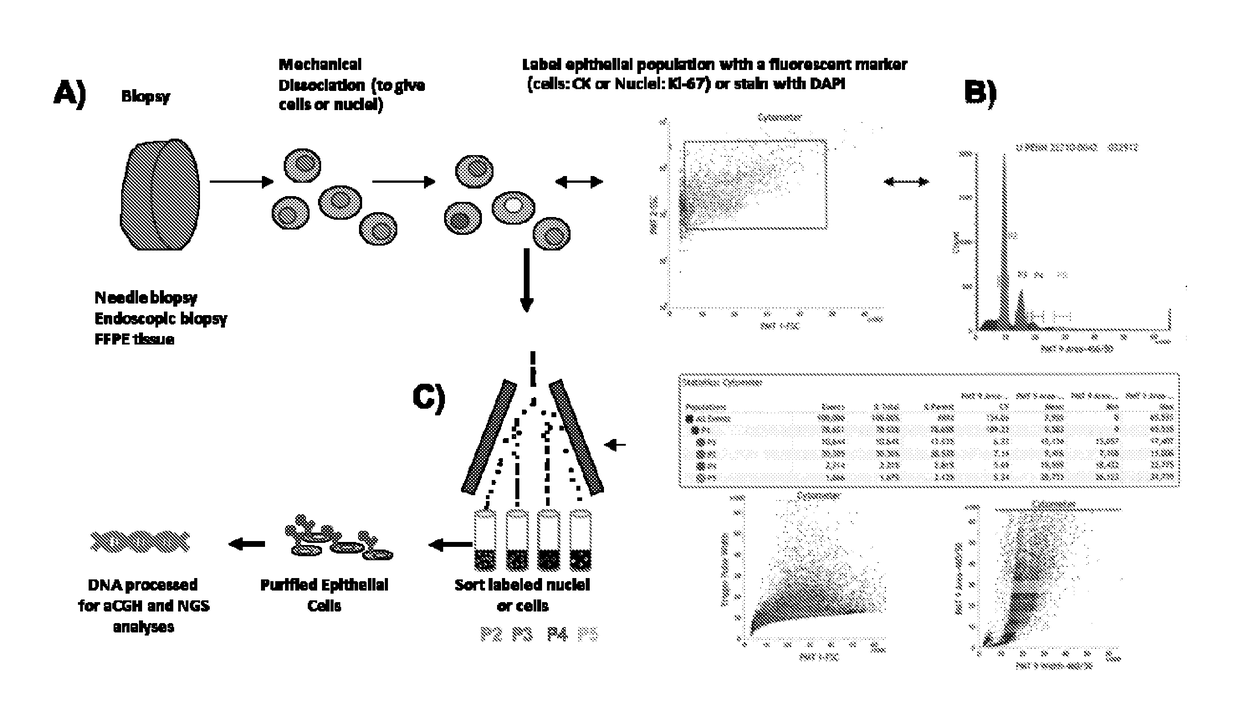
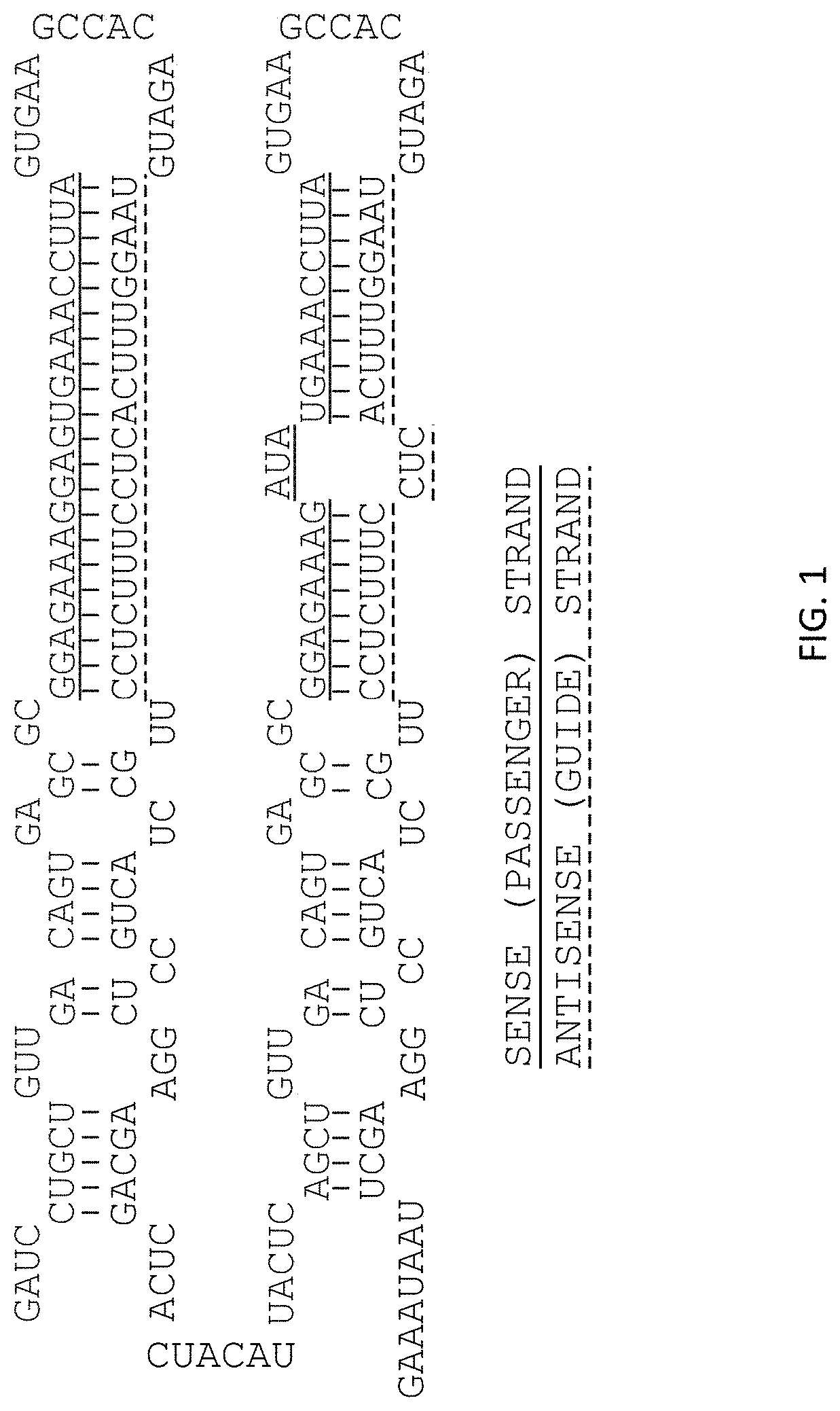
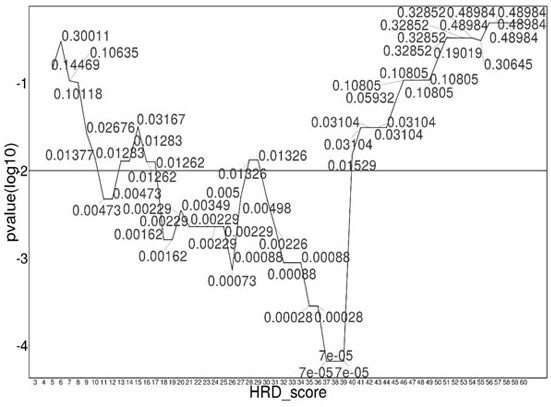
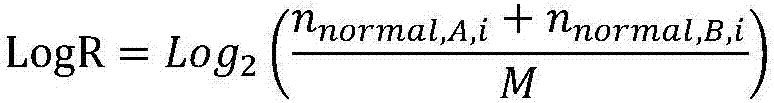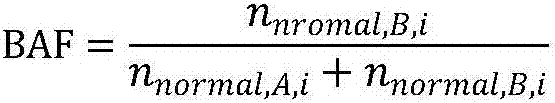Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
32 results about "Homologous Recombination Deficiency" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for predicating homologous recombination deficiency mechanism and method for predicating response of patients to cancer therapy
InactiveCN107287285AInnovativeOvercoming the pitfalls of inaccurate forecastsMicrobiological testing/measurementSequence analysisAbnormal tissue growthPolymerase L
The invention discloses a method for predicating a homologous recombination deficiency (HRD) mechanism and a method for predicating response of patients to cancer therapy and relates to the field of biological information predication. The method comprises the step of judging whether a tumor sample has homologous recombination deficiency or not according to one or more comprehensive values in a large-segment INDEL (Insertion / Deletion) fraction, a copy number variation fraction and a tumor mutation load fraction, wherein the comprehensive values can also comprise a loss of heterozygosity variation fraction. By adopting the method disclosed by the invention, predication of a chromosome large-segment structure, a chromosome gene type number, a chromosome gene copy number, a chromosome variation interval and abnormal loss of heterozygosity and chromosome telomeric imbalance is realized, so that an evaluation range is more complete and HRD can be accurately predicated; the comprehensive values also can be used for determining whether the patients have response to a therapeutic regimen containing one or more of a PARP (Poly Adenosine Diphosphate Ribose Polymerase) inhibitor, an DNA (Deoxyribonucleic Acid) injury inhibitor, a topoisomerase II / II+inhibitor, a topoisomerase I inhibitor and radiotherapy; the method is simple and has wide general applicability.
Owner:SHANGHAI ORIGIMED CO LTD
Single-sample whole-genome allele specific copy number variation prediction method
ActiveCN112802548ALow sequencing depthImprove detection accuracyBiostatisticsProteomicsSingle sampleGenetics
A single-sample whole-genome allele specific copy number variation prediction method comprises the following steps: analyzing and comparing sequencing data of a to-be-detected sample to a reference genome, and extracting tumor purity information, whole-genome replication information and total copy number variation information; performing classification processing according to the total copy number variation information of each segment of the chromosome, converting the total copy number variation information into allele specific copy number variation information, if the total copy number variation information of the chromosome segment is an odd number interval or a zero interval, directly calculating the allele specific copy number variation information, and if the total copy number variation information of the chromosome segment is an odd number interval or a zero interval, directly calculating the allele specific copy number variation information; and if the total copy number variation information of the chromosome segment is a non-zero even interval, obtaining allele specific copy number variation information of the interval through model prediction. According to the method, only a single sample is needed, a normal sample does not need to be paired, the sequencing depth of the needed to-be-detected sample is low, the detection accuracy is high, and the homologous recombination defect of the low-tumor-purity sample can be detected.
Owner:苏州吉因加医学检验有限公司
Method and test kit for determining genome instability on basis of next-generation sequencing technology
ActiveCN111676277AHigh detection sensitivityImprove capture efficiencyMicrobiological testing/measurementBiostatisticsGenomic StabilityGenome instability
The invention relates to a method and a test kit for determining genome instability on the basis of a next-generation sequencing technology, belonging to the technical field of molecular detection. Inorder to overcome the defect that genome structure variation is not considered during detection of homologous recombination defects in the prior art, the invention provides the method and the test kit for determining genome instability on the basis of the next-generation sequencing technology. The method comprises the following steps: breaking a to-be-detected gene, then adding an A connector fora corresponding PCR reaction, carrying out hybridization capture through a designed probe, then amplifying a captured DNA library, sequencing the library, and carrying out evaluating through a software so as to determine the homologous recombination defect state. Compared with the prior art, the method provided by the invention can comprehensively evaluate the HRD state, is reliable in data results and is applicable to popularization.
Owner:臻和(北京)生物科技有限公司 +1
Homologous recombination defect judgment method based on DNA sequencing data
ActiveCN111462823AOvercoming the Difficulty of Data InsufficiencyThe result is easy to judgeBiostatisticsSequence analysisGenomic StabilityGenome instability
The invention discloses a homologous recombination defect judgment method based on DNA sequencing data. The method comprises the following steps: acquiring characteristic attributes; extracting validdata; based on a triple learning method framework, selecting three different base classifiers H1, H2 and H3 by considering good generalization ability, high accuracy and processing efficiency for multi-dimensional feature attributes; performing iterative training on the H1, H2 and H3 to obtain an extended training set, and updating a model to complete a training process; and marking a unmarked sample set U by using the trained model, and completing judgment of the HRD state according to the marking result. According to the method, the limitation that HRD state judgment is carried out by usinglocal characteristics such as a single genome instability state or a small number of genome instability states and the like is solved, the difficulty that the number of samples with clinically known HRD states is extremely small is overcome, learning of multi-characteristic attributes under existing sample data is achieved, and the performance of the HRD judgment method can be improved.
Owner:XI AN JIAOTONG UNIV +1
Methods and materials for assessing homologous recombination deficiency
InactiveUS20140363521A1Raise the possibilityHeavy metal active ingredientsMicrobiological testing/measurementRegimenCancer cell
This document provides methods and materials involved in assessing samples (e.g., cancer cells) for the presence of homologous recombination deficiency (HRD) or an HRD signature. For example, methods and materials for determining whether or not a cell (e.g., a cancer cell) contains an HRD signature are provided. Materials and methods for identifying cells (e.g., cancer cells) having a deficiency in homology directed repair (HDR) as well as materials and methods for identifying cancer patients likely to respond to a particular cancer treatment regimen also are provided.
Owner:MYRIAD GENETICS
HRD score calculation method based on chip detection
PendingCN112397145AWidely applicableLow costMicrobiological testing/measurementSequence analysisTumor SampleHomologous Recombination Deficiency
The invention discloses an HRD score calculation method based on chip detection. The HRD score calculation method comprises the specific operation steps: 1, establishing a genome heterozygosity deletion LOH calculation method; 2, establishing a calculation method of the telomere allele imbalance TAI; 3, establishing a calculation method of large chip end migration LST; 4, establishing an HRD calculation method. The invention provides a genome heterozygosity deletion LOH calculation method, a telomere allelic imbalance TAI calculation method, a large fragment end migration LST calculation method and an HRD calculation method, which can perform HRD score calculation on paraffin samples, and can evaluate HRD of tumor tissues by combining data of an Affymetrix OncoScan CNV FFPE gene chip withan HRD evaluation method; and the homologous recombination defect state of the detected tumor sample can be more accurately judged through the HRD score.
Owner:HENAN CANCER HOSPITAL
Methods for characterizing and treating a cancer type using cancer images
Described herein are methods, systems, devices and computer program products for characterizing or identifying a type of cancer. Also described are methods of treating a characterized or identified chancer. For example, certain methods may be used to characterize a homologous recombination deficiency status of a cancer.
Owner:TESARO INC
Methods and materials for assessing homologous recombination deficiency
InactiveUS20180030544A1Heavy metal active ingredientsMicrobiological testing/measurementRegimenCancer cell
This document provides methods and materials involved in assessing samples (e.g., cancer cells) for the presence of homologous recombination deficiency (HRD) or an HRD signature. For example, methods and materials for determining whether or not a cell (e.g., a cancer cell) contains an HRD signature are provided. Materials and methods for identifying cells (e.g., cancer cells) having a deficiency in homology directed repair (HDR) as well as materials and methods for identifying cancer patients likely to respond to a particular cancer treatment regimen also are provided.
Owner:MYRIAD GENETICS
Genome recombination fingerprint for characterizing hHRD homologous recombination deficiency and identification method thereof
The invention relates to a novel method for identifying homologous recombination deficiency and application thereof. More particularly, the present invention relates to a characteristic genome recombination fingerprint which is related to hHRD type homologous recombination repair deficiency and is resolved by using high throughput genome re-sequencing, bioinformatics analysis and statistical correlation analysis. The characteristic hHRD recombination fingerprint is caused by the functional deficiency of a specific homologous recombination repair mechanism (namely CRL4WDR70-H2B mono-ubiquitination pathway), and comprises the total frequency of genome recombination in a single sample, the composition of chromosome structure variation types and site-specific copy number variation. The recombinant fingerprint is used for identifying infectious diseases or tumors with the characteristic mutant fingerprint, and is used for guiding targeted drug treatment of PARP inhibitors. The invention relates to the method and application thereof for diseases including but not limited to breast cancer, ovarian cancer, endometrial (like) cancer, ovarian clear cell cancer, prostate cancer, pancreatic cancer, skin cancer and gastric cancer with such characteristics, as well as hepatitis B virus infection or related liver fibrosis cirrhosis, liver cancer and cholangiocarcinoma.
Owner:成都吉诺迈尔生物科技有限公司 +1
Identification method of group of genomic characteristic mutation fingerprints associated with homologous recombination repair defects
The present invention discloses a genomic characteristic mutation fingerprint related to hHRD type homologous recombination repair defects and comprises a combination characteristic of a single nucleotide variation (SNV) and insertion deletion (Indel) genomic fingerprint, and an identification method thereof. The fingerprint characteristics can be subjected to high-throughput whole-genome resequencing, bioinformatics software analysis and statistical correlation analysis and are used to identify infectious diseases or tumors with the characteristic mutation fingerprints, especially hepatitis Bvirus infection and related diseases, including liver cirrhosis, liver fibrosis and liver cancer. The mutation fingerprint can also be used to guide methods and uses of treatments of targeted drugs targeting the homologous recombination defects on a variety of the various tumors with the hHRD homologous recombination repair defects, including but not limited to breast cancer, ovarian cancer, endometrial cancer, endometrioid carcinoma, ovarian clear cell carcinoma, prostate cancer, gastric cancer, skin cancer, hepatitis B virus infection or related liver cirrhosis, liver cancer and bile duct cell cancer.
Owner:THE WEST CHINA SECOND UNIV HOSPITAL OF SICHUAN +1
Compositions and methods for targeting and treating homologous recombination-deficient tumors
InactiveUS20190133980A1Antineoplastic agentsHeterocyclic compound active ingredientsDNA Repair InhibitionA-DNA
Owner:YALE UNIV
Method of characterising a DNA sample
Owner:GENOME RES LTD
Homologus Recombination Deficiency-Interstitial Aberration (HRD-IA) Assay
InactiveUS20180291460A1Useful in treatmentInhibit functioningMicrobiological testing/measurementDNA Repair InhibitionHomologous Recombination Deficiency
Owner:MAYO FOUND FOR MEDICAL EDUCATION & RES
Method and kit for detecting homologous recombination pathway gene mutation based on next-generation sequencing technology
InactiveCN111793678AHigh detection sensitivityImprove capture efficiencyMicrobiological testing/measurementGene MutantHomologous Recombination Deficiency
The invention relates to a method and a kit for detecting homologous recombination pathway gene mutation based on a next-generation sequencing technology, and belongs to the technical field of molecular detection. In order to overcome the technical defects that in the prior art, other genes on a homologous recombination pathway are not considered when homologous recombination defects are detected,the invention provides the method and the kit for detecting homologous recombination pathway gene mutation based on the next-generation sequencing technology. The method comprises the following steps: breaking a gene to be detected, adding a linker A, carrying out corresponding PCR, carrying out hybridization capture by using a designed probe, amplifying the captured DNA library, carrying out library sequencing, and carrying out evaluating by using software to determine the homologous recombination pathway gene mutation. Compared with the prior art, the method is comprehensive in gene detection, reliable in data result and suitable for popularization.
Owner:ZHENYUE BIOTECHNOLOGY JIANGSU CO LTD
Methods for treating cancers
Compositions and methods for prevention of ovarian cancer recurrence and for the treatment of BRCA1 / 2-wild type ovarian cancer are disclosed herein. In some embodiments, the composition comprises an autologous tumor cell vaccine comprising cells genetically modified for furin knockdown and GM-CSF expression. In some embodiments, the method comprises administration of an autologous tumor cell vaccine prior to administration of a combination of the autologous tumor cell vaccine and atezolizumab. Also disclosed herein are methods for treating a cancer in an individual comprising a wild-type BRCA1 gene, a wild-type BRCA2 gene, or a combination thereof, and is identified as homologous recombination deficiency (HRD)-negative.
Owner:GRADALIS
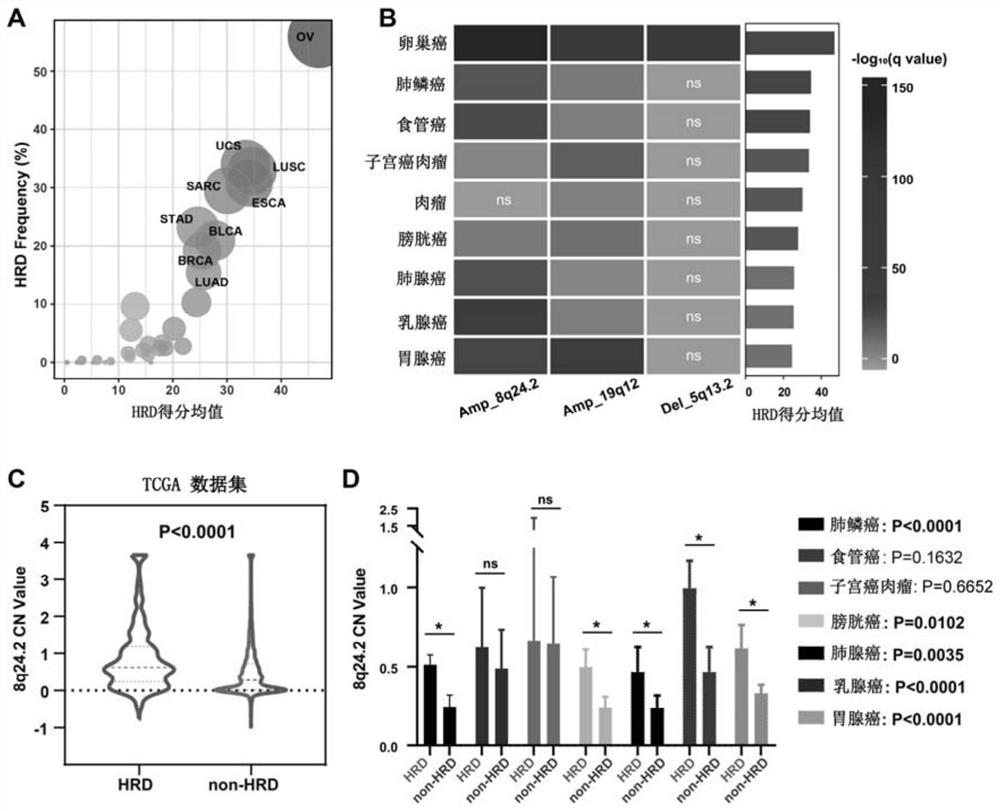
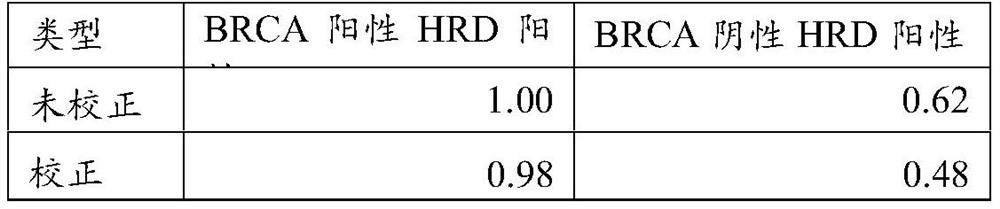
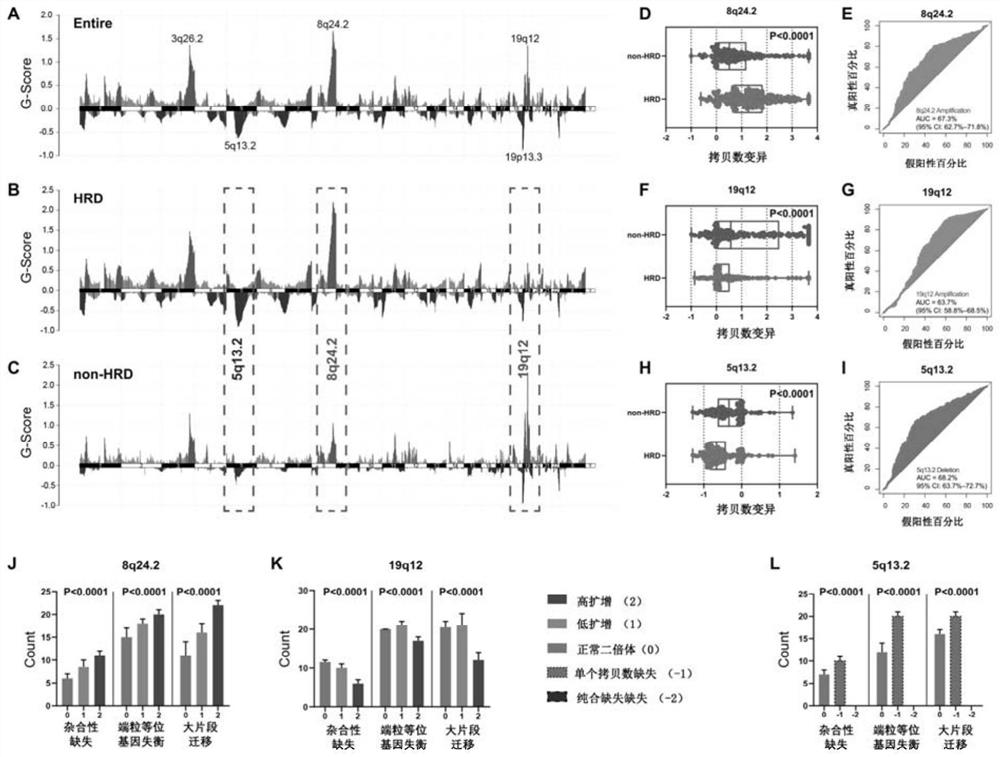
A method and application for predicting homologous recombination deficiency in ovarian cancer based on genomic copy number variation biomarkers
ActiveCN113684277BForecast stabilityNot susceptible to back mutationsMicrobiological testing/measurementDNA/RNA fragmentationTissue biopsyBiomarker (medicine)
The invention relates to the field of biomedicine, in particular to a method and application for predicting homologous recombination deficiency in ovarian cancer based on biomarkers of genome copy number variation. The biomarker is a CNV based on any of chromosome segment 5q13.2, 8q24.2 or 19q12. The method for predicting the homologous recombination deficiency of ovarian cancer by the biomarkers is as follows: collecting surgical resection samples or tissue biopsy samples of ovarian cancer patients; performing DNA sequencing on the samples to obtain corresponding sequencing files; using GISTIC 2.0 to analyze the DNA sequencing files , to obtain the CNV map of the whole tumor genome; predict the HRD status of ovarian cancer based on the CNV of chromosome fragment 5q13.2, 8q24.2 or 19q12. The present invention provides the application of copy number variation CNV at the subchromosomal and gene levels in predicting homologous recombination defects in ovarian cancer patients, so as to screen potential beneficiaries of targeted therapy and bring more clinical benefits to HRD patients.
Owner:NANFANG HOSPITAL OF SOUTHERN MEDICAL UNIV
A method and kit for determining genome instability based on next-generation sequencing technology
ActiveCN111676277BHigh detection sensitivityImprove capture efficiencyMicrobiological testing/measurementBiostatisticsGenomic StabilityGenome instability
Owner:臻和(北京)生物科技有限公司 +1
A detection method and quality control system for homologous recombination defects based on ngs platform
ActiveCN113658638BCorrect accuracySolve the problem of light pollutionProteomicsGenomicsAllele frequencyNucleotide
The invention provides a method for correcting tumor purity in the process of bioinformatics analysis and a method for detecting homologous recombination defects based on an NGS platform. The method of the invention compares the sequencing depth and single nucleotide polymorphism of clinical samples and negative samples in the target region The differences in the allele frequencies of sex loci can be effectively corrected for tumor purity and ploidy, and HRD assessment can be realized.
Owner:SIMCERE DIAGNOSTICS CO LTD +2
Application of gene combination in preparation of products for graded detection of human tumor homologous recombination defect, tumor mutation load and microsatellite instability
PendingCN114854861AImprove reliabilityIncrease credibilityMicrobiological testing/measurementHuman tumorHomologous Recombination Deficiency
The invention relates to the field of grading detection of tumors, in particular to application of a gene combination to preparation of products for grading detection of human tumor homologous recombination defects, tumor mutation loads and microsatellite instability, and the gene combination is composed of a gene set A and a gene fragment set B, the gene combination is obtained from actual high-throughput sequencing data of a first hospital of Beijing University through specific pairing clustering analysis, the data from the real world has higher reliability and credibility, and homologous recombination defect, tumor mutation load and microsatellite instability grading and prediction can be accurately carried out on the pan cancer.
Owner:PEKING UNIV FIRST HOSPITAL
Method and device for correcting homologous recombination defect score and storage medium
ActiveCN114067909AEliminate increaseEliminate the situationProteomicsGenomicsOptimality modelHomologous Recombination Deficiency
The invention discloses a method and device for correcting a homologous recombination defect score and a storage medium. The method comprises the following steps that: a system CNV and a system SNV of a to-be-detected sample are acquired; a WGD value, an original LOH score value, an original TAI score value and an original LST score value of the to-be-detected sample under an optimal model are calculated by utilizing system CNV and system SNV; and for the to-be-detected sample subjected to WGD, the corrected LST score value is equal to the product of (1-k1* WGD value) and the original LST score value, and the corrected TAI score value is equal to the product of (1-k2 * WGD value) and the original LST score value. According to the method and the device for correcting the homologous recombination defect score, the TAI and the LST are corrected by using the WGD value, so that the problem that TAI and LST scores of a whole genome multiplication sample are relatively high is solved, and the sensitivity and the accuracy of homologous recombination defect state evaluation are improved.
Owner:北京吉因加医学检验实验室有限公司 +1
Method and device for evaluating homologous recombination defects of single sample and storage medium
The invention discloses a method and device for evaluating homologous recombination defects of a single sample and a storage medium. The method comprises the following steps of: acquiring SNV mutation sites of a to-be-detected tumor sample and the mutation frequency and mutation site depth of each SNV mutation site, and annotating the SNV mutation sites to obtain a system SNV mutation site set; according to the comparison result of the to-be-detected tumor sample, analyzing segments of the tumor sample with CNV mutation, and the size, including the number of probes and the BAF value, of each segment, and annotating CNV mutation sites to obtain a system CNV mutation site set; and calculating an LOH score value, a TAI score value and an LST score value by utilizing system CNV and SNV results, thereby realizing the homologous recombination defect scoring of the single sample. According to the method, homologous recombination defect evaluation can be achieved only through the to-be-detected tumor sample, and the defect that an existing homologous recombination defect evaluation method depends on a blood cell sample is overcome.
Owner:SHENZHEN GENEPLUS CLINICAL LAB +1
Method for predicting ovarian cancer homologous recombination defect based on genome copy number variation biomarker and application
ActiveCN113684277AForecast stabilityNot susceptible to back mutationsMicrobiological testing/measurementDNA/RNA fragmentationTissue biopsyBiomarker (medicine)
The invention relates to the field of biomedicine, in particular to a method for predicting ovarian cancer homologous recombination defects based on a genome copy number variation biomarker and application. The biomarker is a CNV based on any one of a chromosome segment 5q13. 2, a chromosome segment 8q24.2 or a chromosome segment 19q12. The method for predicting the ovarian cancer homologous recombination defect by using the biomarker comprises the following steps of, collecting an operation excision sample or a tissue biopsy sample of an ovarian cancer patient; performing DNA sequencing on the sample to obtain a corresponding sequencing file; analyzing the DNA sequencing file by using GISTIC 2.0 to obtain a CNV map of a tumor whole genome; and predicting HRD status of ovarian cancer based on the CNV of the chromosome segment 5q13.2, 8q24.2 or 19q12. The invention provides the application of the copy number variation CNV at the subchromosome and gene level in predicting the homologous recombination defect of the ovarian cancer patient, so that potential benefit people of targeted therapy are screened, and more clinical benefits are brought to the HRD patient.
Owner:NANFANG HOSPITAL OF SOUTHERN MEDICAL UNIV
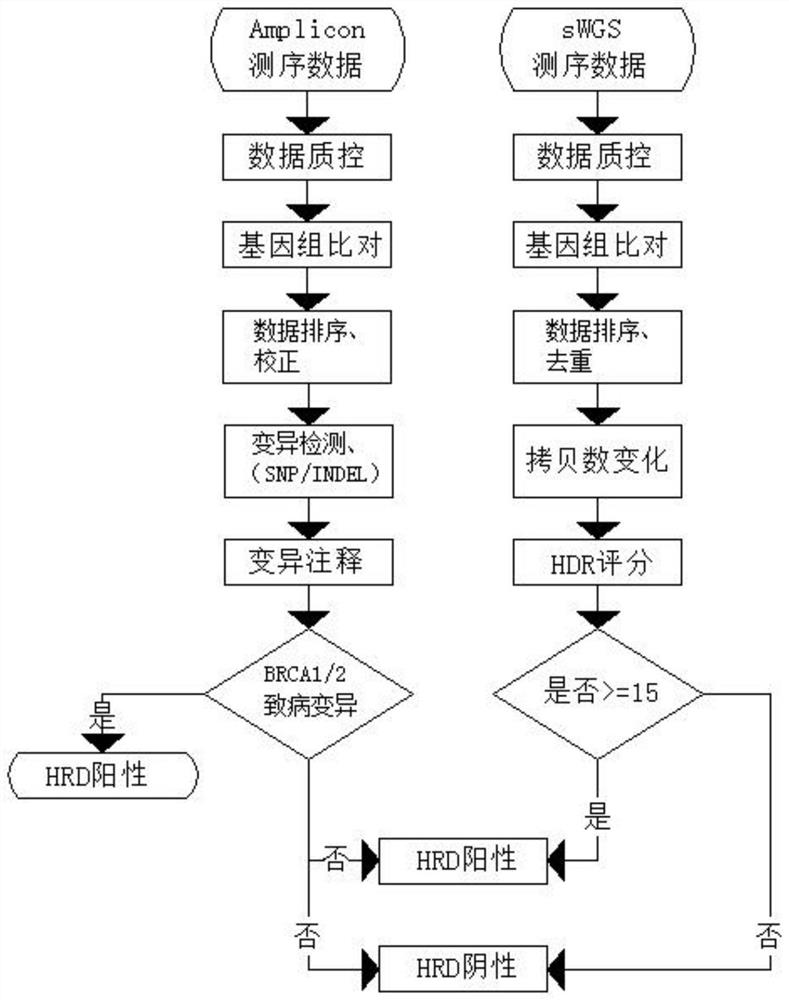
Homologous recombination defect detection method
According to the homologous recombination defect detection method provided by the invention, the detection of accompanying diagnosis of multiple drugs can be simultaneously met, the requirement on the initial quantity of a sample is low, the cost is low, and the detection period is short; the homologous recombination defect detection method comprises the following steps: S1, carrying out total DNA extraction, DNA fragmentation and DNA amplification on a human tissue sample; s2, detecting a part of amplified tissue sample DNA by adopting an SNP chip, and calculating an HRD value; next-generation sequencing is carried out on the other part of amplified tissue sample DNA, and the gene mutation state in an HRD signal channel and accompanying diagnosis of other targeted drugs are evaluated; and S3, integrating an SNP chip detection result and a next-generation sequencing result, and judging which tumor treatment medicine the sample can benefit from.
Owner:YIKON GENOMICS SHANGHAI CO LTD
Homologous recombination deficiency (HRD) detection method and reagent set thereof
PendingCN114096681AMicrobiological testing/measurementProteomicsGeneticsHomologous Recombination Deficiency
The invention provides a method, a system and a reagent set for evaluating the homologous recombination deficiency (HRD) state of an individual. The invention further provides a method, a system and a reagent set for determining therapy according to the HRD state of a human individual.
Owner:ACT GENOMICS IP CO LTD
Homologous recombination defect gene analysis method
PendingCN114300053AImprove the detection rateReduce testing costsData visualisationProteomicsRepetitive SequencesGenetics
The invention discloses a homologous recombination defective gene analysis method, which comprises the following steps of: 1, performing quality control and filtering on low-depth whole genome (sWGS) sequencing data by adopting fastp software, comparing the data to a human reference genome by adopting bwa software, sequencing the data according to a chromosome sequence by adopting gatk software, simultaneously marking and removing a repetitive sequence, and comparing the repetitive sequence with the human reference genome; qdnaseq software is adopted to analyze the copy number change in the genome, and shallowHRD software is adopted to evaluate HRD based on the copy number change of the genome and give a score. By accurately evaluating the instability of the genome, the positive detection rate of the homologous recombination defect can be effectively increased, and the detection cost is reduced; meanwhile, the method has the advantages of low time consumption, capability of being used for single sample analysis and the like, and is combined with BRCA1 / 2 gene variation; meanwhile, the depth requirements of gene variation and instability evaluation are met, so that the homologous recombination defect state of a patient is accurately judged, the detection cost is reduced, and the detection economic benefit is improved.
Owner:苏州绘真医学检验有限公司
A method for determining homologous recombination defects based on DNA sequencing data
ActiveCN111462823BOvercoming the Difficulty of Data InsufficiencyThe result is easy to judgeBiostatisticsSequence analysisGenome instabilityData mining
The invention discloses a method for judging homologous recombination defects based on DNA sequencing data, which obtains characteristic attributes; extracts effective data; Attribute processing efficiency, choosing three different base classifiers H 1 , H 2 , H 3 ; for H 1 , H 2 , H 3 Iterative training is performed to obtain an expanded training set, thereby updating the model and completing the training process; using the trained model to label the unlabeled sample set U, and completing the HRD status determination according to the labeling result. The invention solves the limitation of using a single or a small amount of local features such as genomic instability state to determine the HRD state, overcomes the difficulty that the number of samples with known HRD states is very small in clinical practice, and realizes multi-feature attributes under the existing sample data. It can improve the performance of HRD judgment method.
Owner:XI AN JIAOTONG UNIV +1
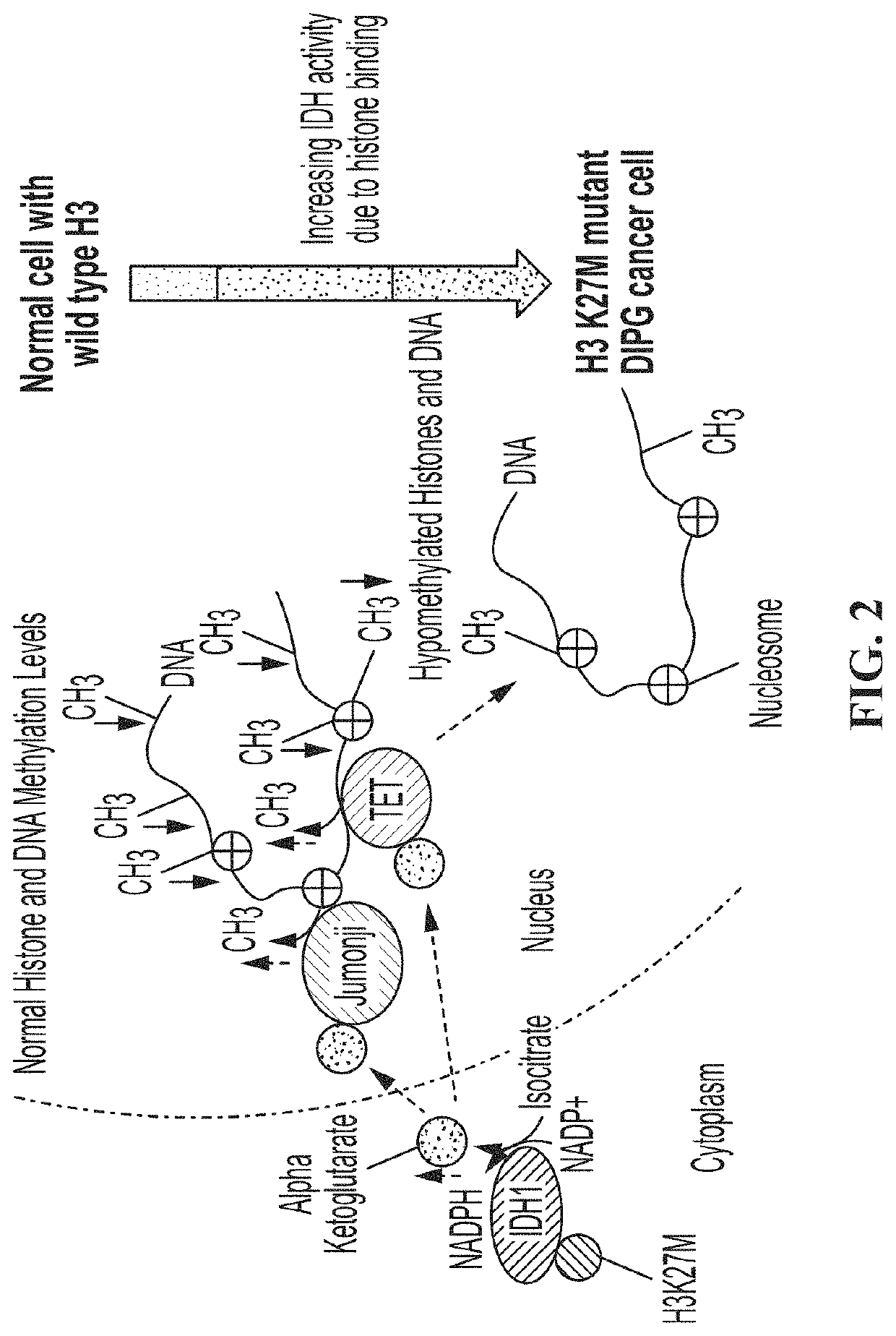
Selective treatment of cancers having histone h3 mutations or aberrant levels of DNA or histone methylation, acetylation or defects in homologous recombination
PendingUS20210369725A1Delay progressSalicyclic acid active ingredientsKetone active ingredientsHistone methylationHistone deacetylase
The invention concerns compositions and a method for treating or delaying the onset, progression, or relapse of a cancer in a subject, the method comprising administering, optionally with radiation therapy, an inhibitor of: (a) DNA-Dependent Protein Kinase catalytic subunit, and an inhibitor of Poly-ADP Ribose Polymerase, wherein the cancer has a histone H3.3 mutation or is a homologous recombination-defective (HR-defective) cancer; or (b) wild-type isocitrate dehydrogenase, wherein the IDH inhibitor is administered in an effective amount to decrease wild-type IDH activity in cells of the cancer, and wherein the cancer carries a histone H3.3 K27M mutation, and / or the cancer has low DNA methylation and / or low histone methylation; or (c) histone acetyltransferase, wherein the cancer carries a histone H3.3 K27M mutation, and / or high histone acetylation; or (d) histone deacetylase, wherein the cancer carries a histone H3.3 K27M mutation, and / or has high histone acetylation; or (e) any combination thereof.
Owner:FLORIDA STATE UNIV RES FOUND INC
A Single-Sample Genome-Wide Method for Predicting Allele-Specific Copy Number Variations
ActiveCN112802548BLow sequencing depthImprove detection accuracyBiostatisticsProteomicsSingle sampleTest sample
A single-sample genome-wide method for predicting allele-specific copy number variations, the method comprising: analyzing the sequencing data of a test sample compared to a reference genome, extracting tumor purity information, genome-wide replication information, and total copy number variations information, and then perform classification processing according to the total copy number variation information of each segment of the chromosome, and convert the total copy number variation information into allele-specific copy number variation information, if the total copy number variation information of the chromosome segment is an odd interval or 0 interval, the allele-specific copy number variation information is directly deduced, and if the total copy number variation information of the chromosome segment is a non-zero even interval, the allele-specific copy number variation of the interval is obtained through model prediction information. The invention only needs a single sample and does not need paired normal samples. The required sample to be tested has a low sequencing depth and high detection accuracy, and can detect homologous recombination defects in samples with low tumor purity.
Owner:苏州吉因加医学检验有限公司
Methods for treating cancers
ActiveUS11400170B2Additional recurrencePeptide/protein ingredientsMammal material medical ingredientsBrca1 geneWild type
Compositions and methods for prevention of ovarian cancer recurrence and for the treatment of BRCA1 / 2-wild type ovarian cancer are disclosed herein. In some embodiments, the composition comprises an autologous tumor cell vaccine comprising cells genetically modified for furin knockdown and GM-CSF expression. In some embodiments, the method comprises administration of an autologous tumor cell vaccine prior to administration of a combination of the autologous tumor cell vaccine and atezolizumab. Also disclosed herein are methods for treating a cancer in an individual comprising a wild-type BRCA1 gene, a wild-type BRCA2 gene, or a combination thereof, and is identified as homologous recombination deficiency (HRD)-negative.
Owner:GRADALIS
Heteroarylmethylene derivatives as DNA polymerase theta inhibitors
Disclosed herein are certain acetamido derivatives that are DNA Polymerase Theta (Polθ) inhibitors of Formula (I). Also, disclosed are pharmaceutical compositions comprising such compounds and methods of treating diseases treatable by inhibition of Polθ such as cancer, including homologous recombination (HR) deficient cancers.
Owner:IDEAYA BIOSCI INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com