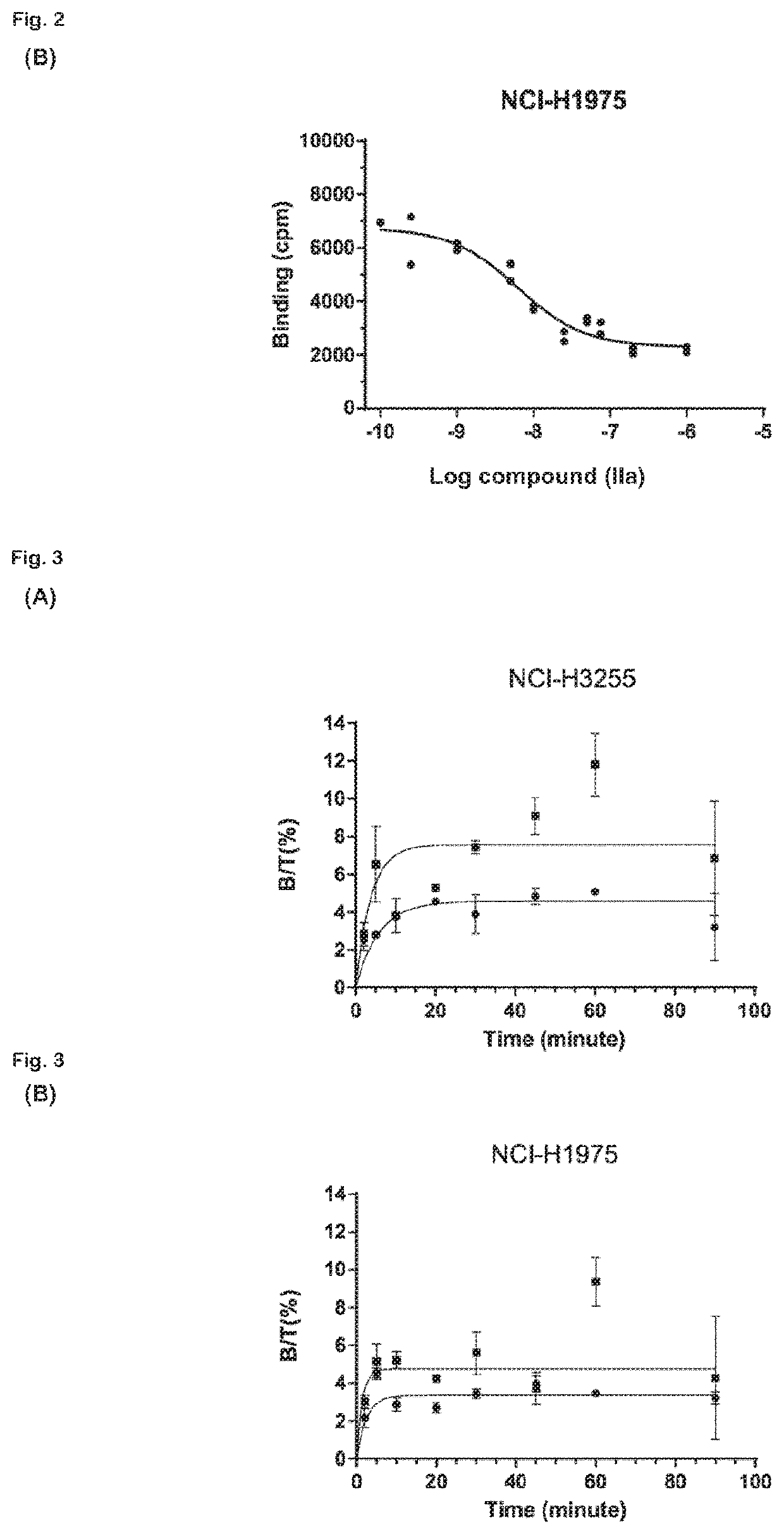
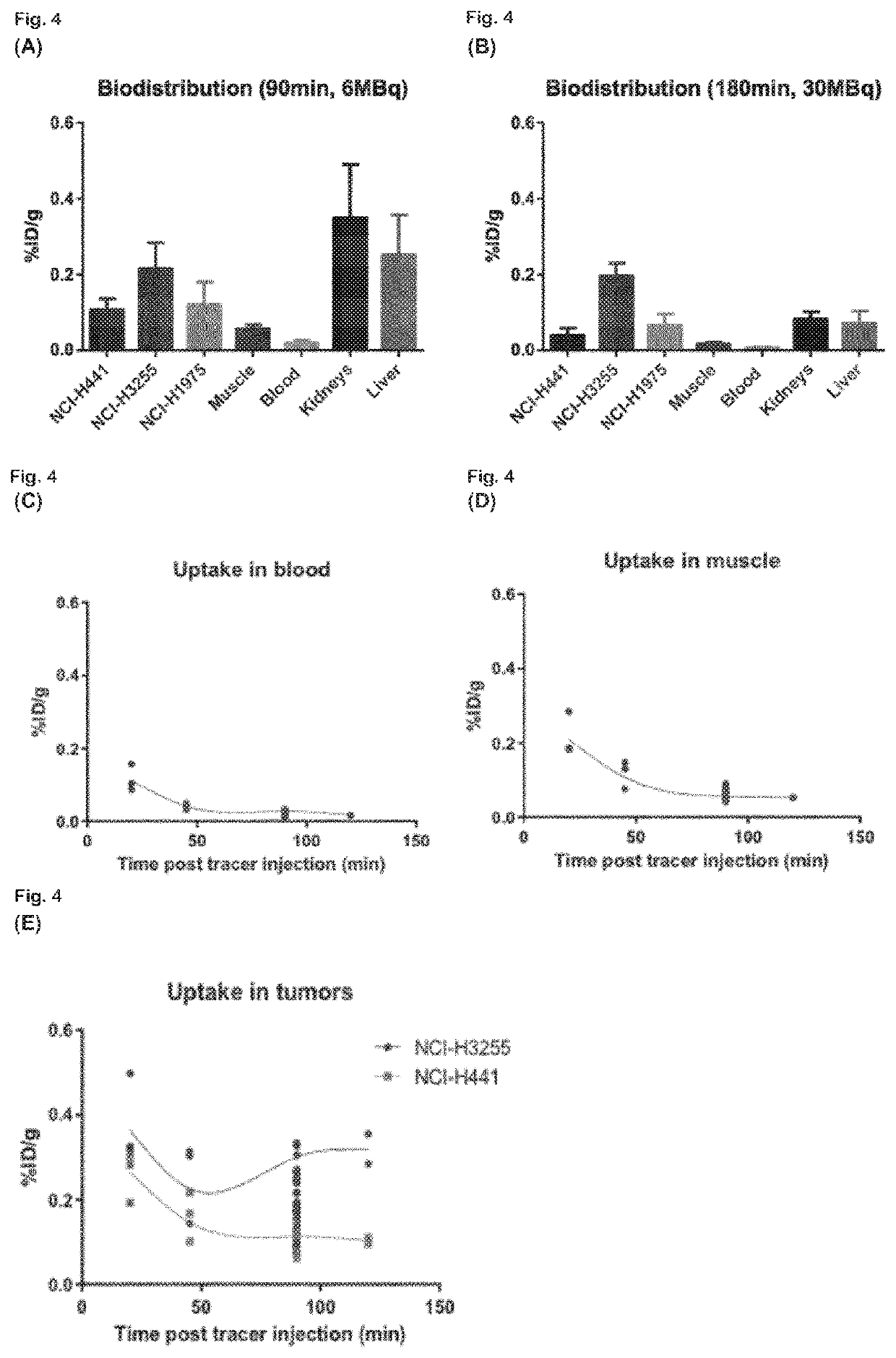
The present invention relates to 18-Fluor radiolabeled macrocyclic quinazoline compounds, which are suitable as positron emission tomography (PET) tracers for imaging epidermal growth factor receptors (EGFR), and their use in in vivo diagnosis, preclinical and clinical tumour imaging, patient stratification on the basis of mutational status of EGFR, and assessing tumour response to therapeutic treatments. The present invention also describes precursor compounds and methods of preparing the radiotracers. The invention is relevant to any cancer that is influenced or driven by deregulated EGFR, such as, but not limited to, non-small cell lung cancer (NSCLC), pancreatic, hepatocellular, oesophageal, gastric, colorectal, prostate, cervical, renal, ovarian, breast cancers, head and neck squamous cell carcinoma, and malignant glioma.
Owner:ONCODESIGN PRECISION MEDICINE (OPM)
Method Of Determining PIK3CA Mutational Status In A Sample
PendingUS20220298565A1Enhance rare allele detectionHigh sensitivityMicrobiological testing/measurementExonFree dna
An ultra-sensitive, specific methodology for detecting PIK3CA mutations in biological samples of cancer patients, comprises a combination of allele-specific, asymmetric rapid PCR and melting analysis in a DNA sample from Circulating Tumor Cells, cell-free DNA in plasma / serum, or Formalin-Fixed Paraffin-Embedded tissues. Using the allele-specific primers for hotspot mutations in exons 9 and 20 (E545K and H1047R), detection can enhance amplification of mutant PIK3CA allele sequence, whereas presence of corresponding competitive blocking unlabeled probes for each exon can avoid non-specific amplification of wild-type PIK3CA sequence increasing the sensitivity and the specificity of method. The mutational detection is completed with melting curve analysis of the unlabeled probe and DNA template of the mutant PIK3CA sequence. Evaluation of PIK3CA mutational status on CTC in peripheral blood and cfDNA in plasma / serum of patients has potential for clinical applications and therapeutic interventions, since presence of PIK3CA mutations is associated with response to molecular targeted therapies.
Owner:PHARMASSIST
A prognostic evaluation system for endometrial cancer incorporating molecular typing and immune scoring
ActiveCN113046438BSignificant difference in prognosisImprove predictive performanceMicrobiological testing/measurementBiological material analysisParanasal Sinus CarcinomaMismatch Repair Protein
Owner:PEKING UNION MEDICAL COLLEGE HOSPITAL CHINESE ACAD OF MEDICAL SCI
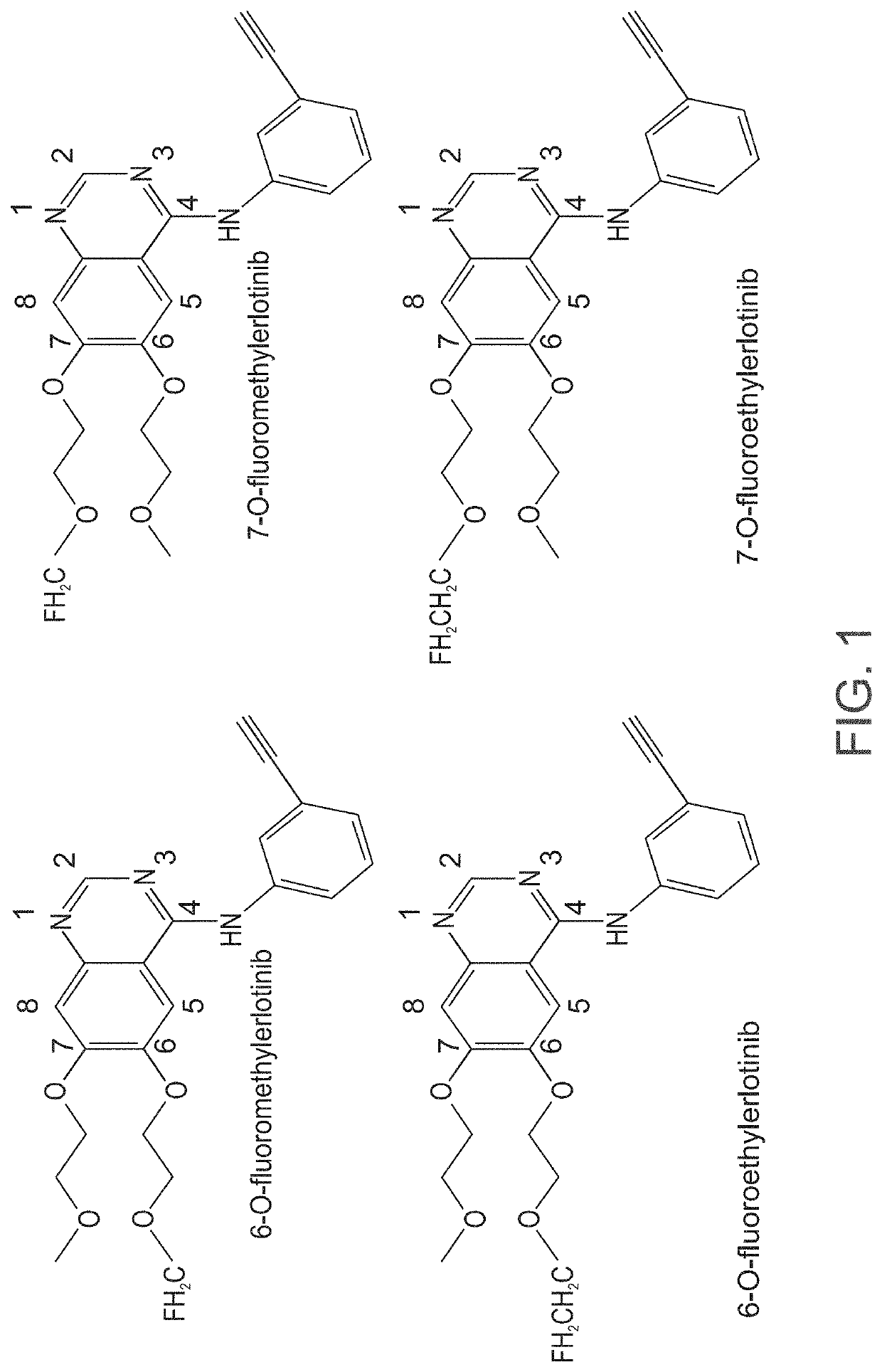
Radiolabeled erlotinib analogs and uses thereof
ActiveUS10710968B2Good choiceEfficiently employedIsotope introduction to heterocyclic compoundsMicrobiological testing/measurementErlotinibHalogen
Radiolabeled compounds which are erlotinib analogs that feature a radioactive halogen and processes of preparing same are disclosed. Uses of these radiolabeled compounds in radioimaging, for identifying and monitoring a level, distribution and / or mutational status of deregulated EGFR, and / or in radiotherapy, are also disclosed.
Owner:HADASIT MEDICAL RES SERVICES & DEVMENT
Method of determining PIK3CA mutational status in a sample
ActiveUS11279979B2Enhance rare allele detectionHigh sensitivityMicrobiological testing/measurementExonFree dna
An ultra-sensitive, specific methodology for detecting PIK3CA mutations in biological samples of cancer patients, comprises a combination of allele-specific, asymmetric rapid PCR and melting analysis in a DNA sample from Circulating Tumor Cells, cell-free DNA in plasma / serum, or Formalin-Fixed Paraffin-Embedded tissues. Using the allele-specific primers for hotspot mutations in exons 9 and 20 (E545K and H1047R), detection can enhance amplification of mutant PIK3CA allele sequence, whereas presence of corresponding competitive blocking unlabeled probes for each exon can avoid non-specific amplification of wild-type PIK3CA sequence increasing the sensitivity and the specificity of method. The mutational detection is completed with melting curve analysis of the unlabeled probe and DNA template of the mutant PIK3CA sequence. Evaluation of PIK3CA mutational status on CTC in peripheral blood and cfDNA in plasma / serum of patients has potential for clinical applications and therapeutic interventions, since presence of PIK3CA mutations is associated with response to molecular targeted therapies.
Owner:PHARMASSIST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com