Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
36 results about "Gene profile" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Gene profile is a measurement of expression of indicated genes. It helps to create a global picture of cellular function and to study the molecular basis of phenotypic differences. It provides valuable insight into the role of differential gene expression in normal biological and disease processes.
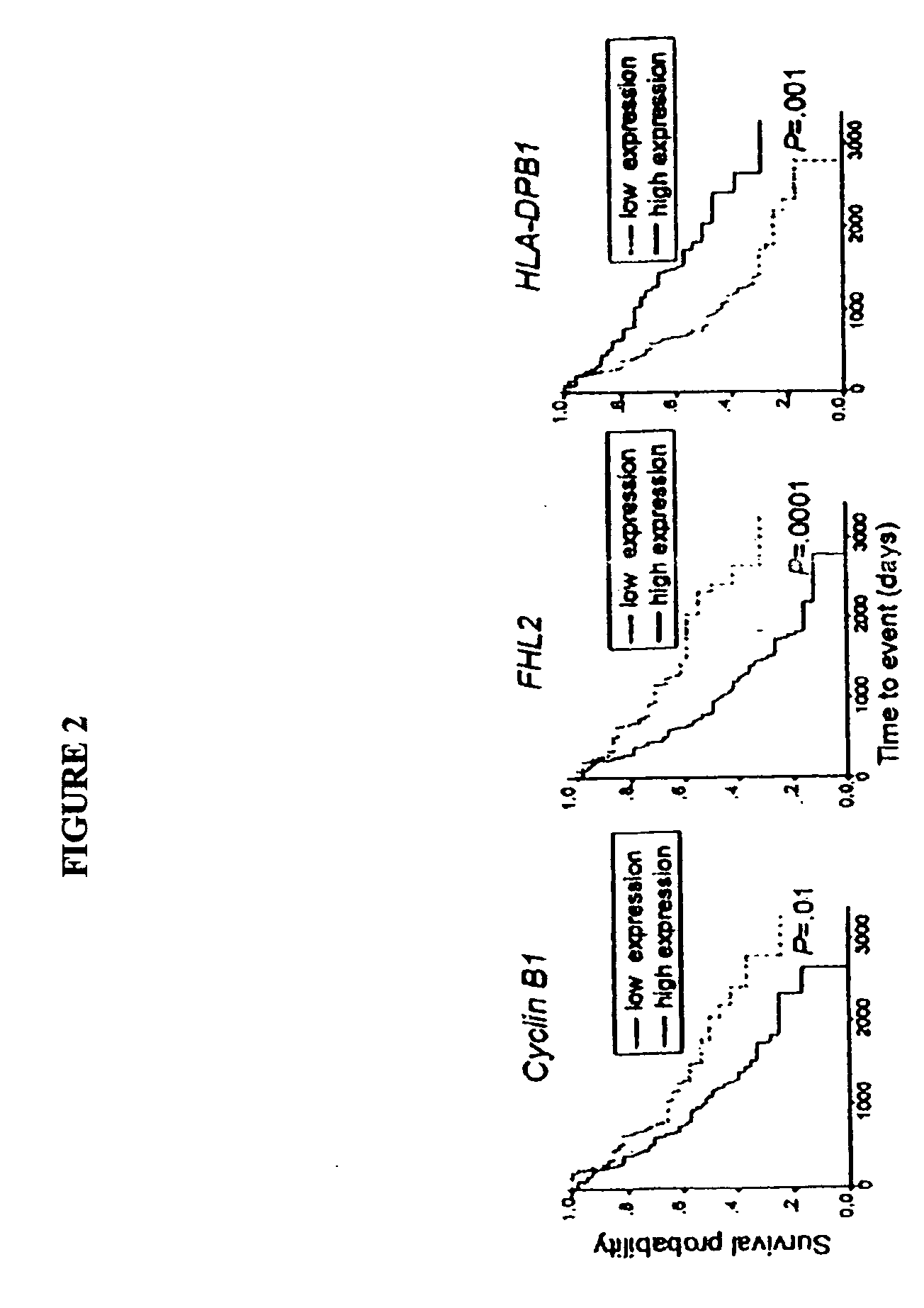
Method of diagnosis of cancer based on gene expression profiles in cells
A method of developing a gene expression profile indicative of the presence or stage of a selected a disease, disorder or genetic pathology in a mammalian subject employs penalized discriminant analysis with recursive feature elimination. A method of diagnosing a cancer in a mammalian subject includes the steps of examining a sample containing the subject's immune cells and detecting a variance in the expression of a statistically significant number of genes, e.g., at least 10 non-tumor genes from those same genes in a characteristic disease or healthy gene expression profile. A significant variance in expression of these genes when compared to a gene expression profile, preferably an average gene expression profile of a normal control, or significant similarities to an average gene profile of subjects with cancer, correlates with a specific type of cancer and / or location of tumor.
Owner:THE WISTAR INST OF ANATOMY & BIOLOGY
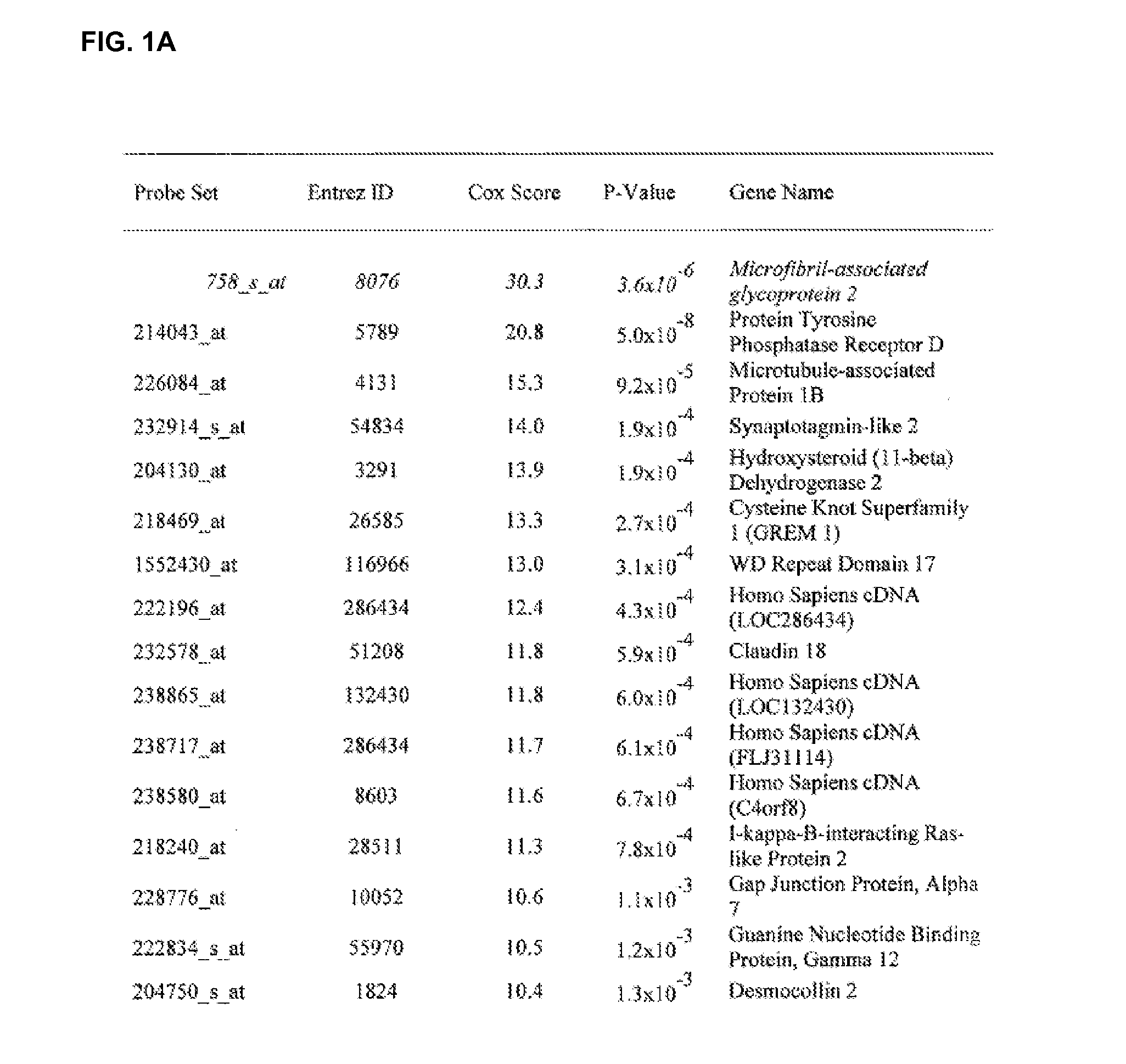
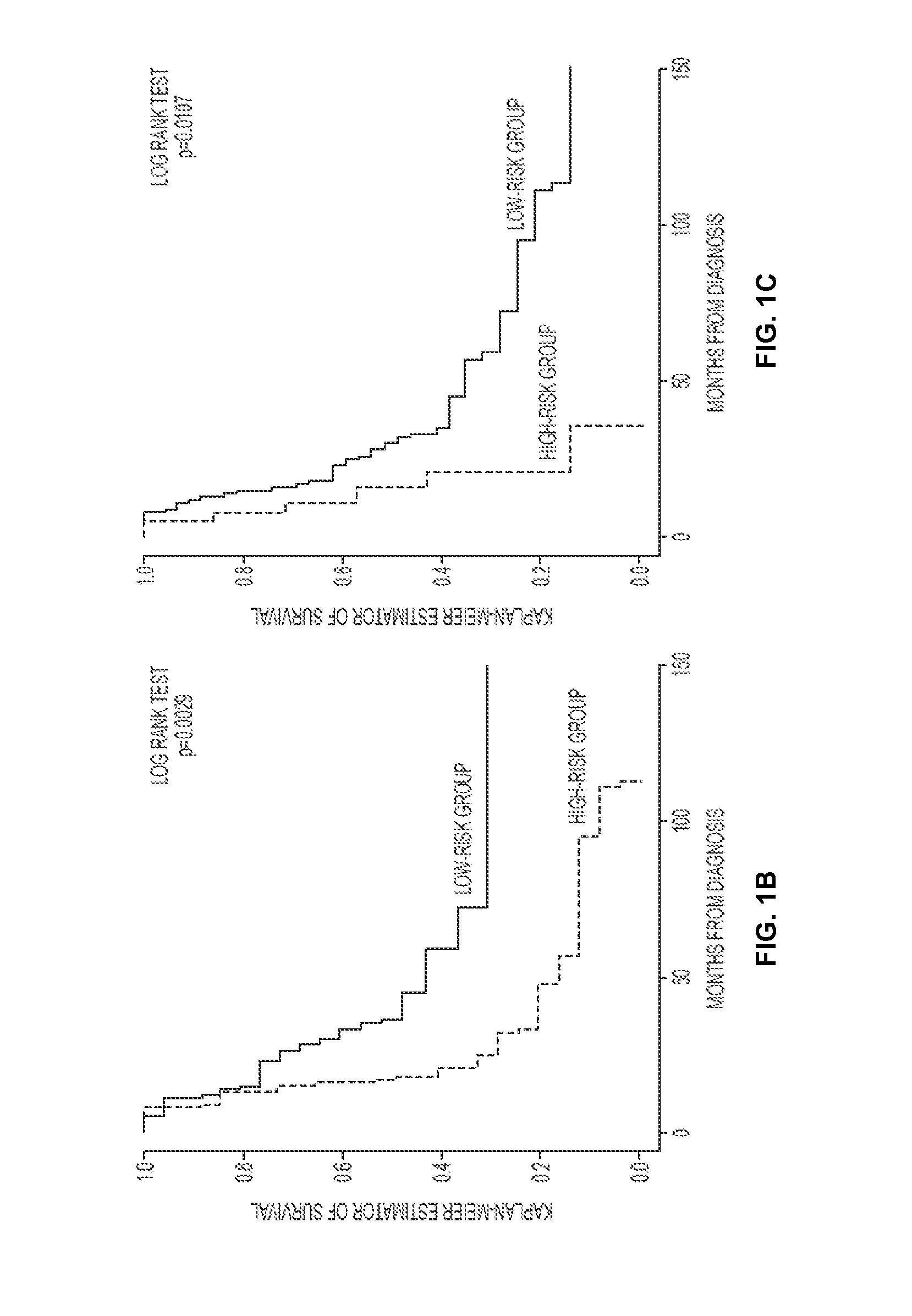
Gene profiles correlating with histology and prognosis
InactiveUS20070026424A1Improved prognosisPoor prognosisMicrobiological testing/measurementCancer deathHistology type
The present invention related to methods and kits for evaluating the histology and prognosis of lung cancer by measuring expression levels of specific gene markers. It is based, at least in part, on the discovery of 99 genes that were found to be differentially expressed among lung cancer subtypes, 30 genes which correlate with a high risk, and 12 genes which correlate with a low risk, of cancer death within 12 months.
Owner:POWELL CHARLES A +1
Determining the chemosensitivity of cells to cytotoxic agents
InactiveUS20050208512A1Bioreactor/fermenter combinationsBiological substance pretreatmentsCancer cellPolynucleotide
Gene expression analysis systems are provided for identifying the chemosensitivity gene profile of a cancer cell, the analysis systems comprising a plurality of polynucleotide probes, wherein each of said polynucleotide probes comprises a polynucleotide sequence that is complementary to a target region of a gene that encodes a protein associated with transport of molecules into and out of cells and that is a marker for the sensitivity or resistance of cancer cells to cytotoxic agents.
Owner:THE OHIO STATES UNIV
Method for preparing and using personal and genetic profiles
ActiveUS7089498B1Microbiological testing/measurementData visualisationChemical structureGenetic data
Method and system for preparing a personal genetic profile includes collecting genetic data from an individual, assigning the data to a coordinate system, storing the data, and providing access for retrieval by the individual from whom the genetic data were collected, after receipt of an Identifier that adequately authenticates the identity of the data requester. Locations of genetic markers are provided as three-dimensional coordinates, described with matrix relationships that are consistent with the primary and secondary chemical structure of molecular constituents of a DNA chain for the individual.
Owner:THOMAS XAVIER +9
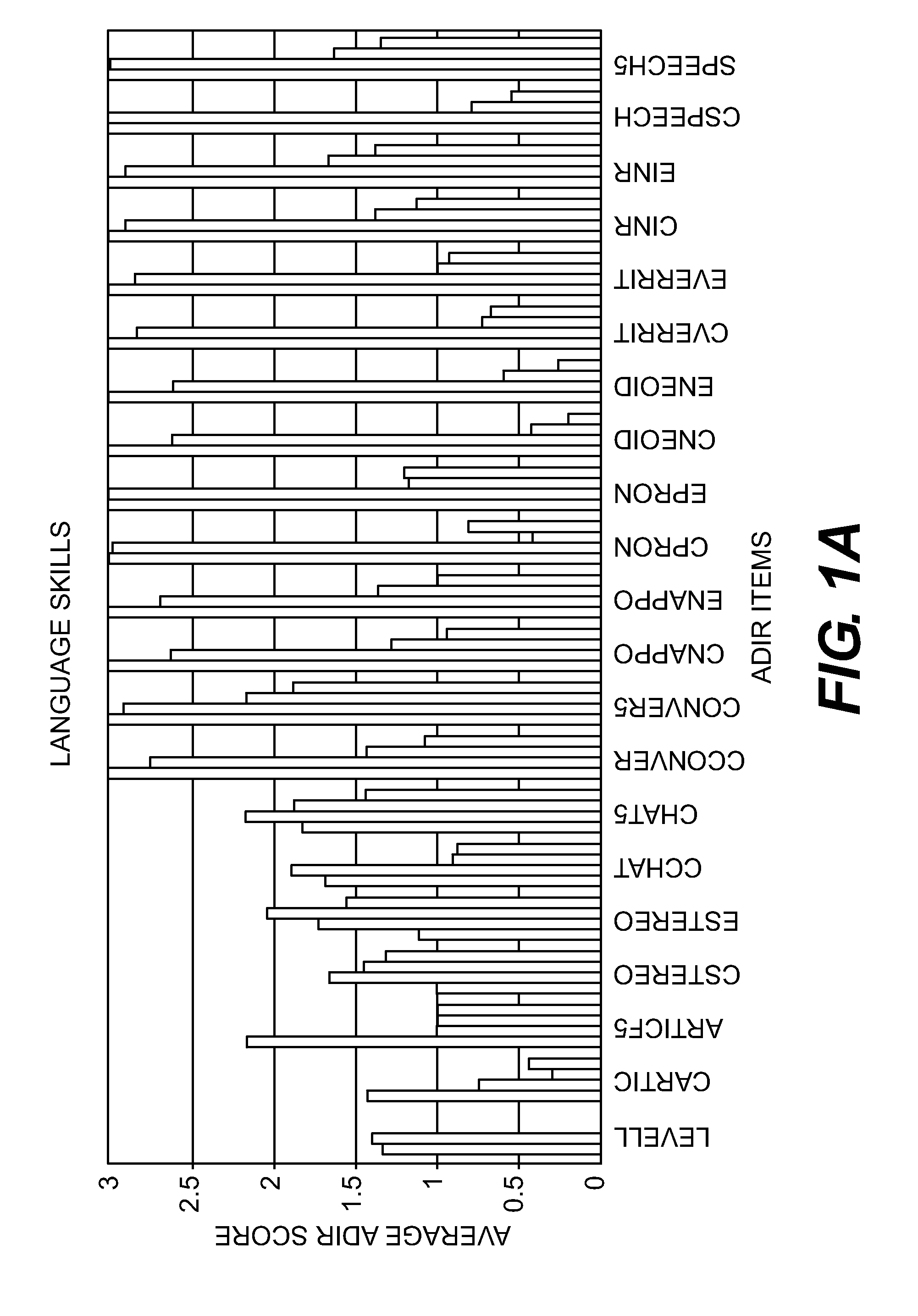
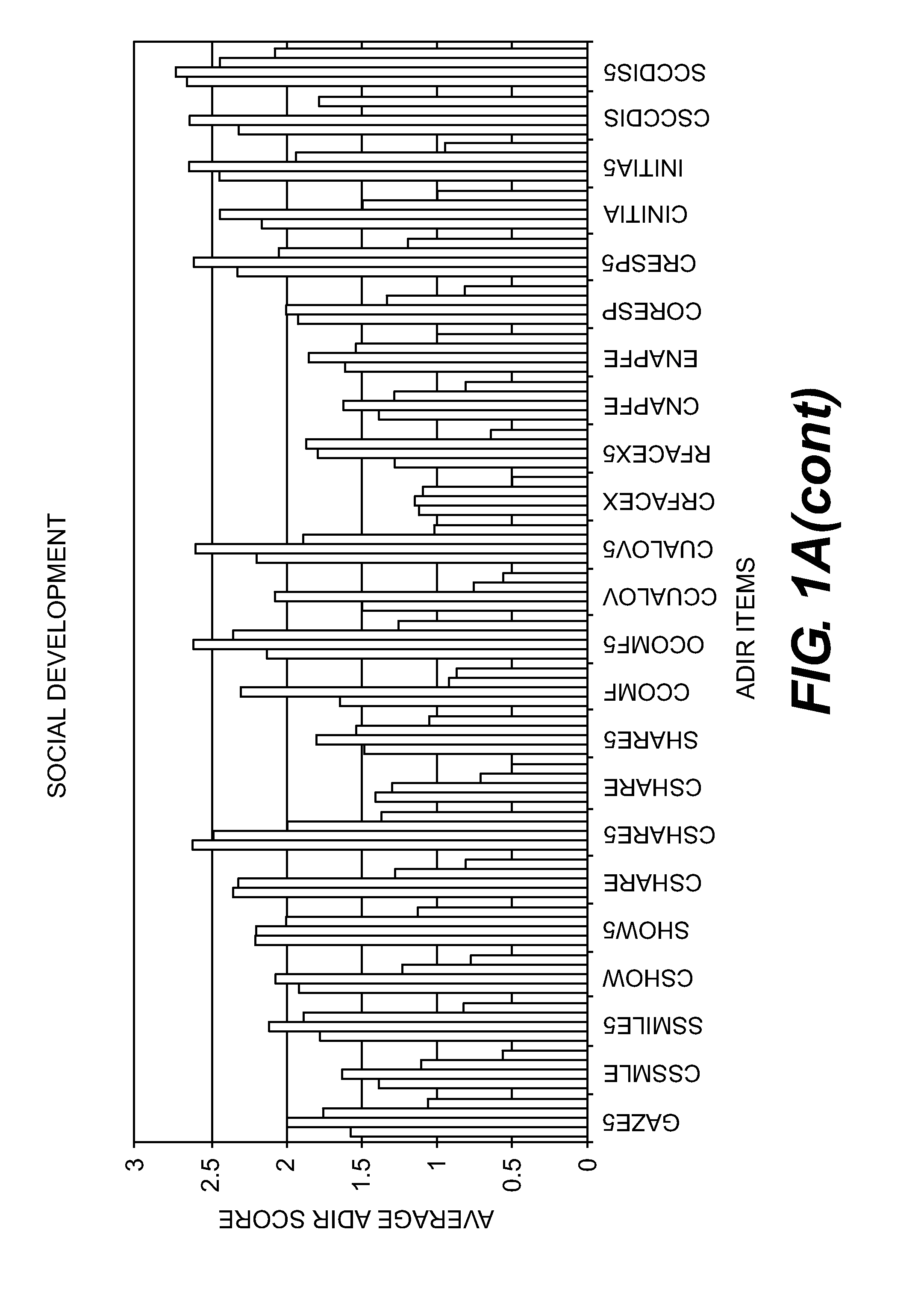
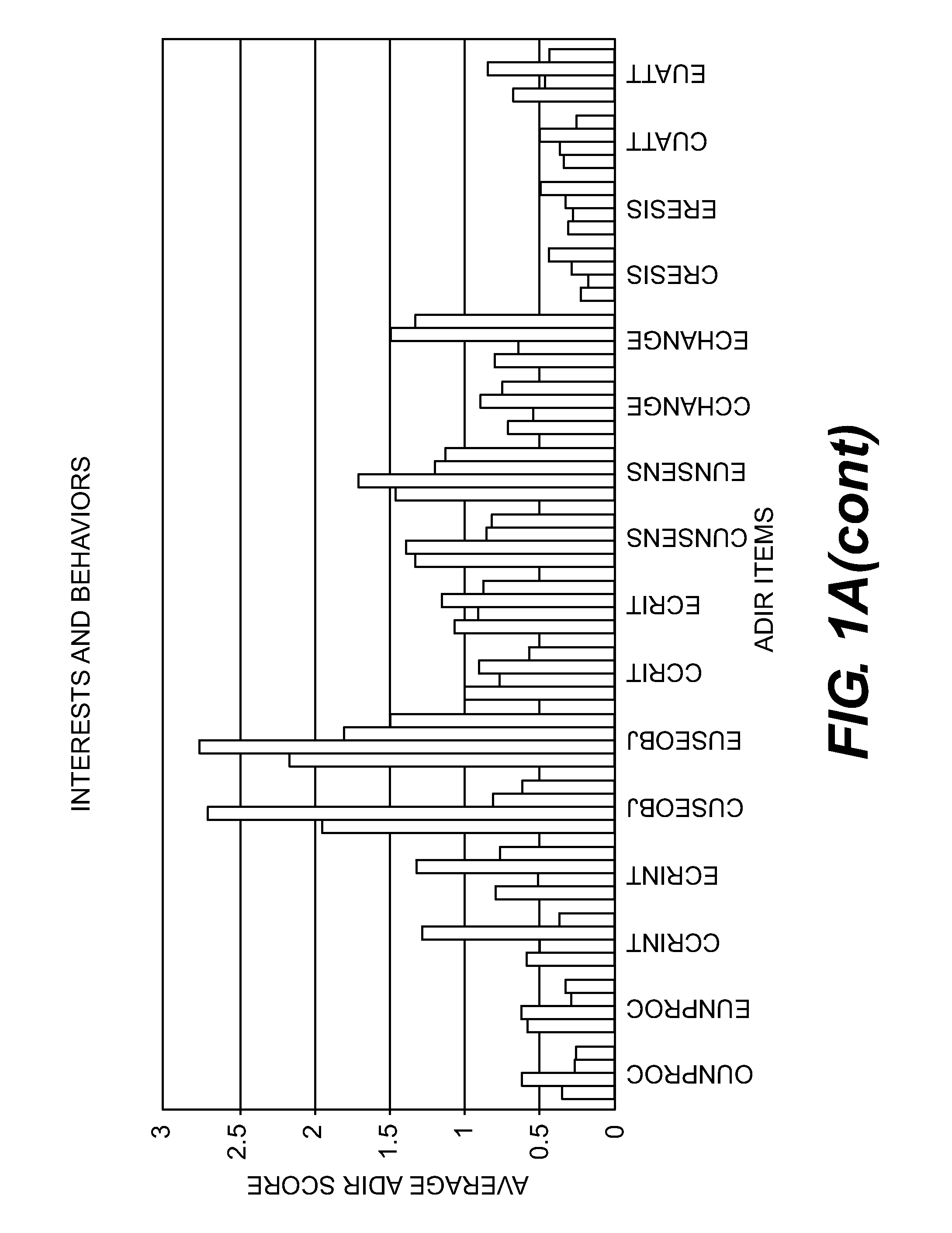
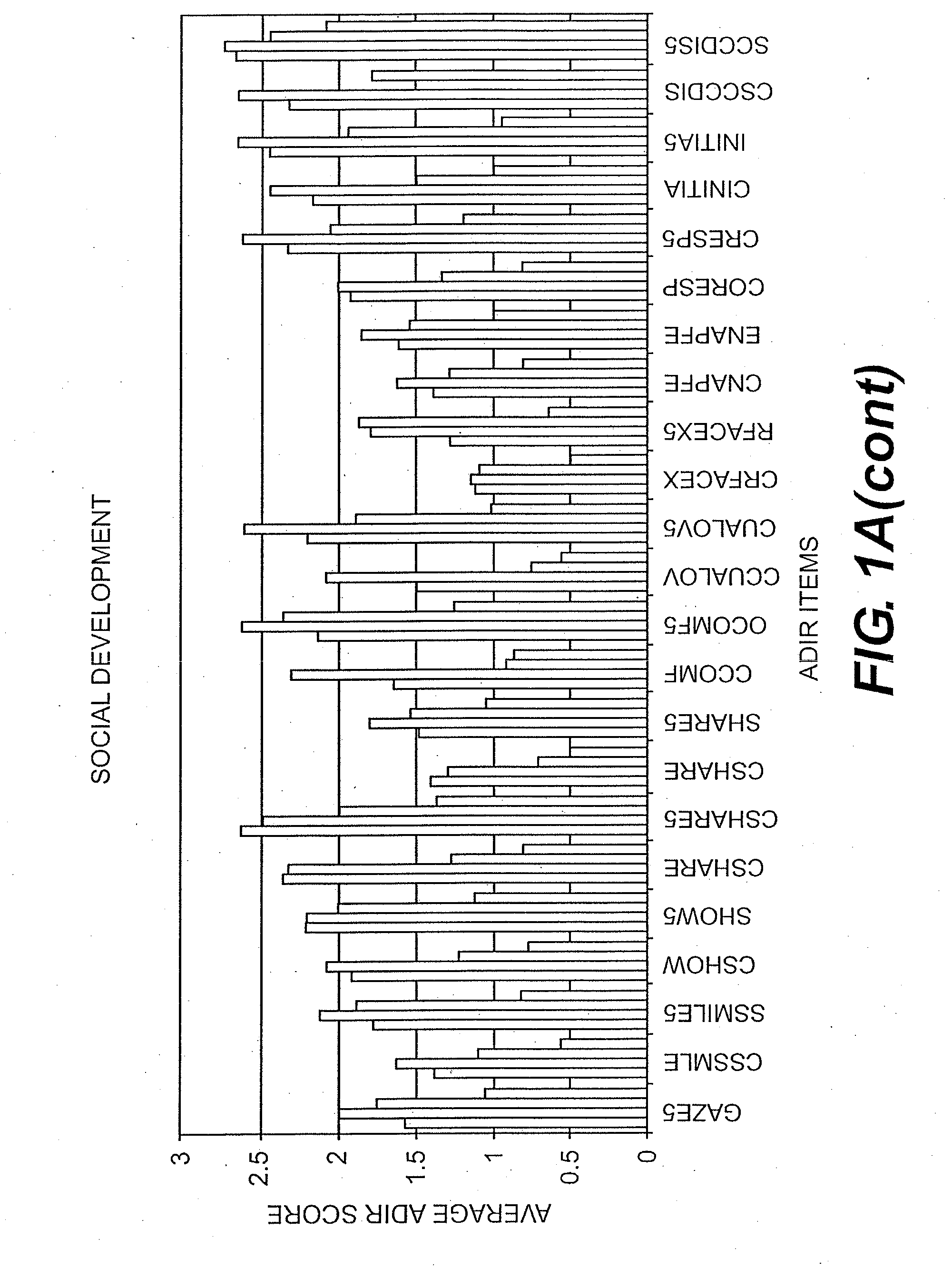
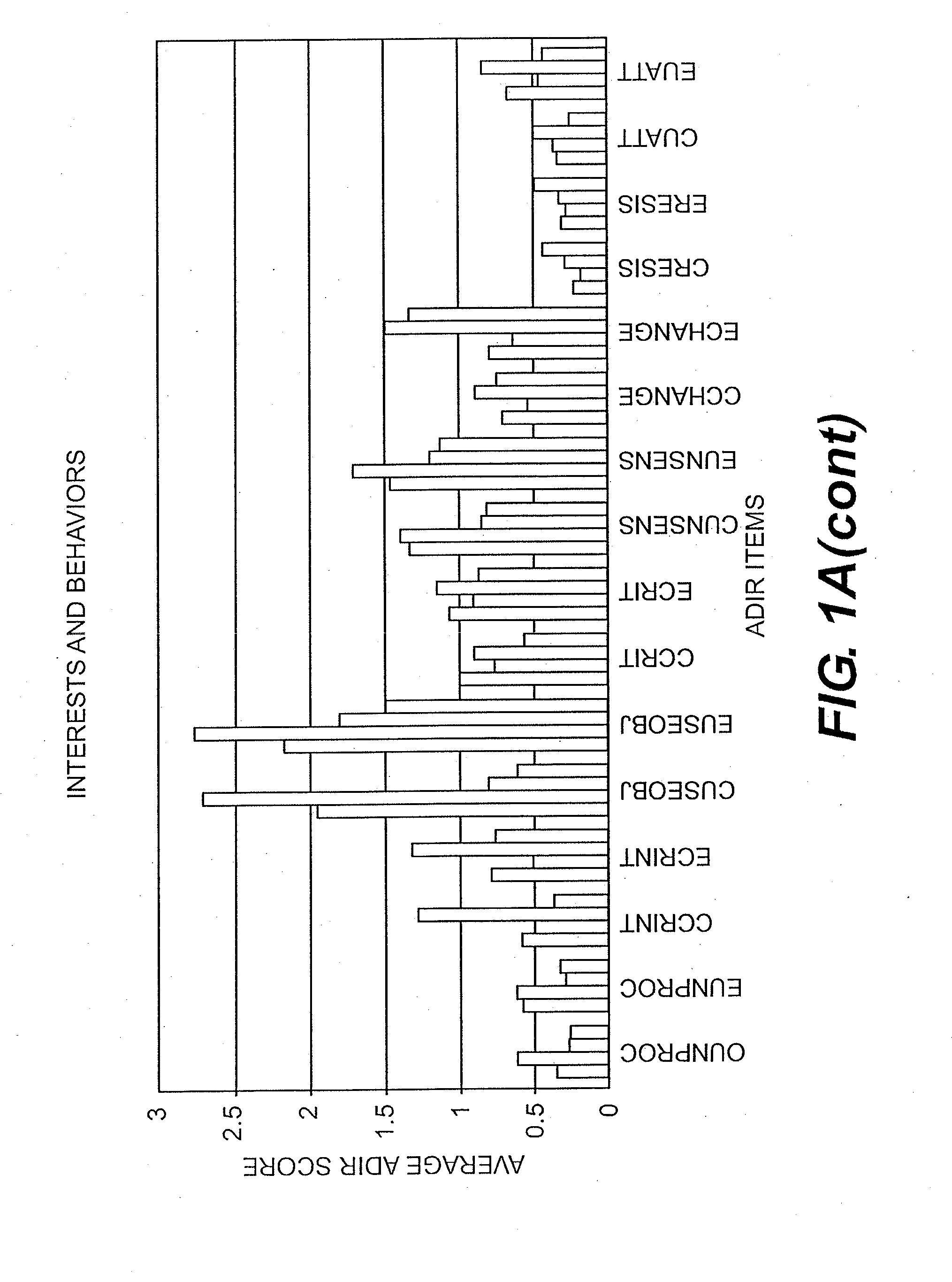
Compositions and Methods for Identifying Autism Spectrum Disorders
The compositions and methods described are directed to gene chips having a plurality of different oligonucleotides with specificity for genes associated with autism spectrum disorders. The invention further provides methods of identifying gene profiles for neurological and psychiatric conditions including autism spectrum disorders, methods of treating such conditions, and methods of identifying therapeutics for the treatment of such neurological and psychiatric conditions.
Owner:GEORGE WASHINGTON UNIVERSITY
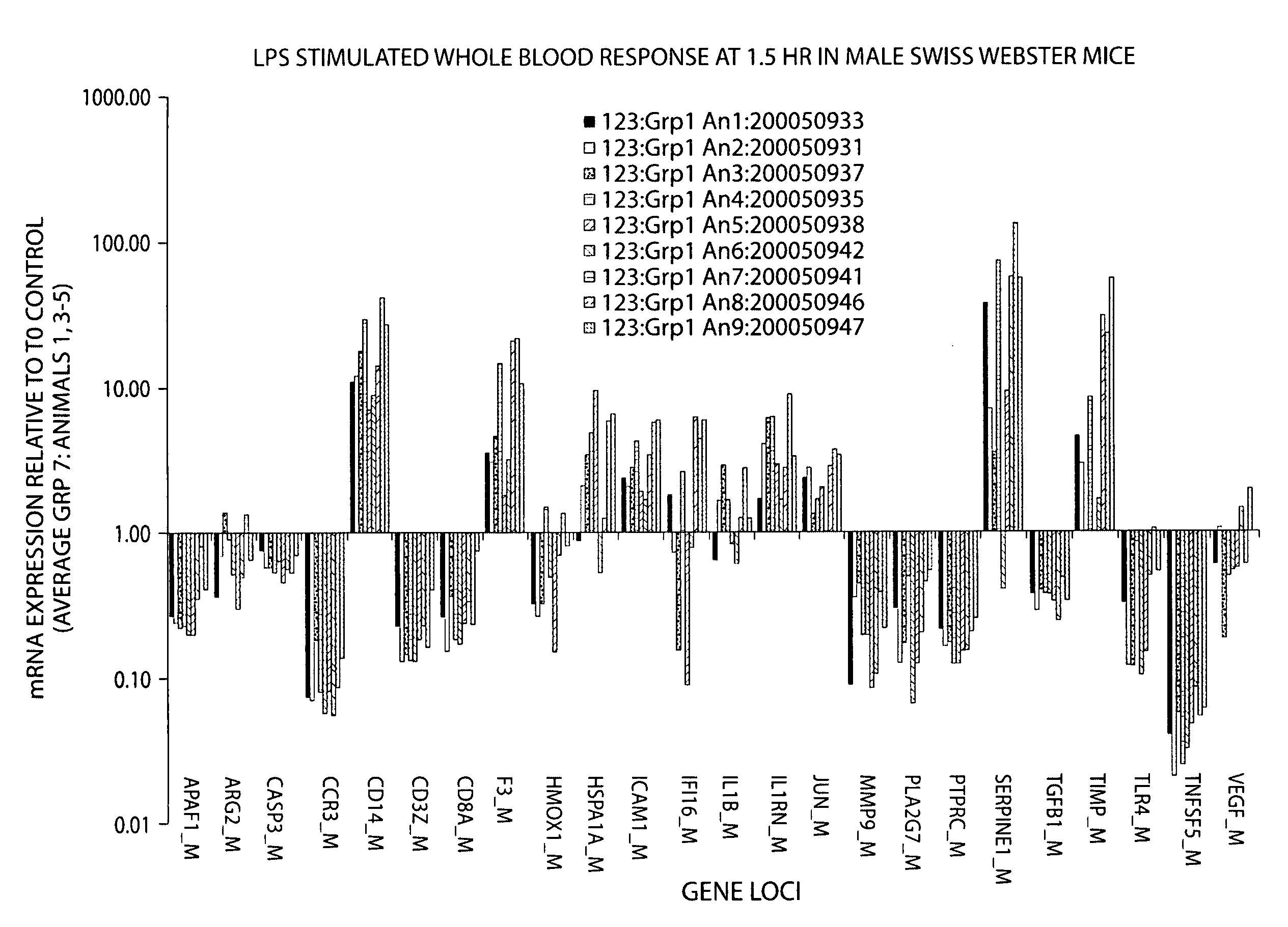
Assessment of effect of an agent on a human biological condition using rodent gene expression panels
InactiveUS20080227655A1Microbiological testing/measurementLibrary screeningEfficacyGene expression profiling
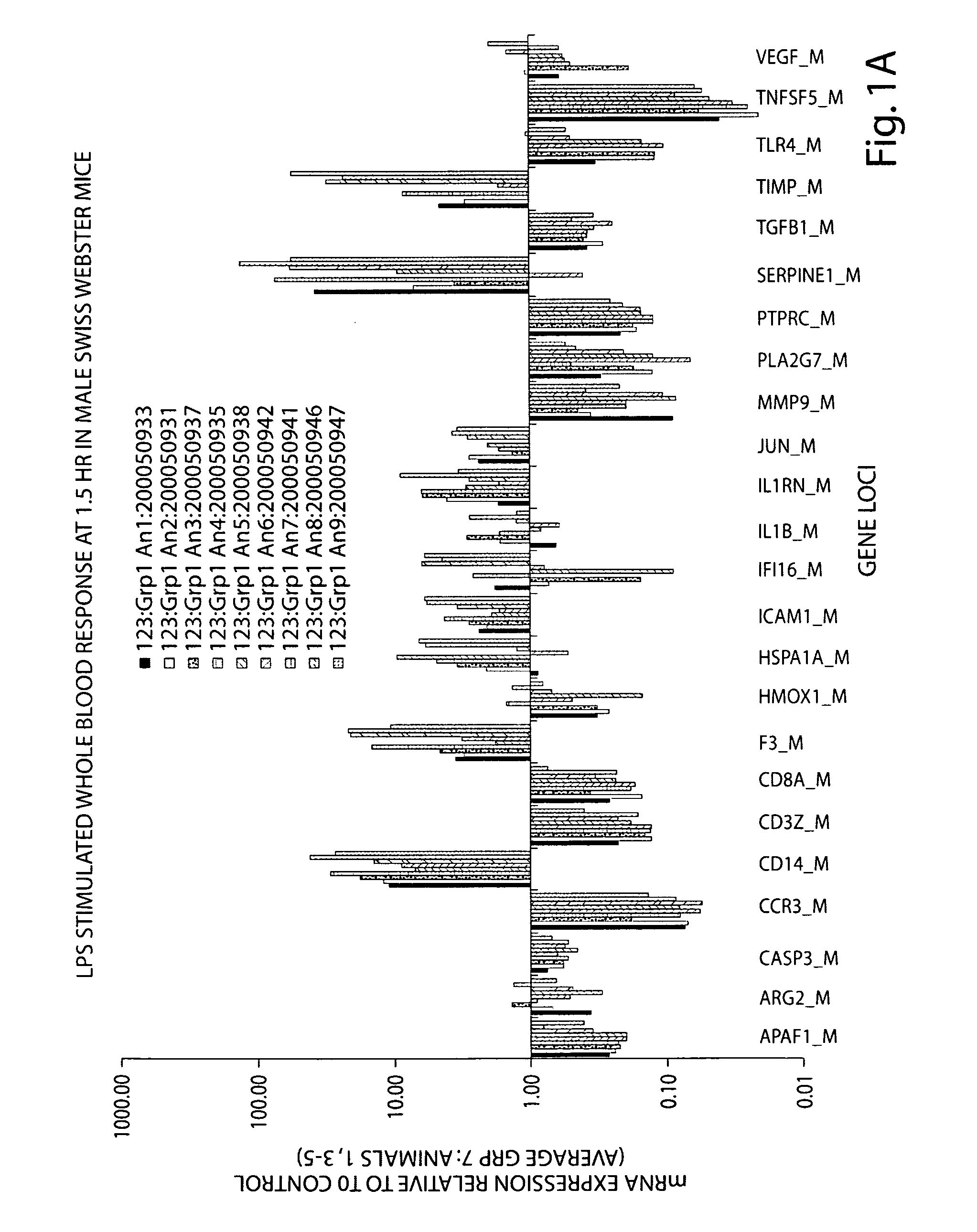
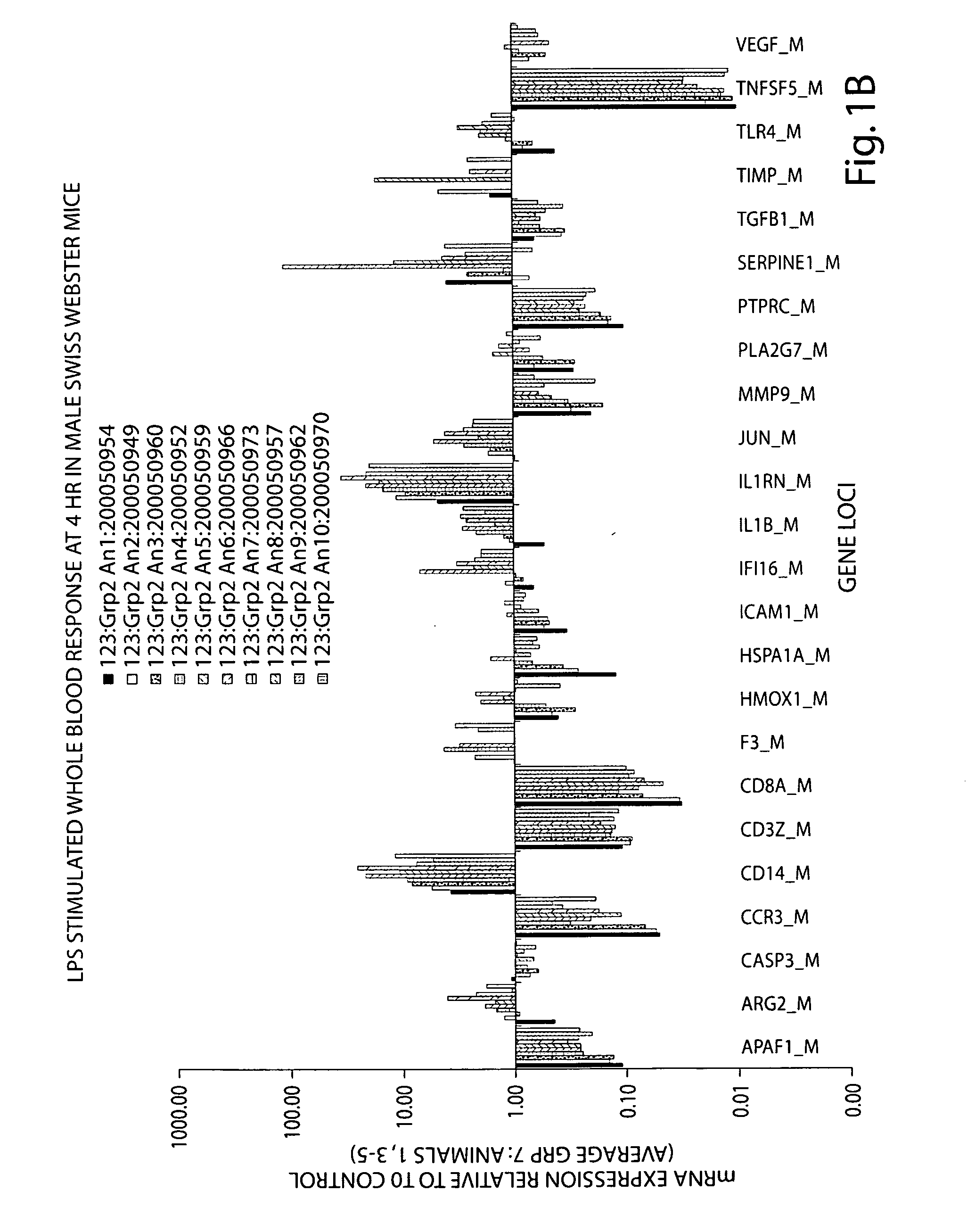
Rodent gene expression data, in particular, gene expression profiles, are created and used to predict the efficacy of therapeutic agents on human biological conditions. Gene Profile data sets are derived from rodent subject samples and include quantitative, substantially repeatable measures of a distinct amount of RNA or protein constituent(s) in a signature panel selected such that measurement of the constituent(s) enables measurement of a biological condition of interest in both human and rodent subjects. Such profile data sets may be used to predict the therapeutic efficacy of a therapeutic agent in humans.
Owner:BEVILACQUA MICHAEL +5
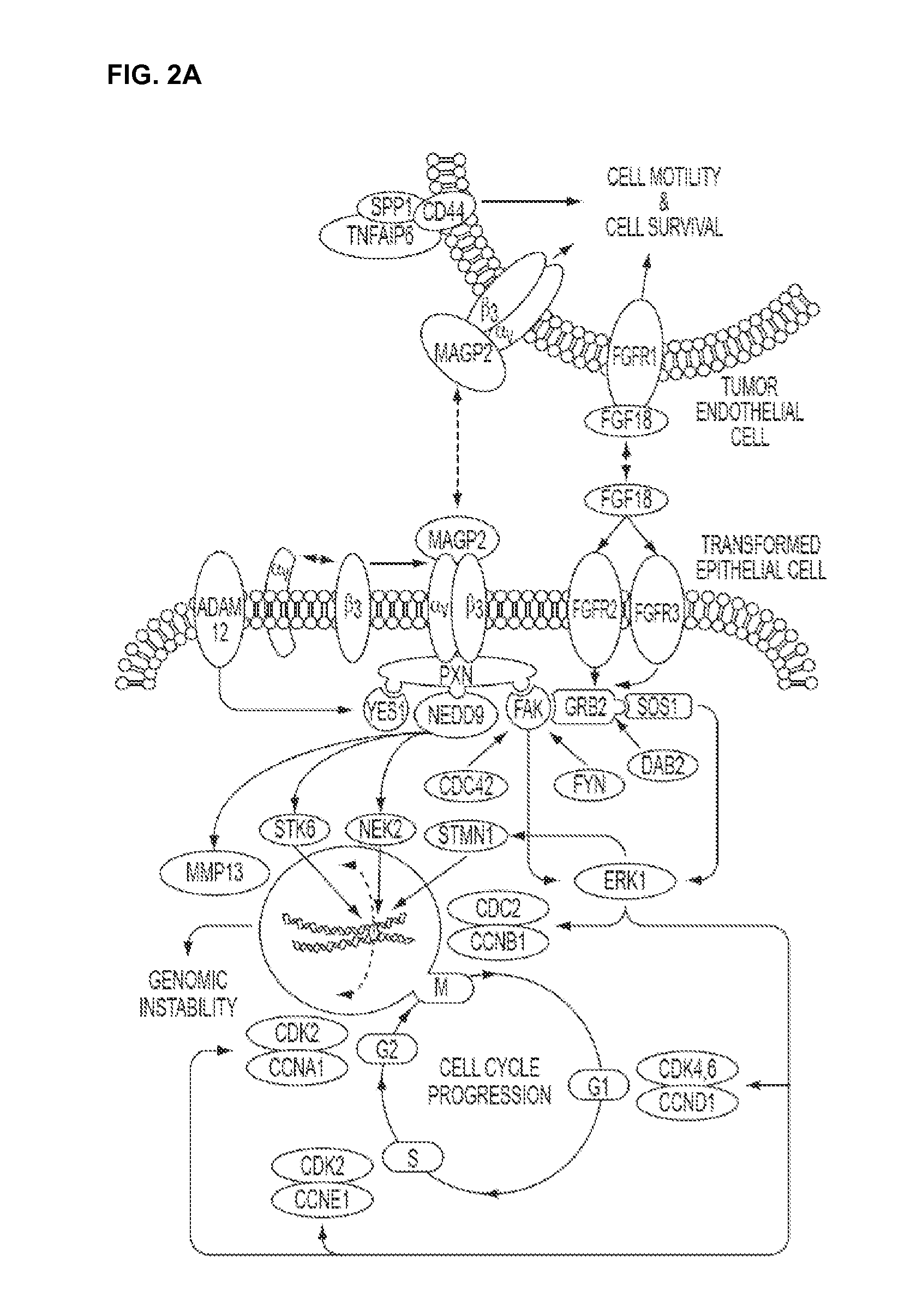
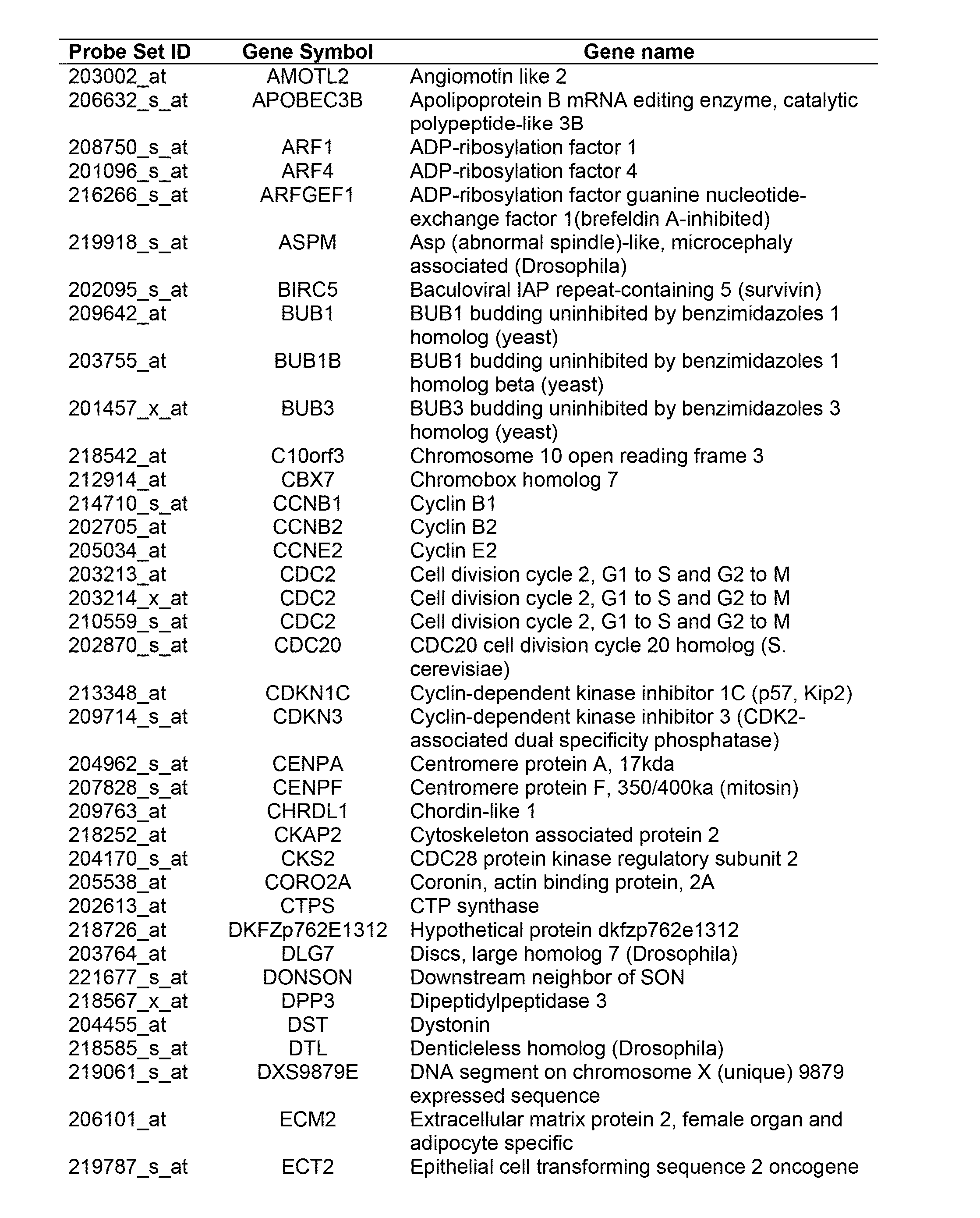
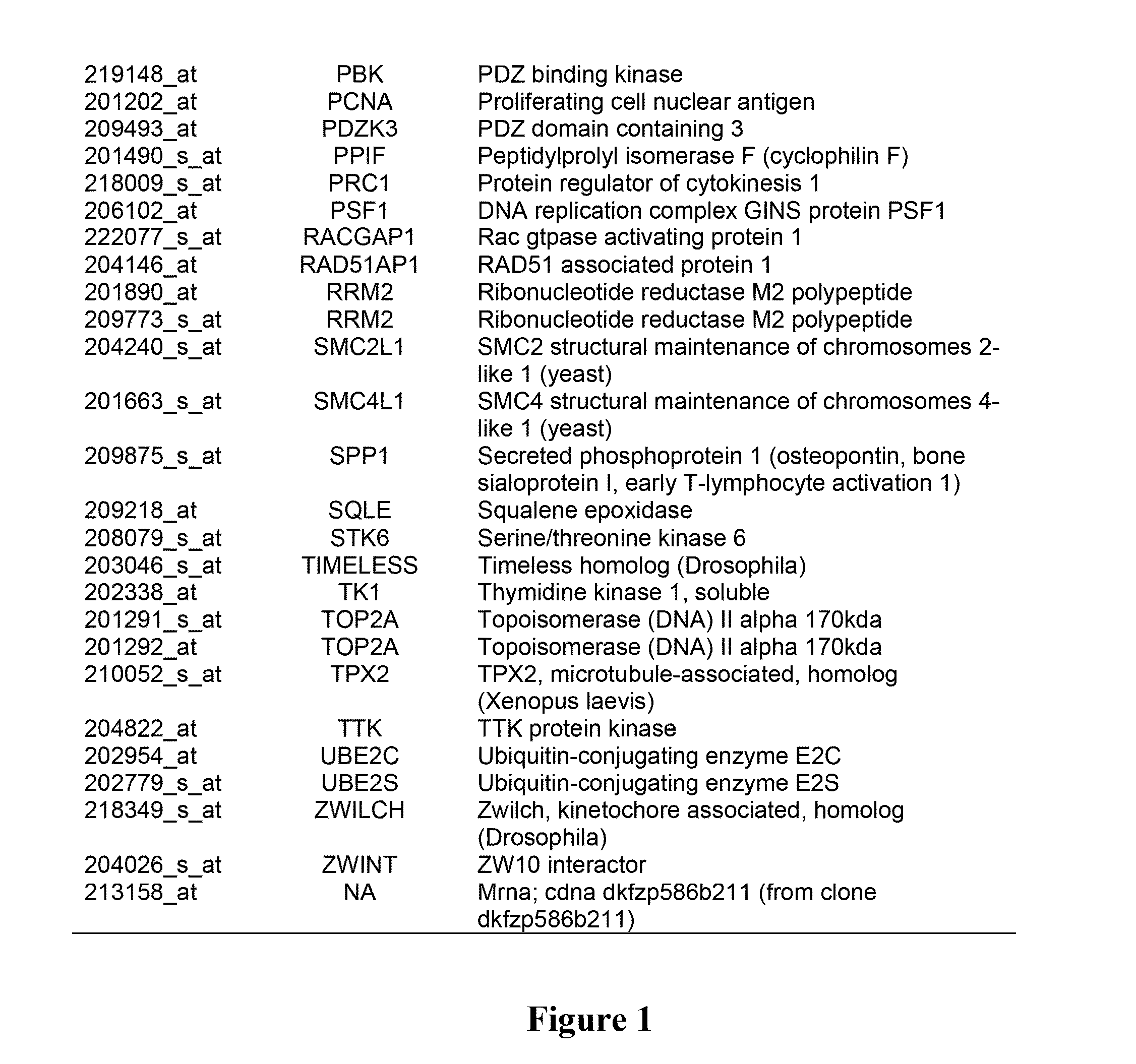
Pro-angiogenic genes in ovarian tumor endothelial cell isolates
InactiveUS20100286237A1Reduce and eliminate ovarian cancerDevelopment of therapyOrganic active ingredientsMicrobiological testing/measurementCell fractionationCell tumor
A gene profiling signature for ovarian tumor endothelial cells is disclosed herein. The gene signature can be used to diagnosis or prognosis an ovarian tumor, identify agents to treat an ovarian tumor, to predict the metastatic potential of an ovarian tumor and to determine the effectiveness of ovarian tumor treatments. Thus, methods are provided for identifying agents that can be used to treat ovarian cancer, for determining the effectiveness of an ovarian tumor treatment, or to diagnose or prognose an ovarian tumor. Methods of treatment are also disclosed which include administering a composition that includes a specific binding agent that specifically binds to one of the disclosed ovarian endothelial cell tumor-associated molecules and inhibits ovarian tumor in the subject.
Owner:US DEPT OF HEALTH & HUMAN SERVICES +1
Method of diagnosis of cancer based on gene expression profiles in cells
A method of developing a gene expression profile indicative of the presence or stage of a selected a disease, disorder or genetic pathology in a mammalian subject employs penalized discriminant analysis with recursive feature elimination. A method of diagnosing a cancer in a mammalian subject includes the steps of examining a sample containing the subject's immune cells and detecting a variance in the expression of a statistically significant number of genes, e.g., at least 10 non-tumor genes from those same genes in a characteristic disease or healthy gene expression profile. A significant variance in expression of these genes when compared to a gene expression profile, preferably an average gene expression profile of a normal control, or significant similarities to an average gene profile of subjects with cancer, correlates with a specific type of cancer and / or location of tumor.
Owner:THE WISTAR INST OF ANATOMY & BIOLOGY
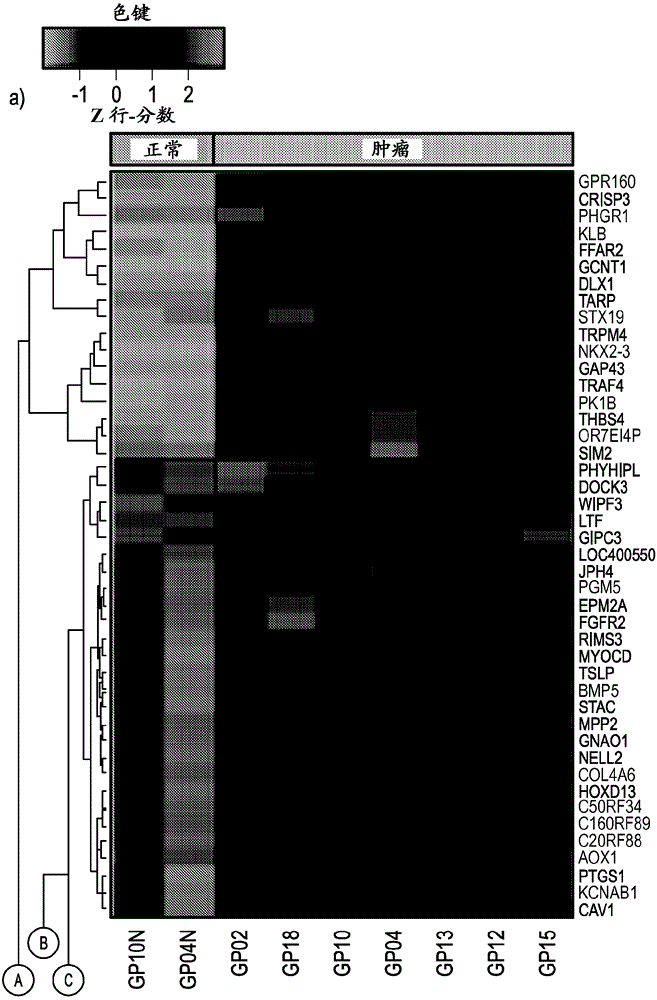
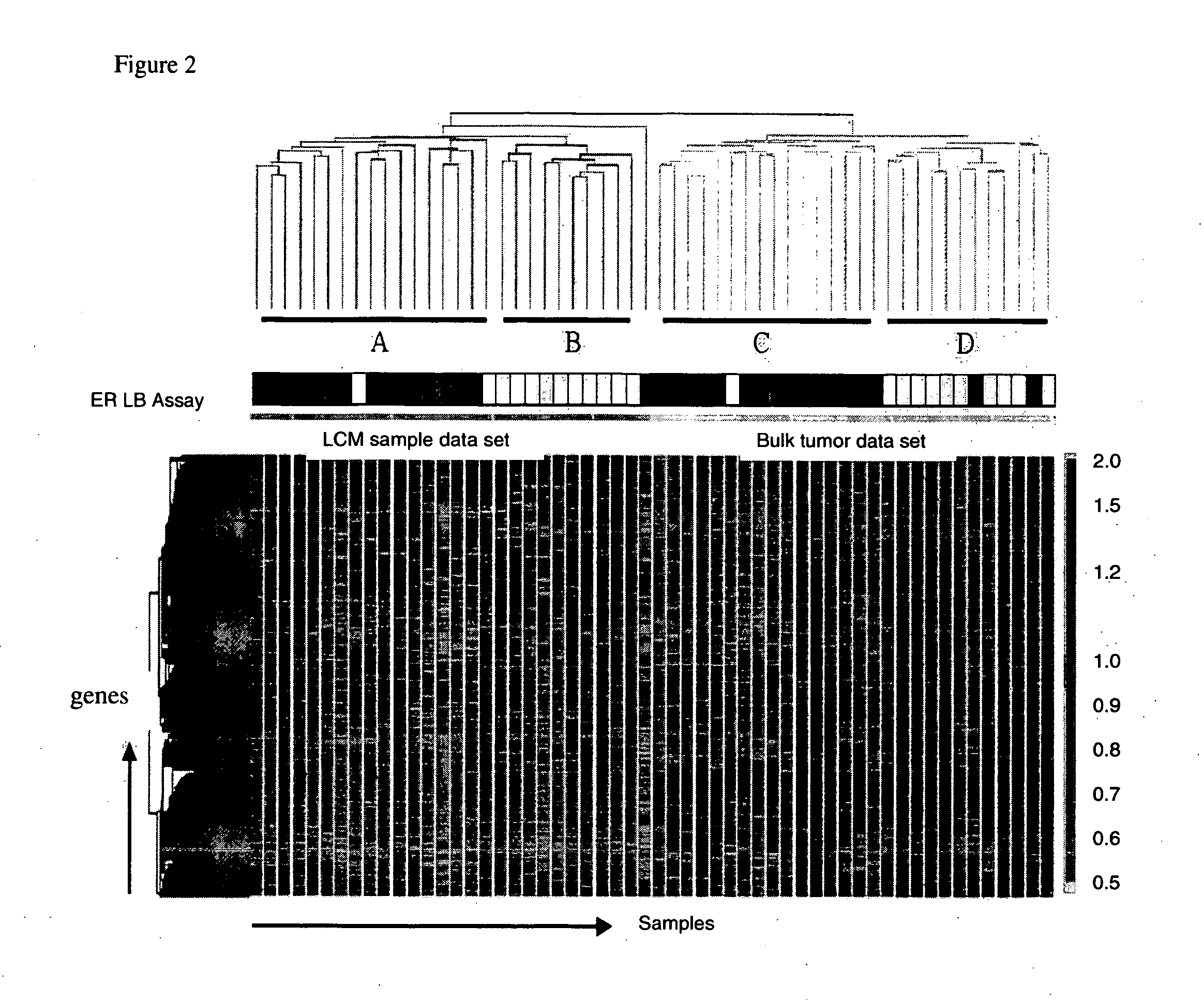
Laser microdissection and microarray analysis of breast tumors reveal estrogen receptor related genes and pathways
InactiveCN101965190APeptide/protein ingredientsMicrobiological testing/measurementPathway analysisLaser micro dissection
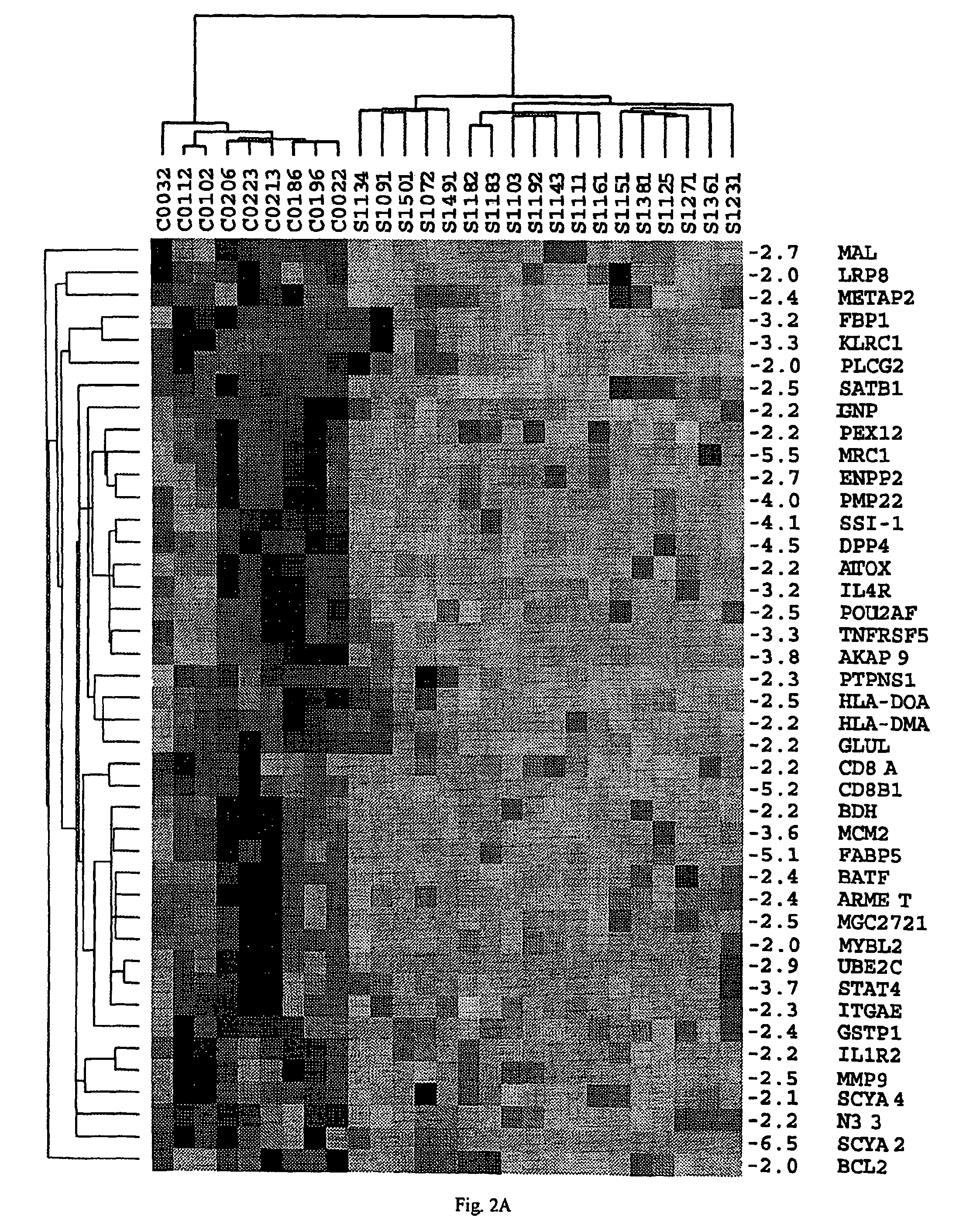
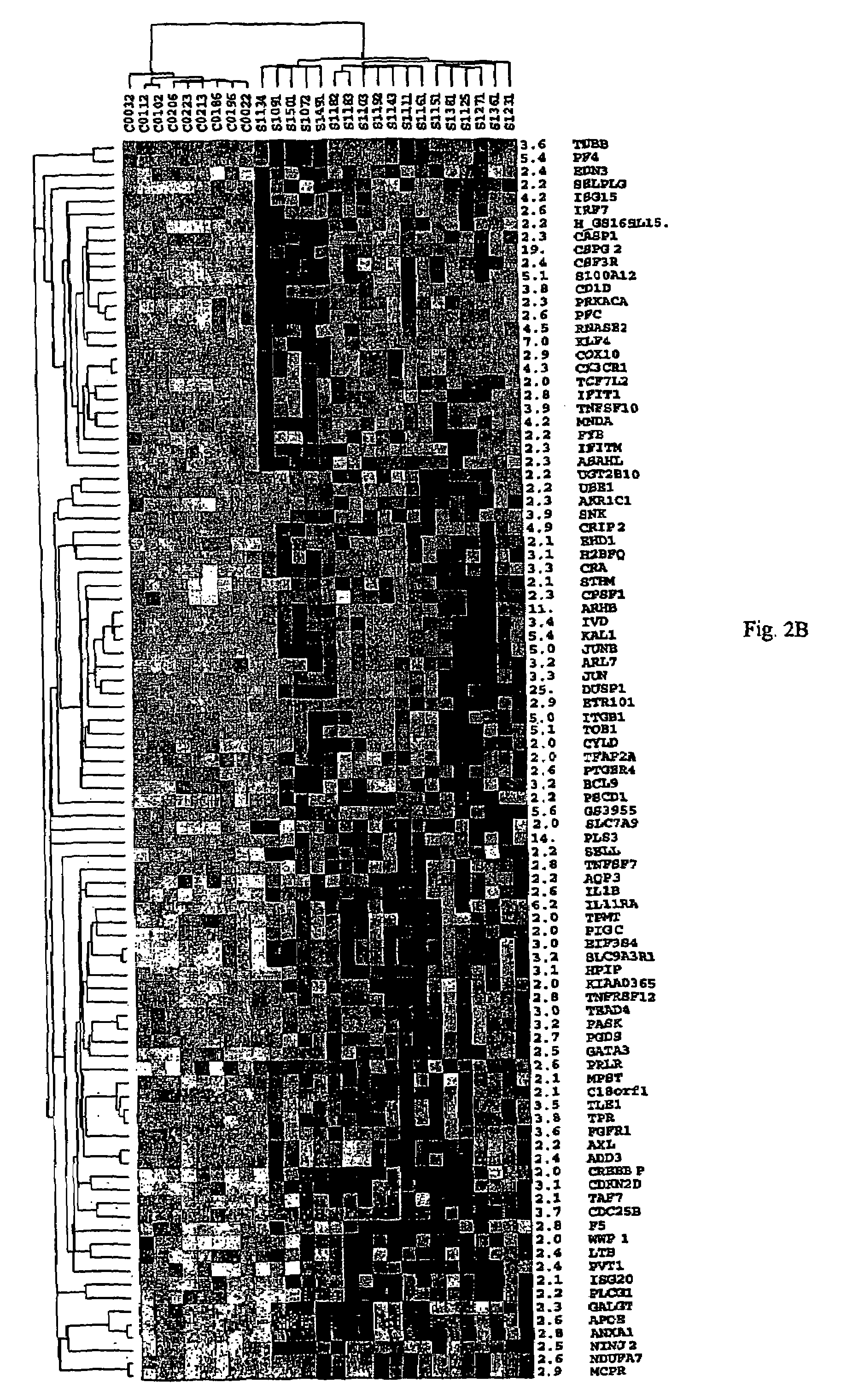
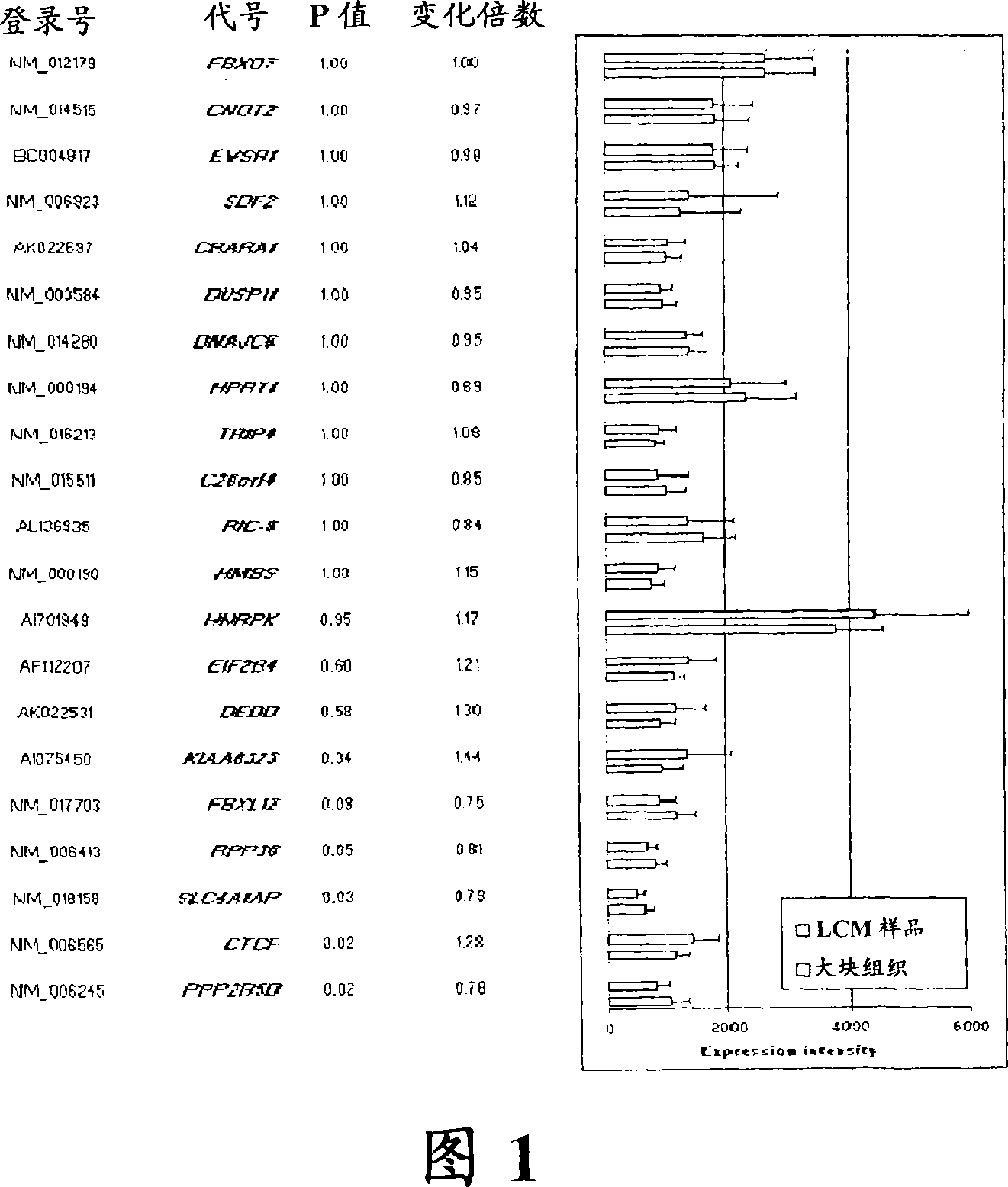
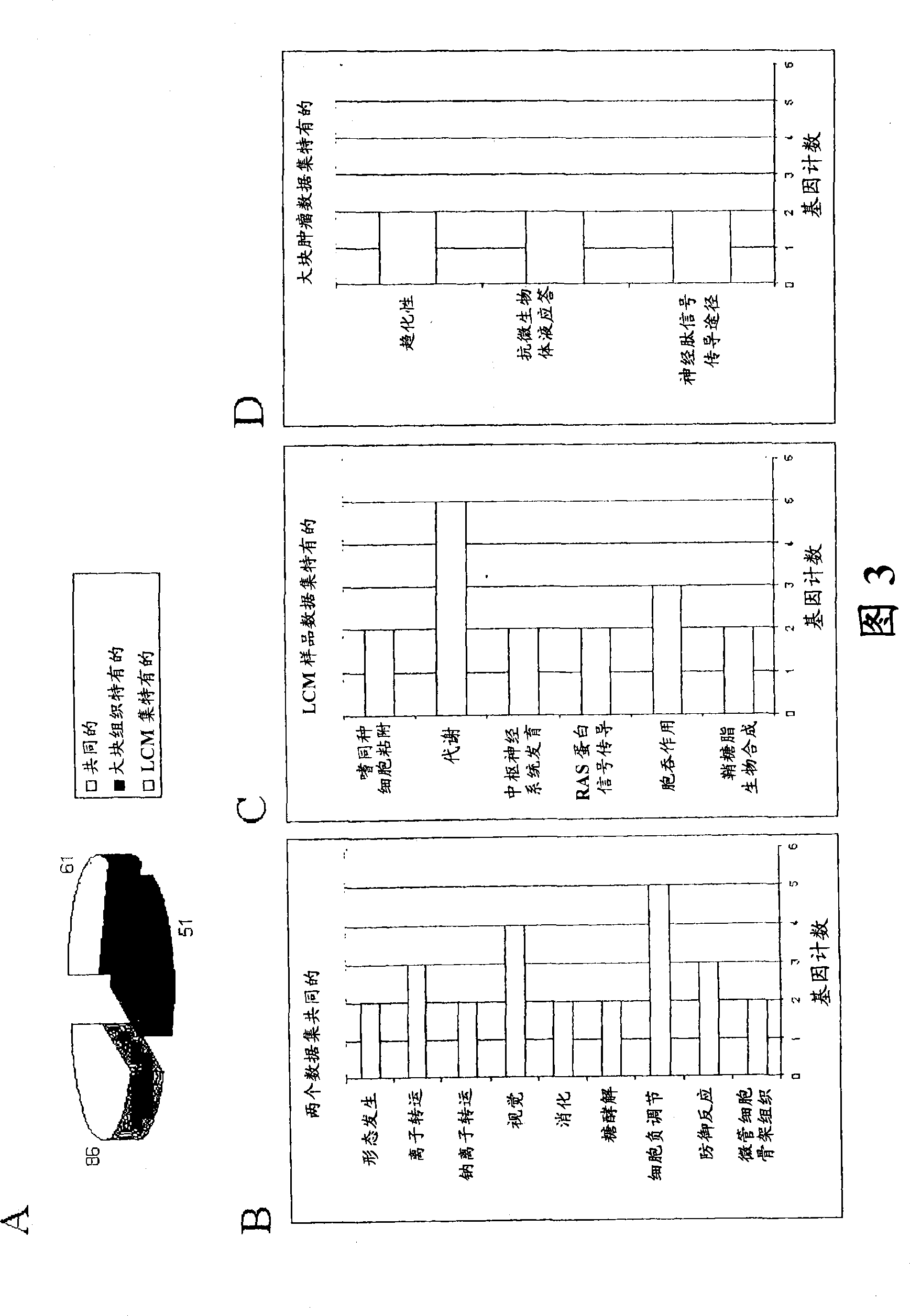
About 70% to 80% of breast cancers express estrogen receptor-a (ERa), and estrogens play important roles in the development and growth of hormone-dependent tumors. Together with lymph node metastasis, tumor size and histological grade, ER status is considered one of the prognostic factors in breast cancer, and an indicator for hormonal treatment. 147 genes and 112 genes with significant P- value and having significant differential expression between ER+ and ER- tumors were identified from the LCM data set and bulk tissue data set, respectively. 61 genes were found to be common in both data sets, while 85 genes were unique to the LCM data set and 51 genes were present only in the bulk tumor data set. Pathway analysis with the 85 genes using Gene Ontology suggested that genes involved in endocytosis, ceramide generation, Ras / ERK / Ark cascade, and JAT- STAT pathway may play roles related to ER. The gene profiling with LCM-captured tumpr cells provides a unique approach to characterize and study epithelial tumor cells and to gain an insight into signaling pathways associated with ER.
Owner:VERIDEX LCC
Application of group of genes related to ovarian cancer prognosis
ActiveCN110551819AAvoid over-medicationAchieve the goal of personalized medicineMicrobiological testing/measurementDNA/RNA fragmentationTreatment choicesWilms' tumor
Based on the discovery of a group of 11 prognostic related gene profiles in serous ovarian cancer and the detection of the expression level of the 11 prognostic related gene profiles in clinical tumorsamples, the prognostic score of a patient is calculated according to the prognostic correlation coefficient of a group of the genes to evaluate the clinical prognosis of ovarian cancer patients andrelated application of the clinical prognosis. The system can assist in evaluating the response of the ovarian cancer patients to treatment interventions, thus whether the patients benefit from chemical or targeted treatments or not is judged, treatment choice is conducted, excessive medical treatment is avoided, and the purpose of personalized treatment is achieved. According to the system and different detection technology platforms, corresponding 11 gene expression measurement kits are designed and developed.
Owner:BERKELEY NANJING MEDICAL RES CO LTD
Biomarkers for the efficacy of calcitonin and parathyroid hormone treatment
InactiveCN1905894APeptide/protein ingredientsMicrobiological testing/measurementRecombinant salmon calcitoninAnabolic Effect
A mufti-organ gene profiling analysis of the results of an administration to a subject of salmon calcitonin or a parathyroid hormone analogue provides biomarkers of calcitonin treatment efficacy and parathyroid hormone or parathyroid hormone analogue treatment efficacy. Among the biomarkers are the expression profiles of the genes for Y-box binding protein, BMPs, FGFs, IGFs, VEGF, &x3B1;-2-HS glycoprotein (AHSG), OSF, nuclear receptors (steroid / thyroid family) and others. The results obtained support the anabolic effect of salmon calcitonin on bone metabolism.
Owner:NOVARTIS AG
Renal cell carcinoma biomarkers
InactiveUS20120009201A1Easy to findOrganic active ingredientsCompound screeningTumor BiomarkersProtein expression profile
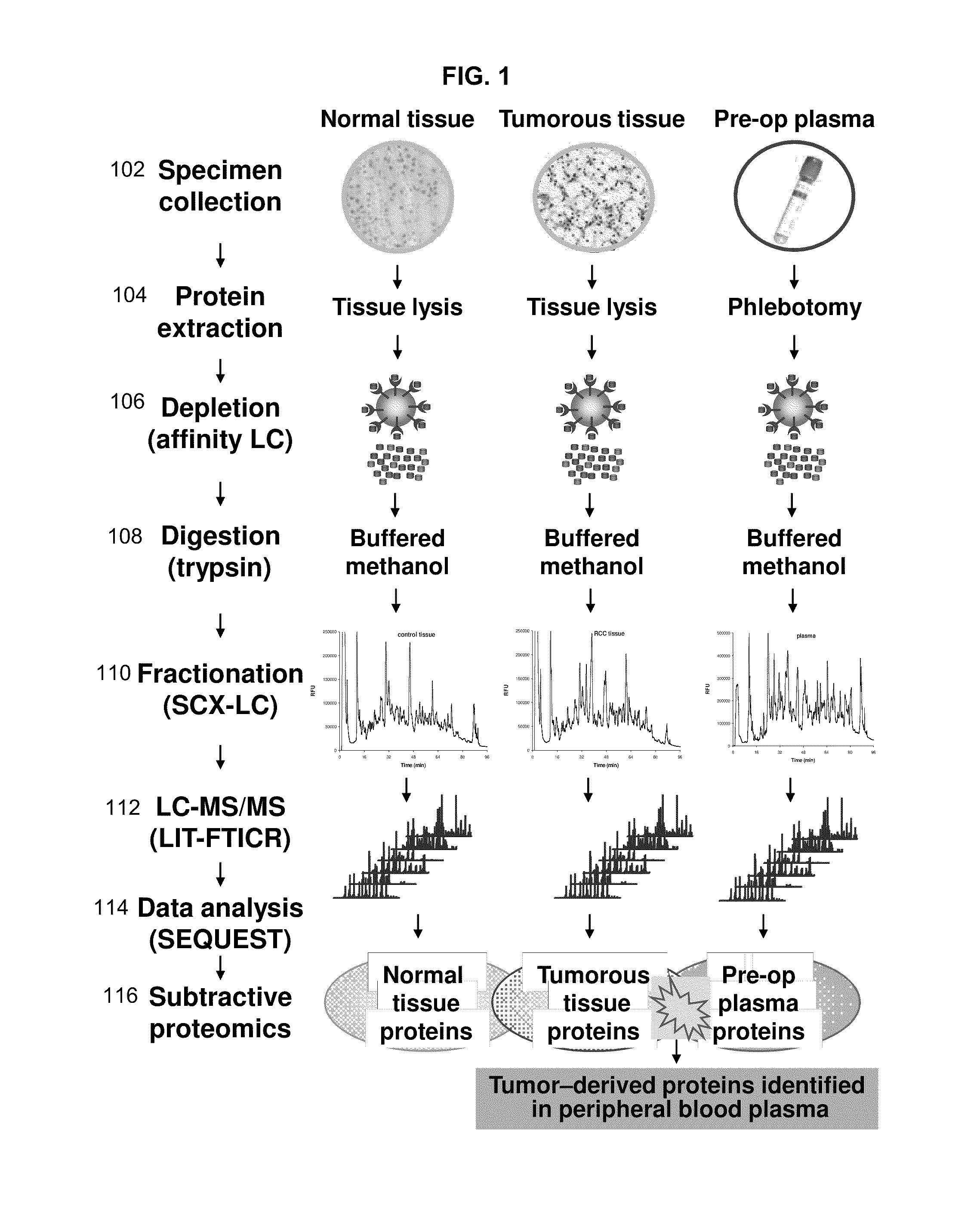
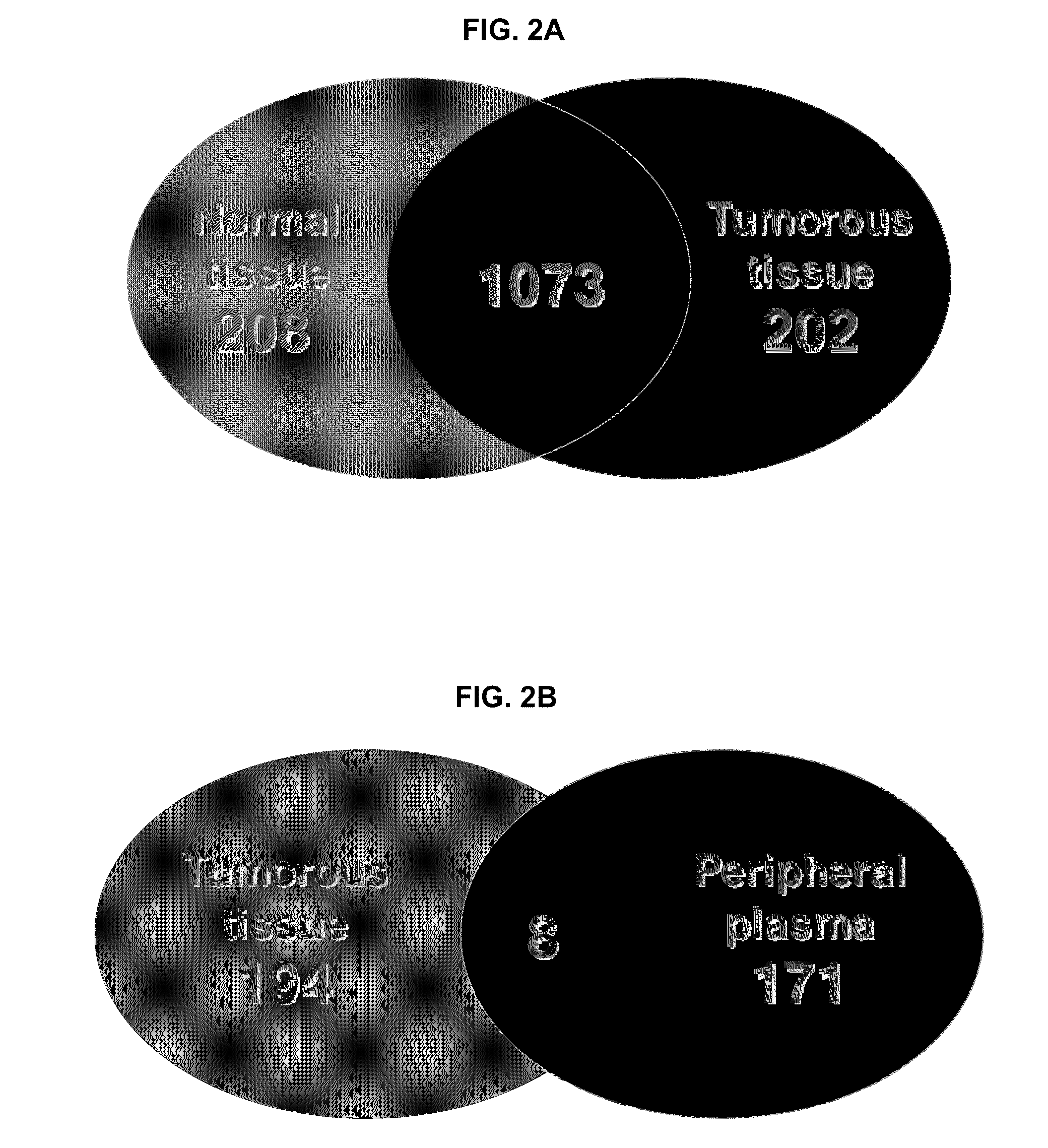
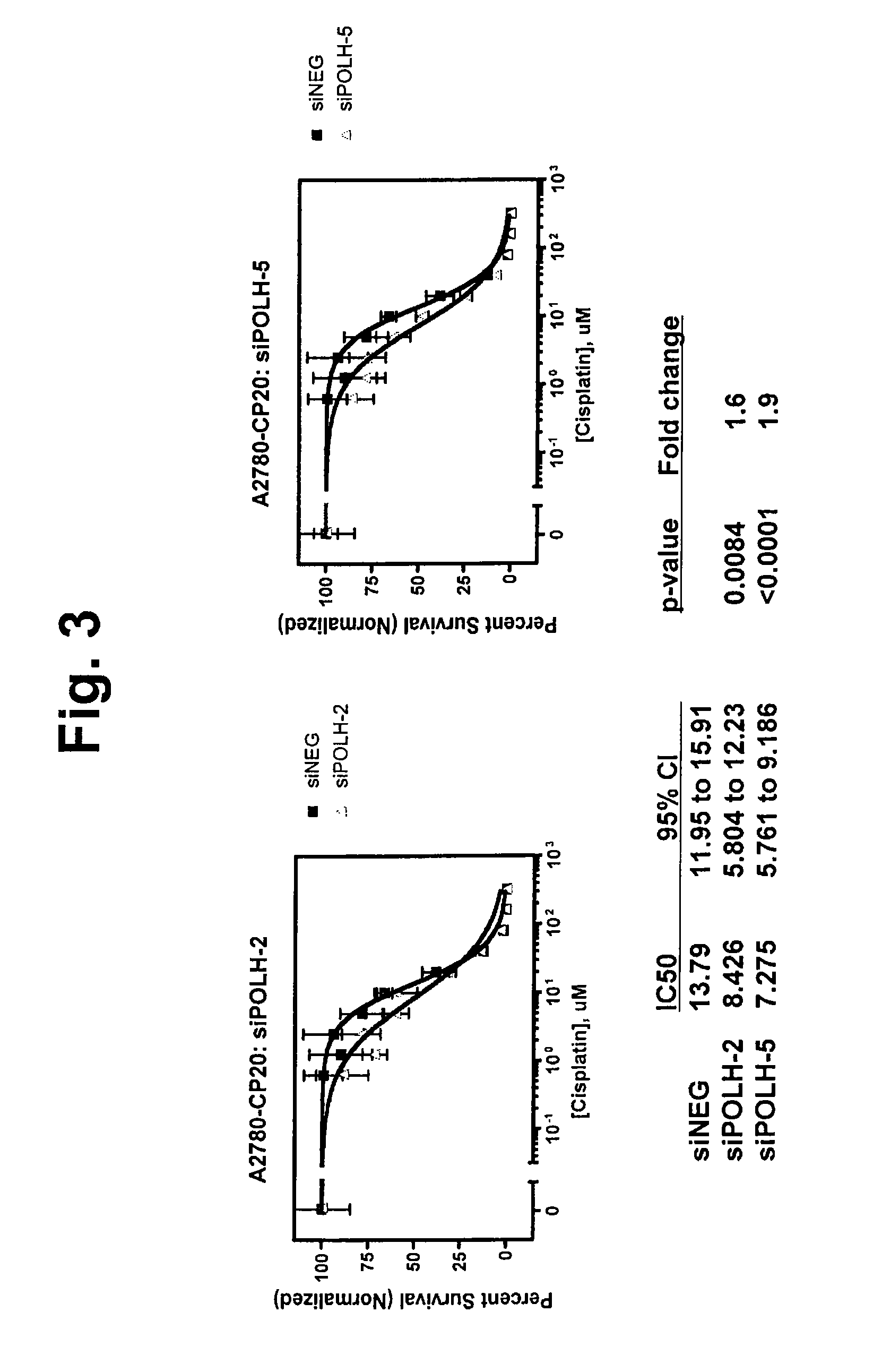
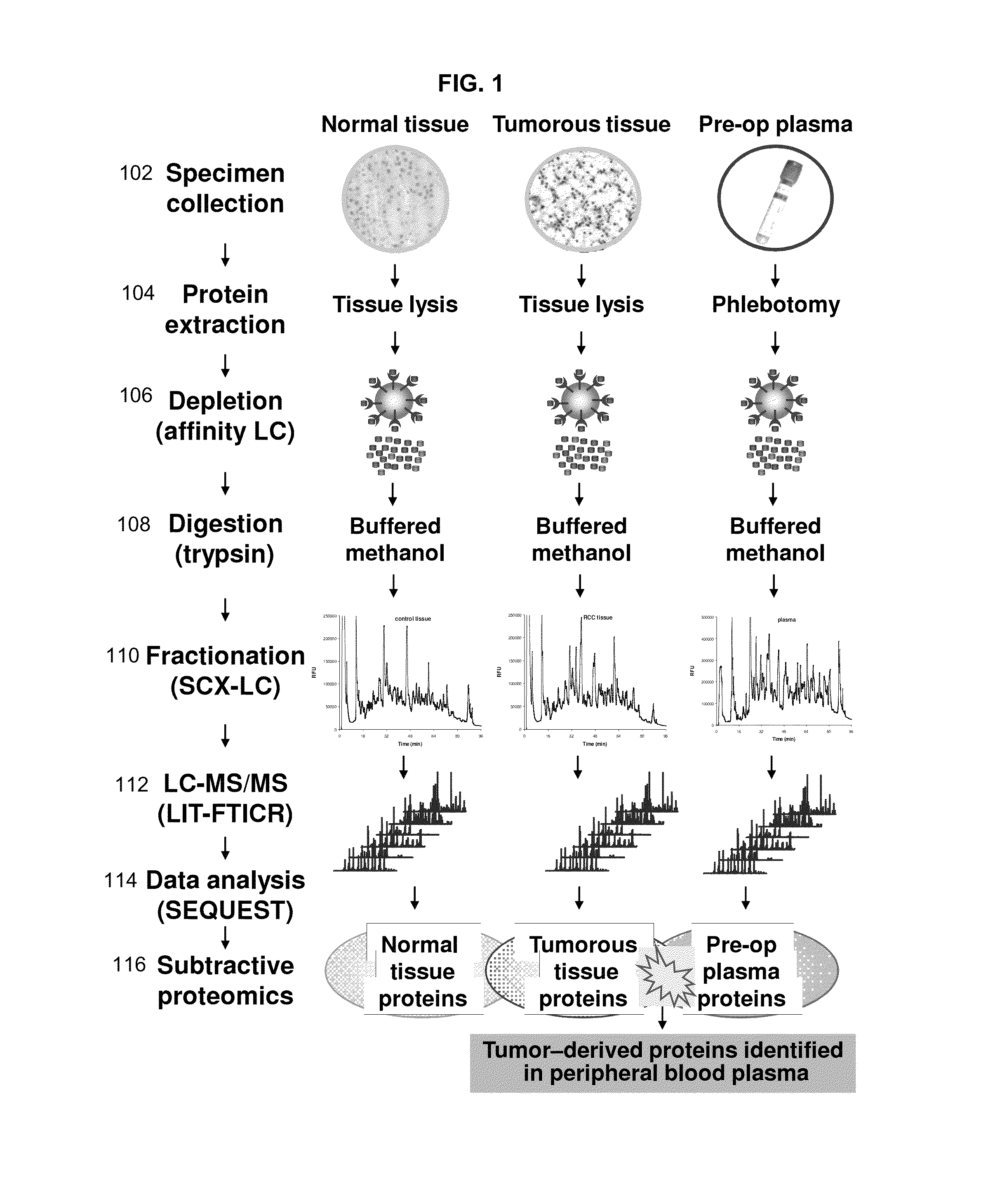
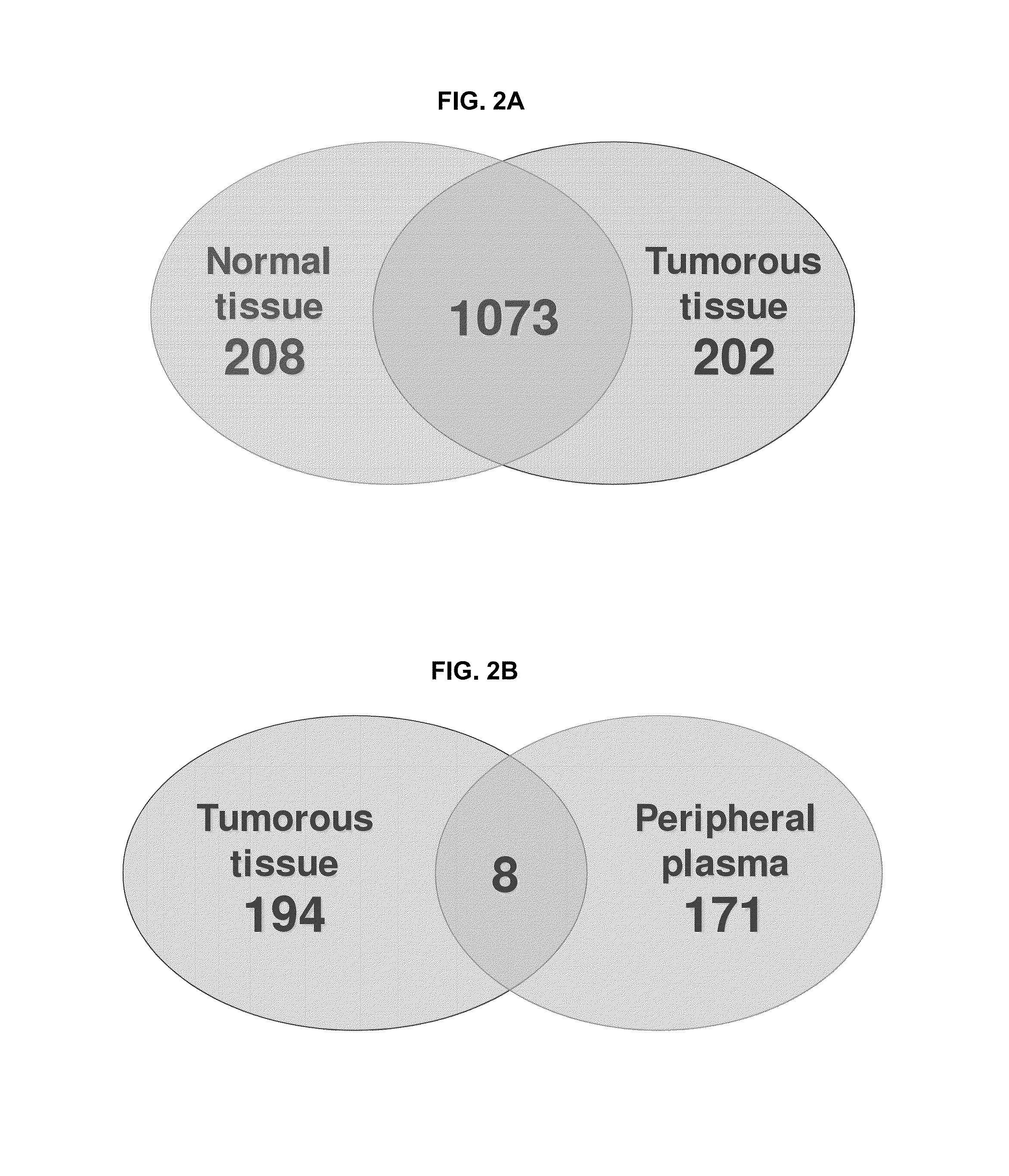
Disclosed herein is a method of identifying a tumor biomarker. In one example, a tumor biomarker is identified by obtaining a peripheral biological fluid sample from a subject with a tumor as well as a tumor sample and an adjacent non-tumor sample from such subject. A protein expression profile is detected in the peripheral biological fluid sample, tumor sample and adjacent non-tumor sample. The protein expression profiles of the peripheral biological fluid sample, tumor sample and adjacent non-tumor sample are then compared, wherein an increase in expression of a specific protein in the tumor sample and peripheral biological fluid sample but not in the adjacent non-tumor sample indicates that the specific protein is a biomarker of the tumor. Also disclosed herein is a gene profiling signature that can be used to diagnosis a subject with renal cell carcinoma (RCC) or to identify agents with therapeutic potential to treat RCC. Thus, methods of diagnosing a subject with RCC are disclosed. Methods are also provided for identifying agents that alter an activity of a RCC biomarker.
Owner:UNITED STATES OF AMERICA
Gene expression profile for predicting ovarian cancer patient survival
InactiveUS20100292303A1Reduce biological activityAltered expressionOrganic active ingredientsOrganic compound preparationPatient survivalFhit gene
A gene profiling signature for predicting ovarian cancer patient survival is disclosed herein. The gene signature can be used to diagnosis or prognosis ovarian cancer, identify agents to treat an ovarian tumor, to predict the metastatic potential of an ovarian tumor and to determine the effectiveness of ovarian tumor treatments. Thus, methods are provided for diagnosing and prognosing an ovarian tumor, such as ovarian cancer, in a subject. Methods are also provided for identifying agents that can be used to treat an ovarian tumor, for determining the effectiveness of an ovarian tumor treatment, or to predict the metastatic potential of an ovarian tumor. Methods of treatment are also disclosed which include administering a composition that includes a specific binding agent that specifically binds to one of the disclosed ovarian survival factor-associated molecules and ovarian tumor in the subject.
Owner:BIRRER MICHAEL J +3
Method of diagnosing early stage non-small cell lung cancer
InactiveUS20130252831A1Easy to predictMicrobiological testing/measurementLibrary screeningPrincipal component analysisSmoking status
A “malignancy-risk” (MR) gene signature score was developed with abundant proliferative genes using principal component analysis. This MR gene signature was shown to be a predictive and prognostic factor of overall survival in early-stage NSCLC. The malignancy-risk signature showed a significant association with OS, with poor survival seen in patients having a higher MR score and better survival seen in patients having a low MR score. As a prognostic factor, the MR gene signature showed a positive correlation with TNM stage, histologic grade, and smoking status. Combination of the MR signature with each clinical parameter often showed the best survival in the low MR group with good clinical outcome. The MR gene profile, tested with a PCA scoring method, discriminated overall survival in lung cancer patients was a predictor independent of pathological staging and other clinical parameters.
Owner:H LEE MOFFITT CANCER CENT & RES INST INC
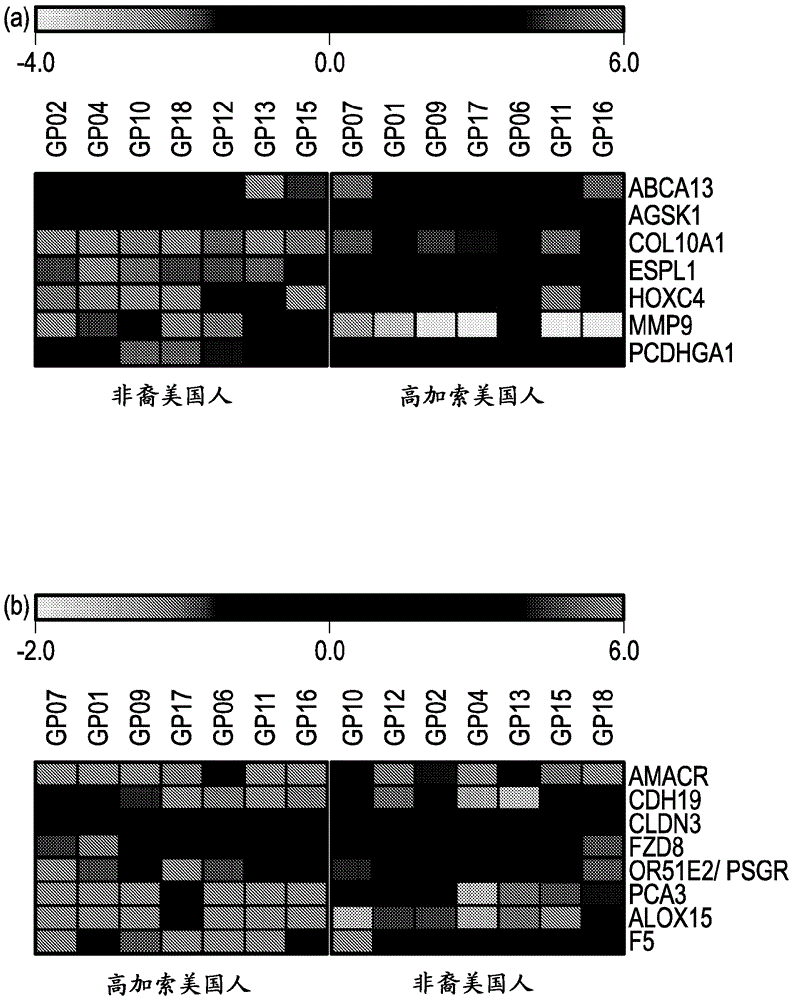
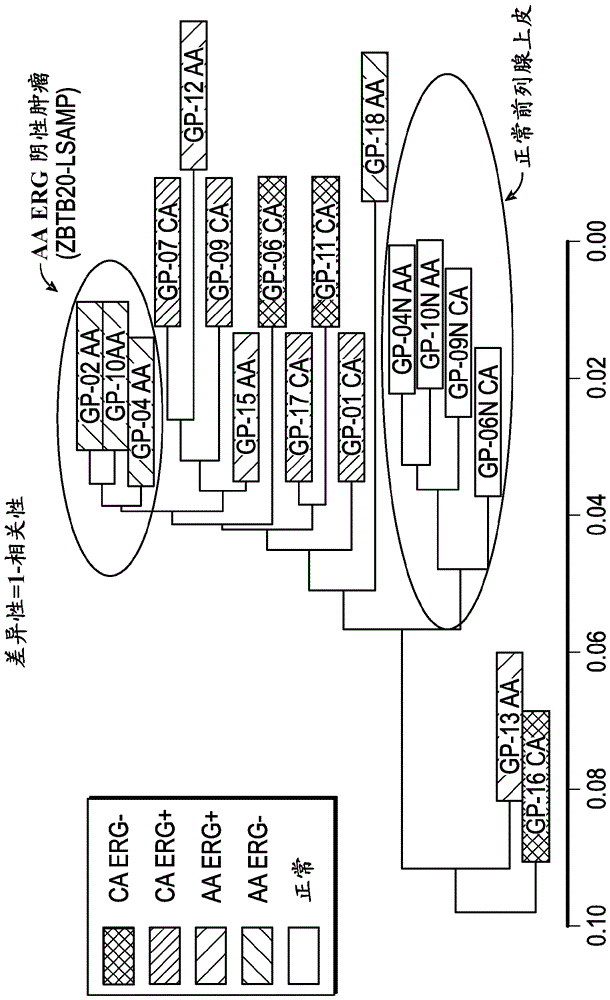
Prostate cancer gene profiles and methods of using the same
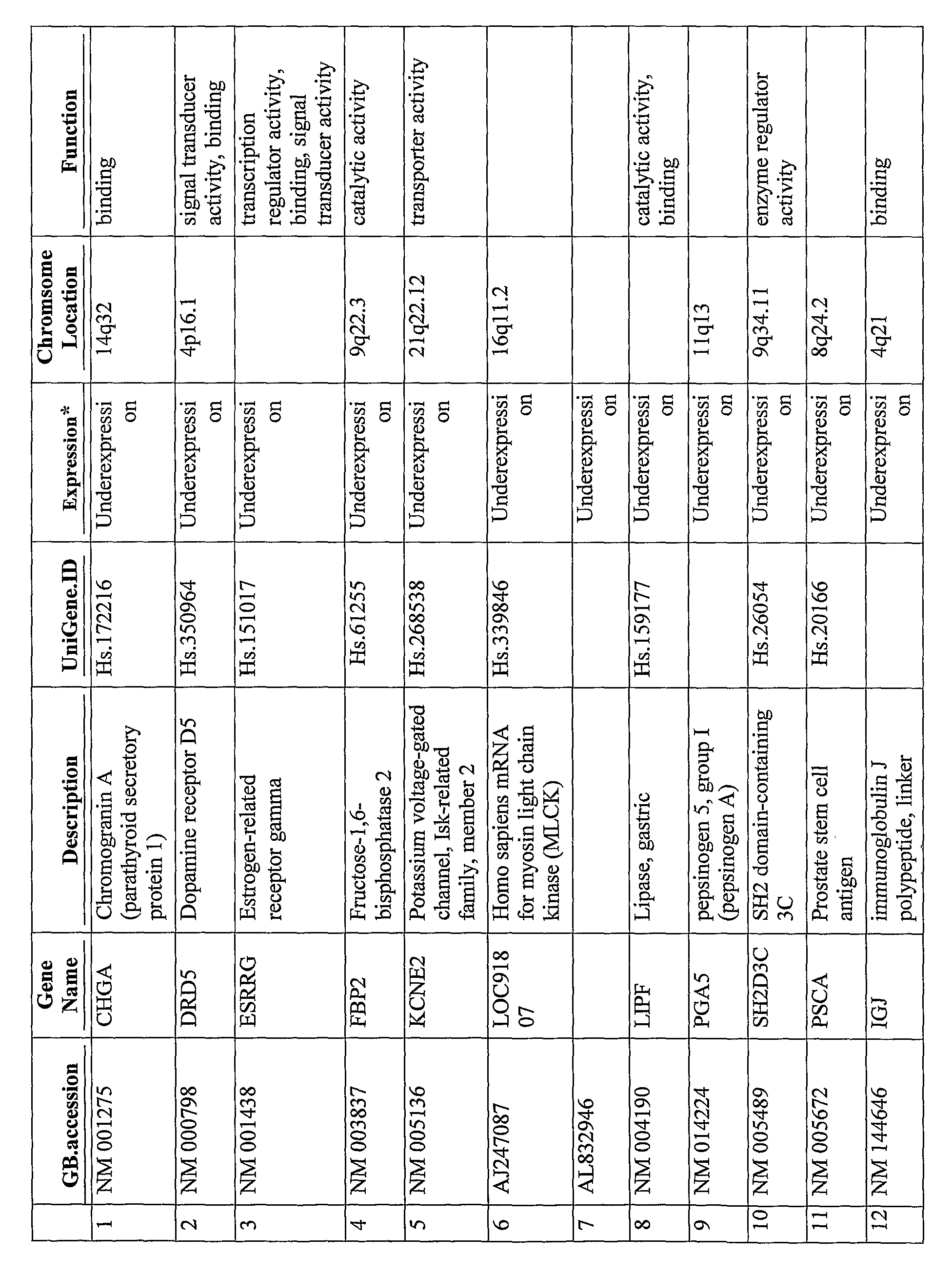
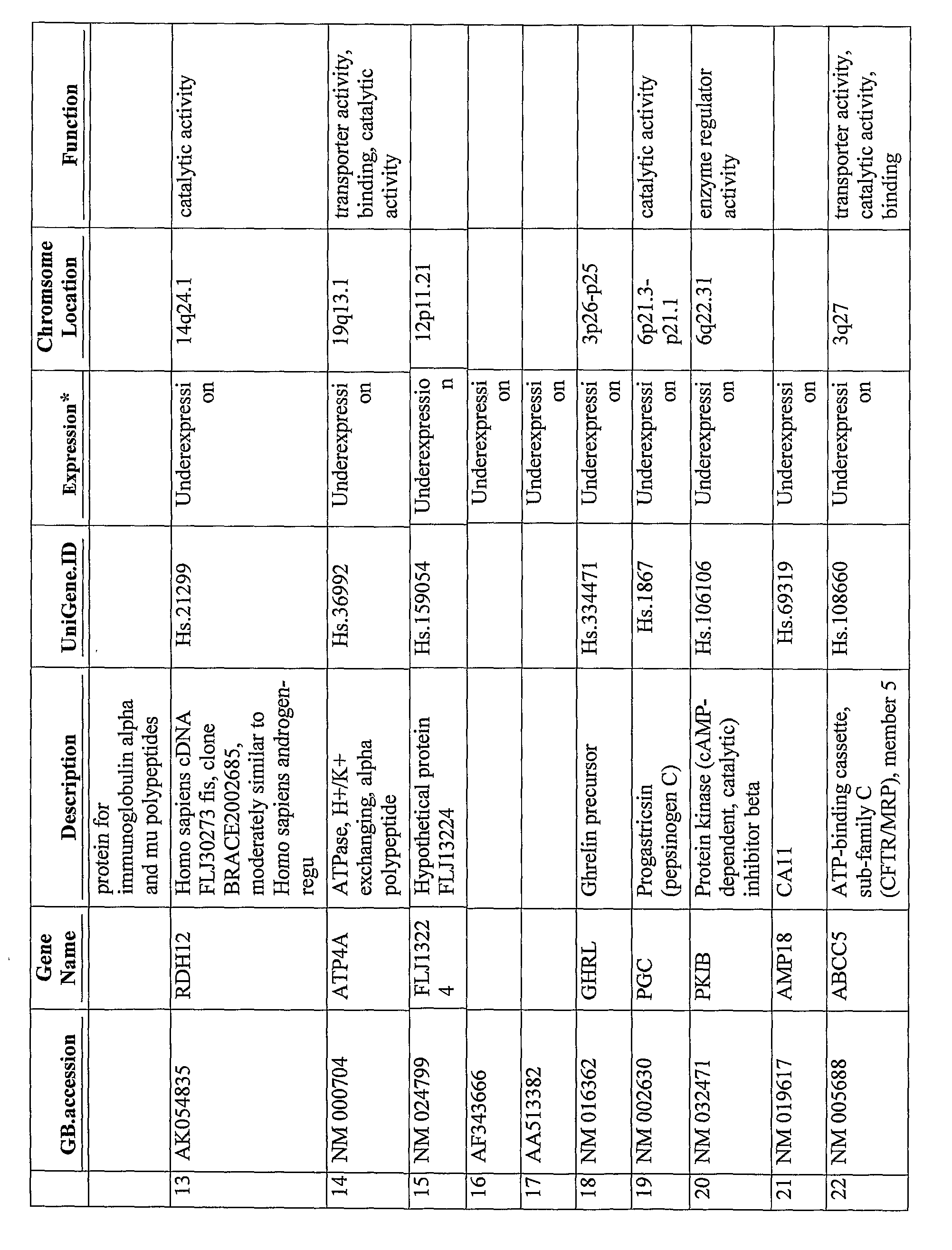
The present disclosure provides gene expression profiles that are associated with prostate cancer, including certain gene expression profiles that differentiate between subjects of African and Caucasian descent and other gene expression profiles that are common to subjects of both African and Caucasian descent. The gene expression profiles can be measured at the nucleic acid or protein level and used to stratify prostate cancer based on ethnicity or the severity or aggressiveness of prostate cancer. The gene expression profiles can also be used to identify a subject for prostate cancer treatment. Also provided are kits for diagnosing and prognosing prostate cancer and an array comprising probes for detecting the unique gene expression profiles associated with prostate cancer in subjects of African or Caucasian descent.
Owner:THE HENRY M JACKSON FOUND FOR THE ADVANCEMENT OF MILITARY MEDICINE INC +1
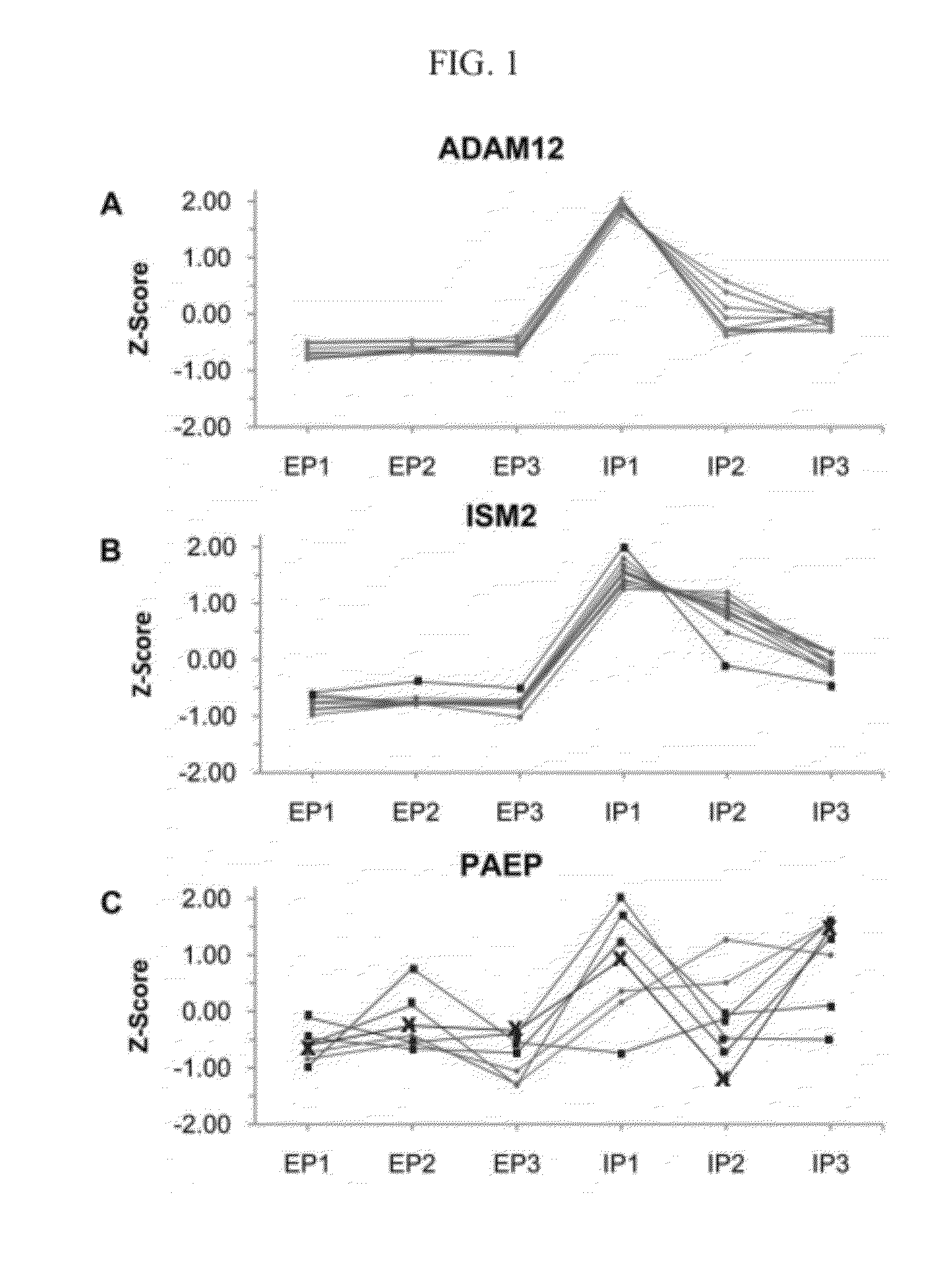
Methods and compositions for diagnosis of ectopic pregnancy
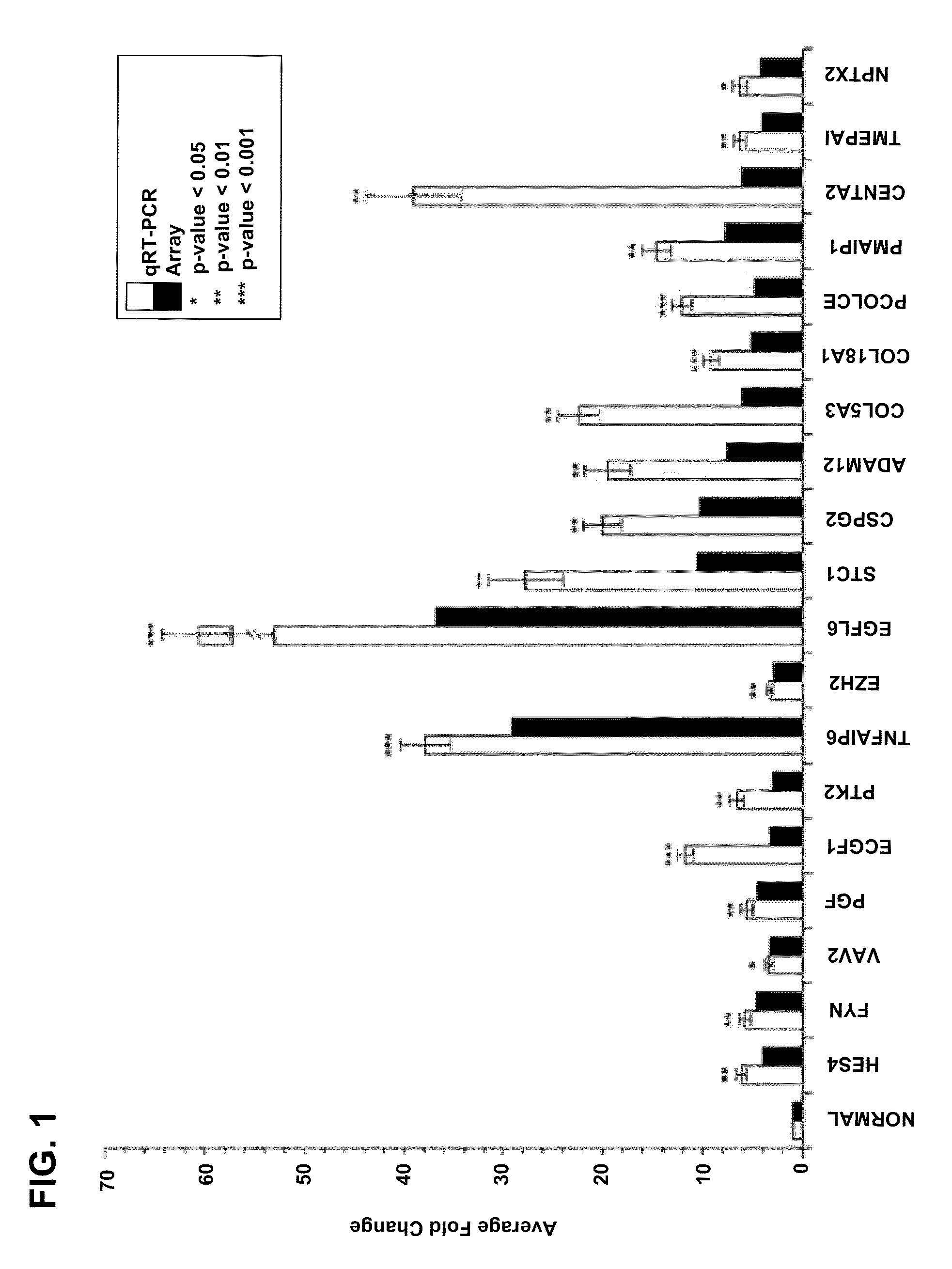
Methods and compositions are provided for diagnosing ectopic pregnancy in a mammalian subject by detecting changes in expression of ISM2, ADAM12, PST1, PSG7, PST11, PSG9, PSG2 and other genes identified therein, including combinations thereof. A selected gene, gene transcript or protein / peptide expression product, or profiles or signatures formed by combinations of same, detected in a biological fluid of a subject, enables comparison of the corresponding genes, proteins or profiles from that of a reference or control having a normal intrauterine pregnancy. Detection of characteristic changes in the gene profile or protein expression signature of the subject is correlated with a diagnosis of ectopic pregnancy. Various compositions for use in such diagnosis include PCR primer-probe sets or ligands, labeled or immobilized, which are capable of detecting the changes in expression or translation of these targets.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA +1
Methods to identify fat and lean animals using class predictors
InactiveUS20090217398A1Accurate identificationOrganic active ingredientsBioreactor/fermenter combinationsPhysiologyAnimal use
A combination comprising two or more polynucleotides that are differentially expressed in fat animals compared to lean animals or two or more proteins produced by the expression of such polynucleotides is disclosed. The combination and probes based upon the combination are used for formulating a prognosis that an animal is likely to become fat, developing a diagnosis that an animal is fat, screening substances to determine if they are useful for modulating the amount of adipose tissue on an animal, and detecting the differential expression of one or more genes differentially expressed in fat animals compared to lean animals in a sample. Methods for using class predictor gene profiles to identify fat and lean animals are also disclosed.
Owner:HILLS PET NUTRITION INC
Laser microdissection and microarray analysis of breast tumors reveal estrogen receptor related genes and pathways
InactiveUS20080305959A1Sugar derivativesMicrobiological testing/measurementPathway analysisEstrogen receptor
About 70% to 80% of breast cancers express estrogen receptor-α (ERα), and estrogens play important roles in the development and growth of hormone-dependent tumors. Together with lymph node metastasis, tumor size and histological grade, ER status is considered one of the prognostic factors in breast cancer, and an indicator for hormonal treatment. 147 genes and 112 genes with significant P-value and having significant differential expression between ER+ and ER− tumors were identified from the LCM data set and bulk tissue data set, respectively. 61 genes were found to be common in both data sets, while 85 genes were unique to the LCM data set and 51 genes were present only in the bulk tumor data set. Pathway analysis with the 85 genes using Gene Ontology suggested that genes involved in endocytosis, ceramide generation, Ras / ERK / Ark cascade, and JAT-STAT pathway may play roles related to ER. The gene profiling with LCM-captured tumor cells provides a unique approach to characterize and study epithelial tumor cells and to gain an insight into signaling pathways associated with ER.
Owner:VERIDEX LCC
Diagnostic marker for lung adenocarcinoma based on metabolic gene profile
PendingCN110055328AHigh AUC valueMicrobiological testing/measurementDNA/RNA fragmentationAdenocarcinomaASAH1
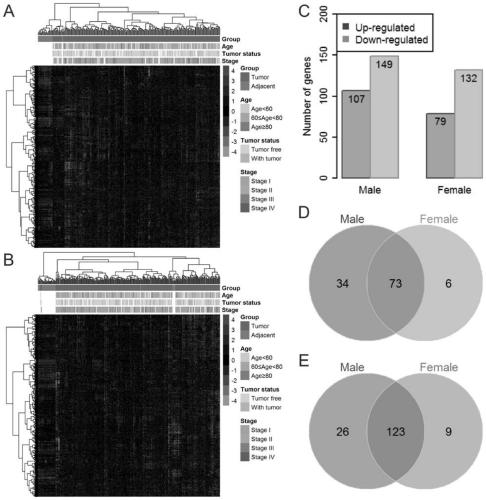
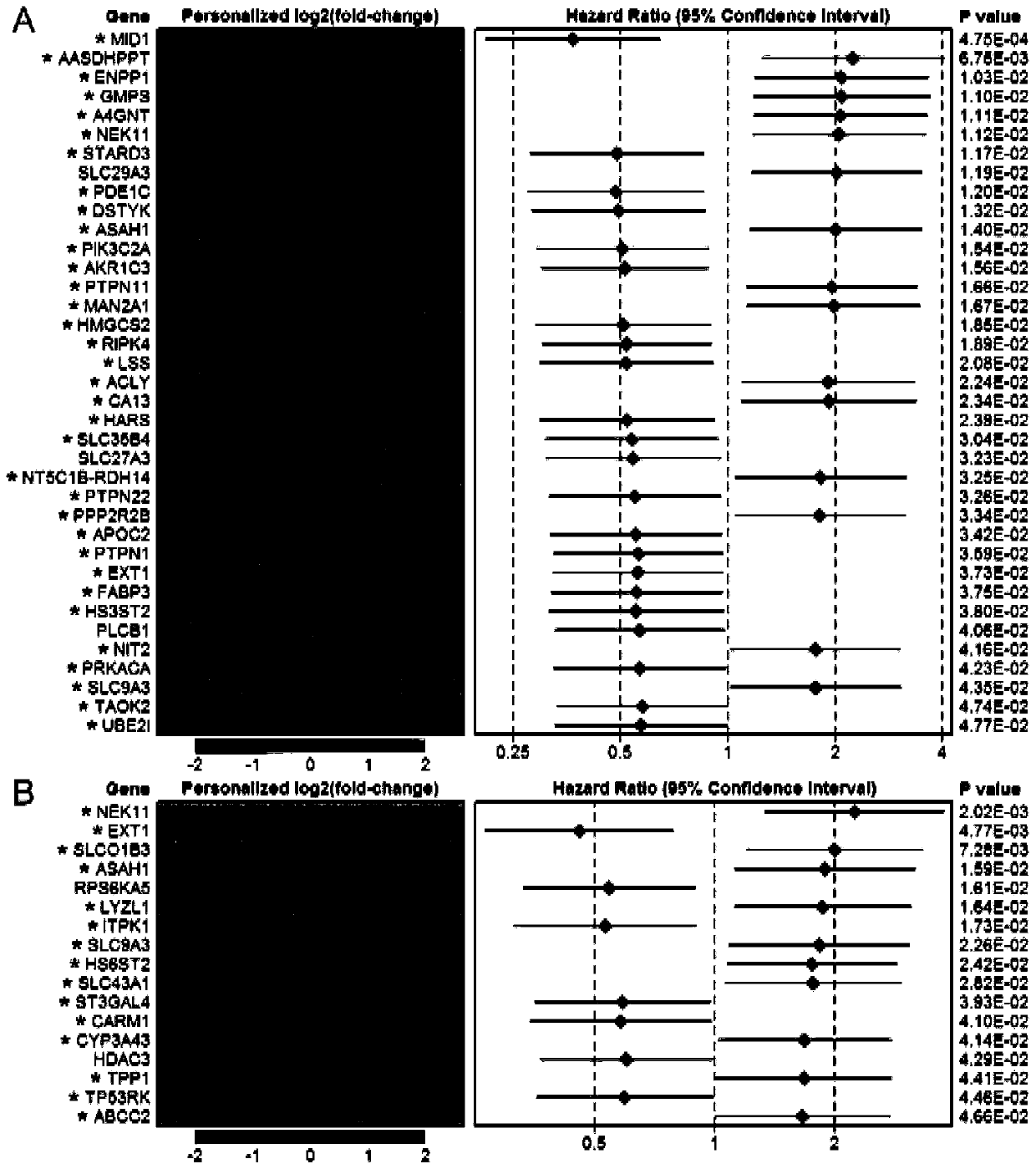
The invention relates to the technical field of lung cancer diagnosis, in particular to a diagnostic marker for lung adenocarcinoma based on a metabolic gene profile. Application of TAOK2 and ASAH1 asdiagnostic markers for the lung adenocarcinoma is provided. An embodiment demonstrates the holistic metabolic difference between the male and the female in the lung adenocarcinoma. 34 risk metabolismgenes identified in the male and 15 risk metabolism genes in the female all have higher AUC values, indicating that these genes, alone or in combination, can be used as the diagnostic marker for thelung adenocarcinoma. The TAOK2 has the highest AUC value of 1.000 in the male risk metabolism genes, and the TAOK2 expression is down-regulated in male patients. The ASAH1 has the highest AUC value of0.985 in the female risk metabolism genes, and the ASAH1 expression is up-regulated in female patients. The diagnostic marker contributes to the study of the difference of the diagnostic marker for the lung adenocarcinoma between the male and the female.
Owner:FIRST AFFILIATED HOSPITAL OF KUNMING MEDICAL UNIV
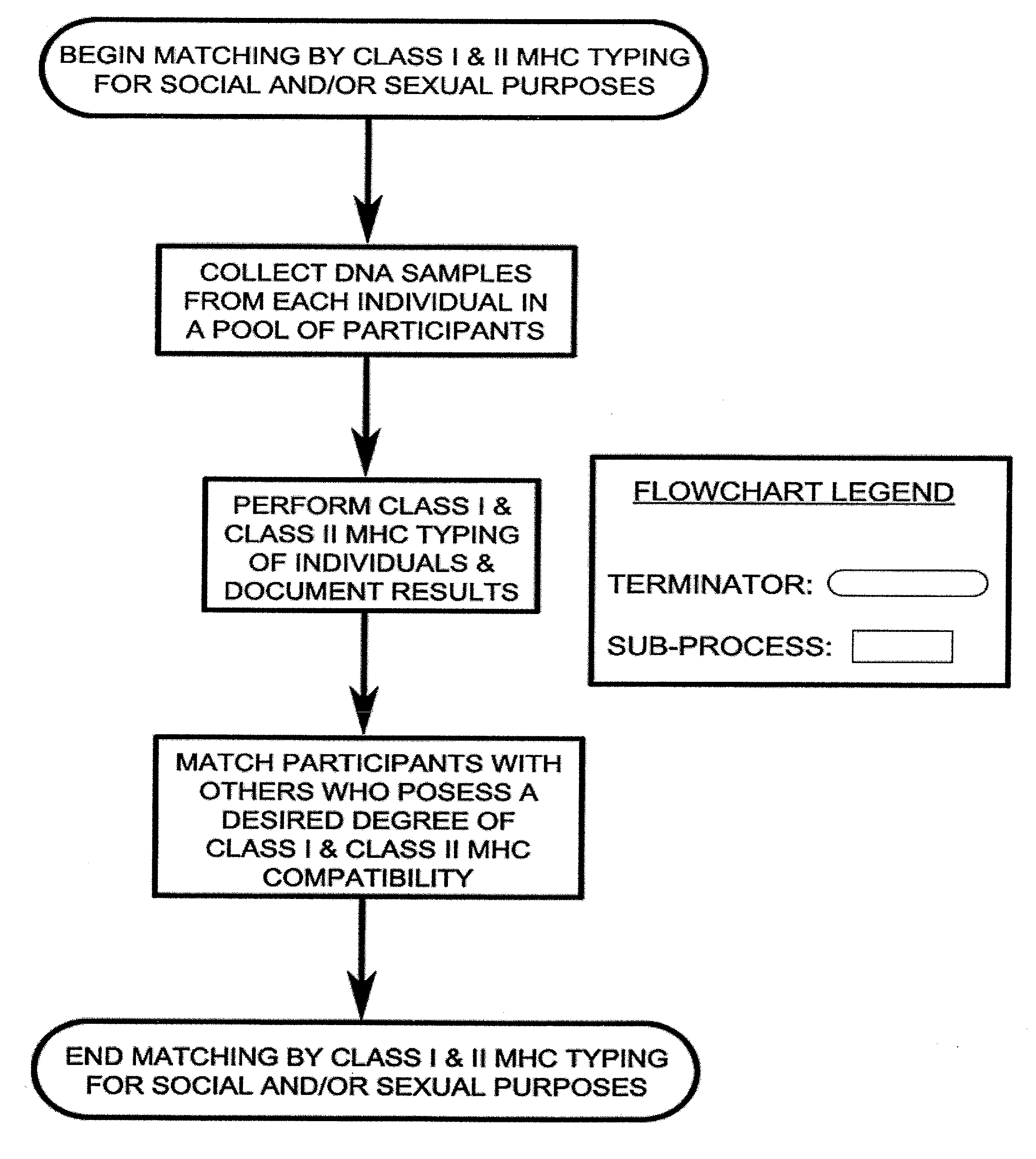
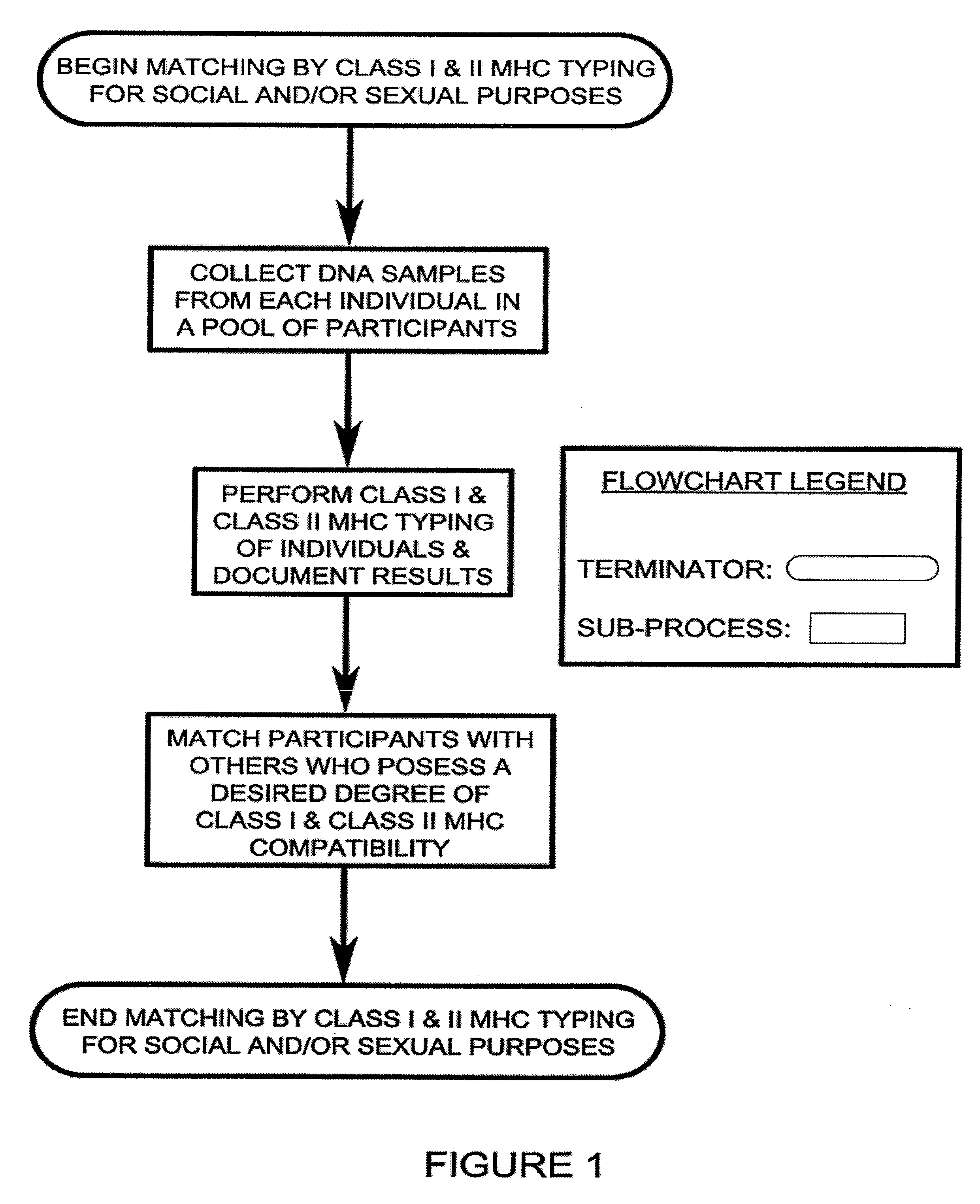
Class I and Class II MHC Profiling for Social and Sexual Matching of Human Partners
An improved process of matching people with one another, for social and / or sexual purposes, based on the profiles of the participants' Class I and Class II MHC genes. The quality of this human matching is inversely proportional to the number of common alleles of said genetic profiles of the individuals. Higher quality matches of this type result in greater sexual attraction, more attractive physical odor, and offspring with more robust immune systems among matched participants.
Owner:HOLZLE ERIC
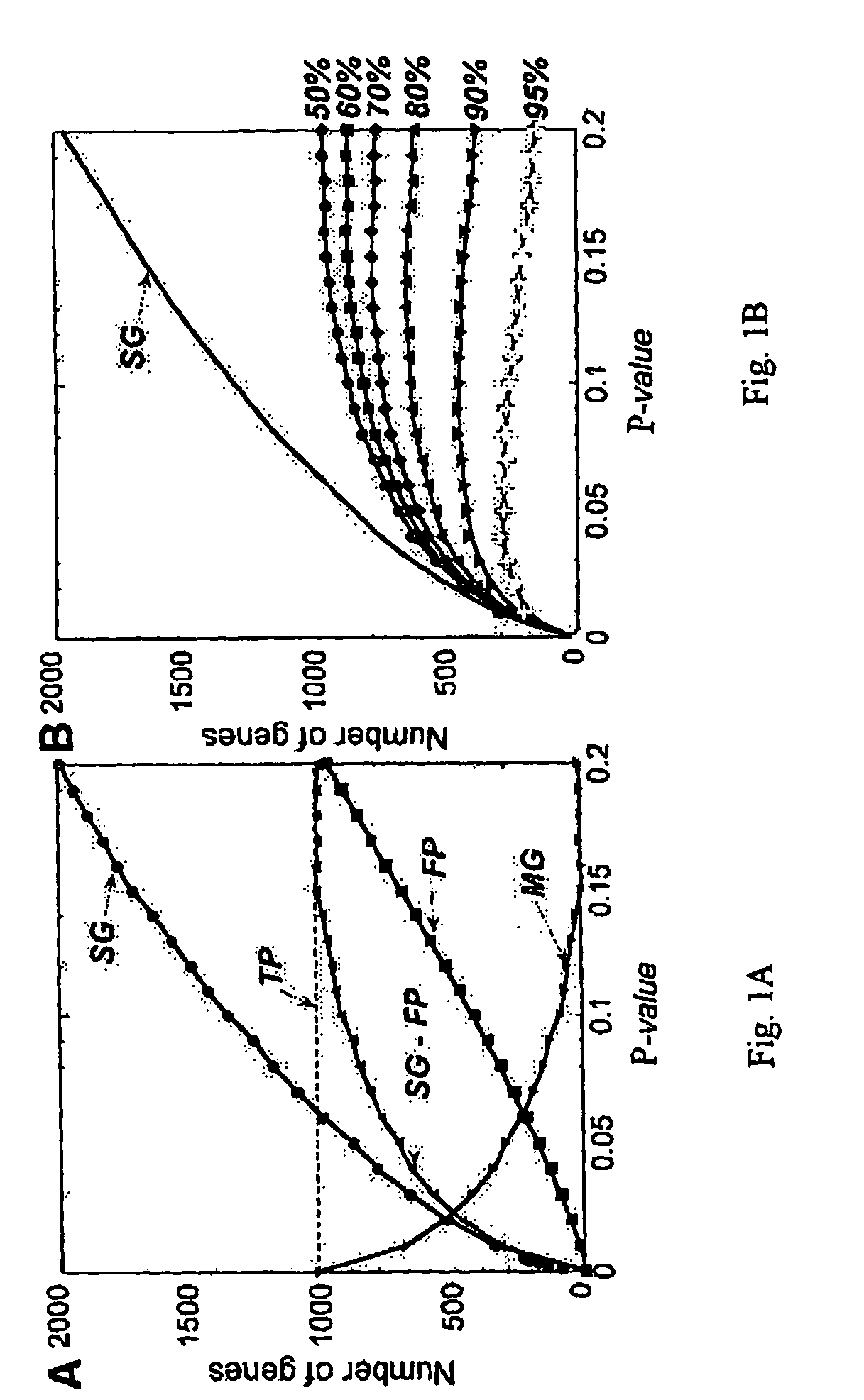
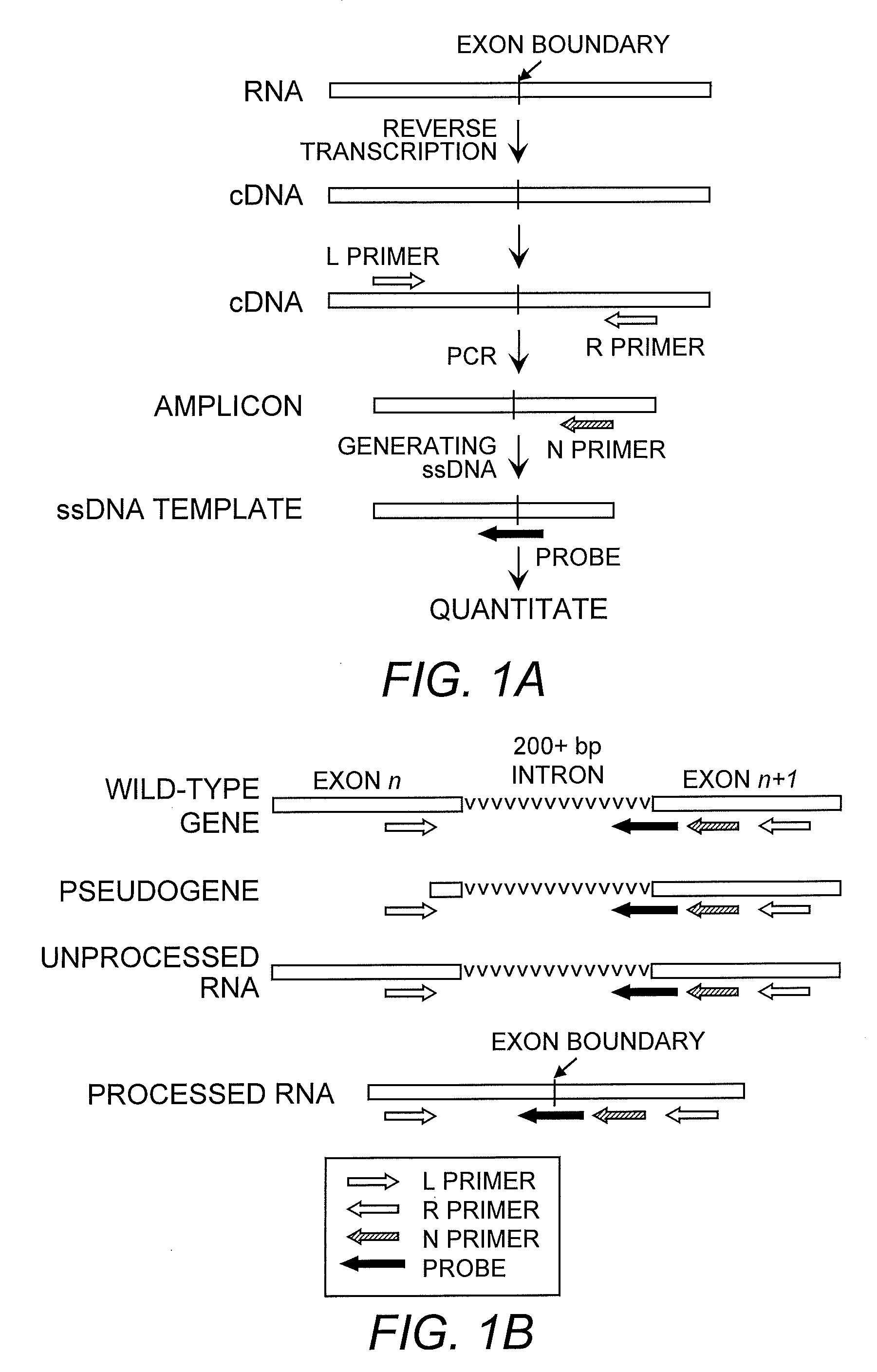
Method for high-throughput gene expression profile analysis
The present invention embraces a method for high-throughput gene profiling with high specificity and sensitivity. With this system, >1000 mRNA species can be co-amplified using gene-specific primers from a single cell. The primers are designed to amplify sequences of desirable length, which are in different exons. The exons can be either adjacent and separated by a large intron or include more than two exons. The amplified sequences are then analyzed by microarray with probes hybridizing to neighboring exons.
Owner:UNIV OF MEDICINE & DENTISTRY OF NEW JERSEY
Biomarkers for the efficacy of calcitonin and parathyroid hormone treatment
InactiveUS20070099828A1Peptide/protein ingredientsCalcitoninsRecombinant salmon calcitoninAnabolic Effect
A mufti-organ gene profiling analysis of the results of an administration to a subject of salmon calcitonin or a parathyroid hormone analogue provides biomarkers of calcitonin treatment efficacy and parathyroid hormone or parathyroid hormone analogue treatment efficacy. Among the biomarkers are the expression profiles of the genes for Y-box binding protein, BMPs, FGFs, IGFs, VEGF, &x3B1;-2-HS glycoprotein (AHSG), OSF, nuclear receptors (steroid / thyroid family) and others. The results obtained support the anabolic effect of salmon calcitonin on bone metabolism.
Owner:BOBADILLA MARIA
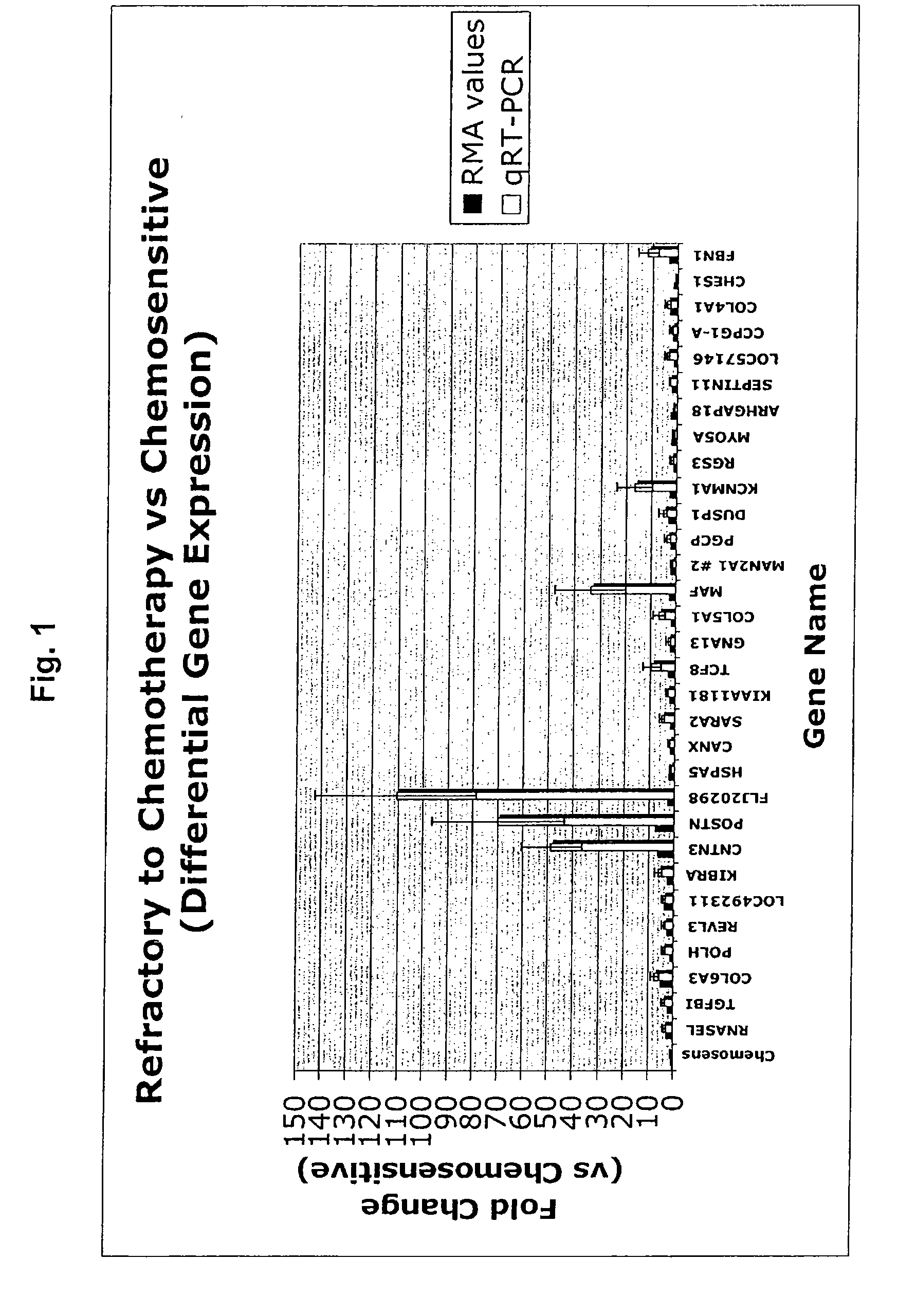
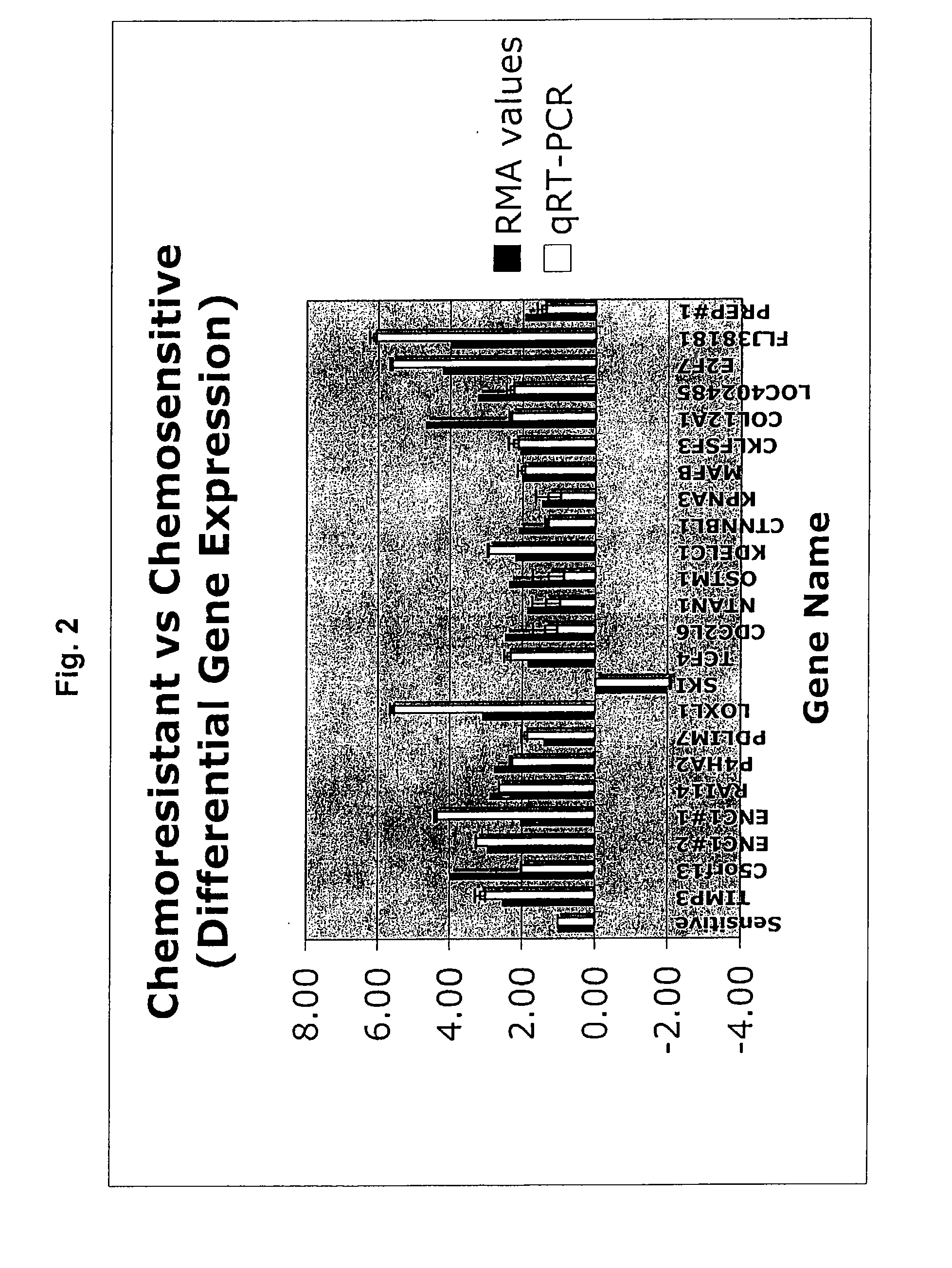
Gene expression profile that predicts ovarian cancer subject response to chemotherapy
InactiveUS20110178154A1High sensitivityReduce sensitivityOrganic active ingredientsMicrobiological testing/measurementOncologyOvarian cancer
A gene profiling signature is disclosed herein. The gene signature can predict whether a subject with ovarian cancer will be chemorefractory, chemoresistant or chemosensitive. Thus, methods are disclosed for determining whether a subject with ovarian cancer is sensitive to treatment with a chemotherapeutic agent. Methods are also provided for increasing sensitivity to the chemotherapeutic agent if the presence of differential expression indicates that the ovarian cancer has a decreased sensitivity to chemotherapeutic agent.
Owner:BIRRER MICHAEL J +3
Renal cell carcinoma biomarkers
Disclosed herein is a method of identifying a tumor biomarker. In one example, a tumor biomarker is identified by obtaining a peripheral biological fluid sample from a subject with a tumor as well as a tumor sample and an adjacent non-tumor sample from such subject. A protein expression profile is detected in the peripheral biological fluid sample, tumor sample and adjacent non-tumor sample. The protein expression profiles of the peripheral biological fluid sample, tumor sample and adjacent non-tumor sample are then compared, wherein an increase in expression of a specific protein in the tumor sample and peripheral biological fluid sample but not in the adjacent non-tumor sample indicates that the specific protein is a biomarker of the tumor. Also disclosed herein is a gene profiling signature that can be used to diagnosis a subject with renal cell carcinoma (RCC) or to identify agents with therapeutic potential to treat RCC. Thus, methods of diagnosing a subject with RCC are disclosed. Methods are also provided for identifying agents that alter an activity of a RCC biomarker.
Owner:UNITED STATES OF AMERICA
Compositions and Methods for Identifying Autism Spectrum Disorders
InactiveUS20140213469A1Microbiological testing/measurementLibrary screeningAutism spectrum disorderGene
The compositions and methods described are directed to gene chips having a plurality of different oligonucleotides with specificity for genes associated with autism spectrum disorders. The invention further provides methods of identifying gene profiles for neurological and psychiatric conditions including autism spectrum disorders, methods of treating such conditions, and methods of identifying therapeutics for the treatment of such neurological and psychiatric conditions.
Owner:GEORGE WASHINGTON UNIVERSITY
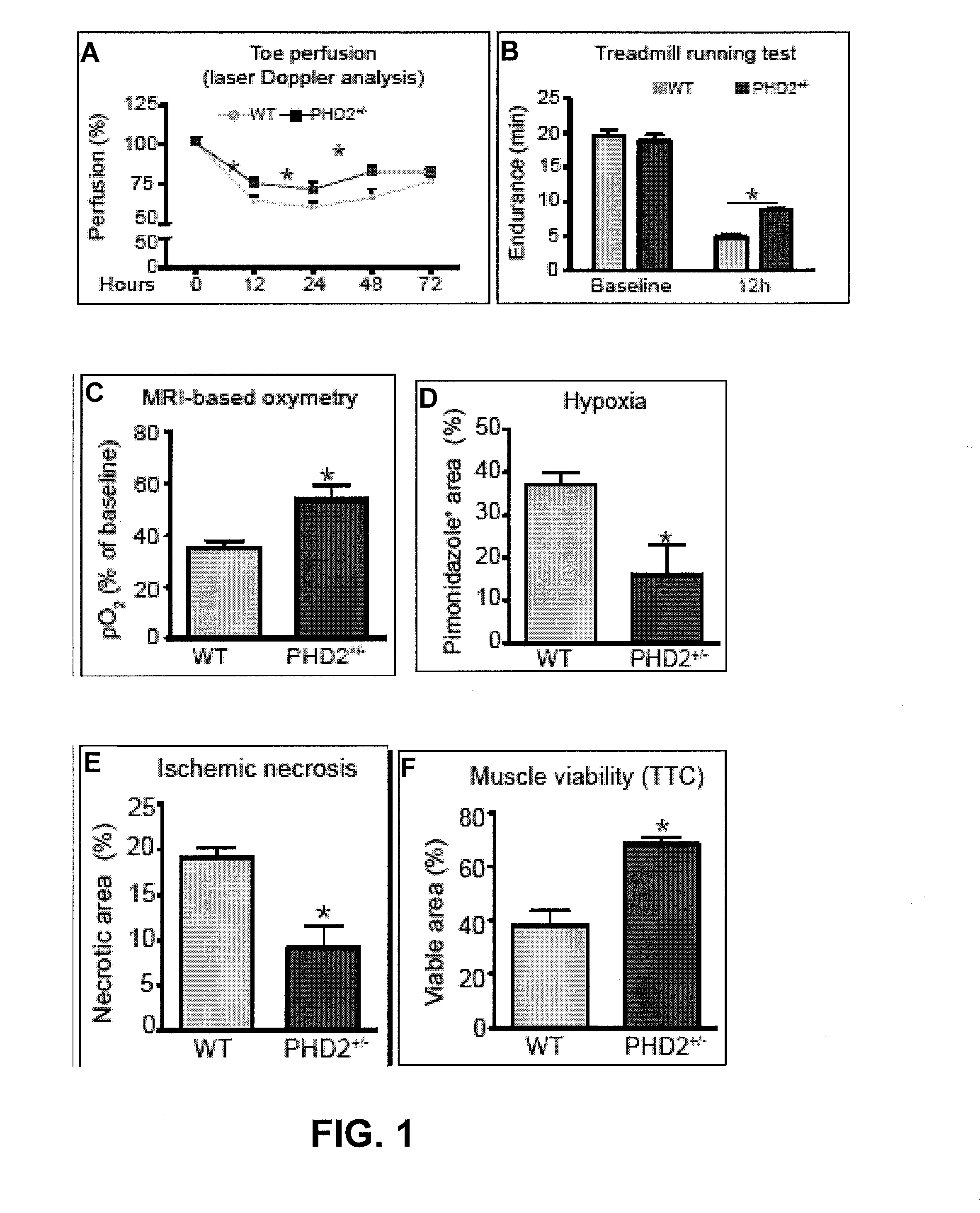
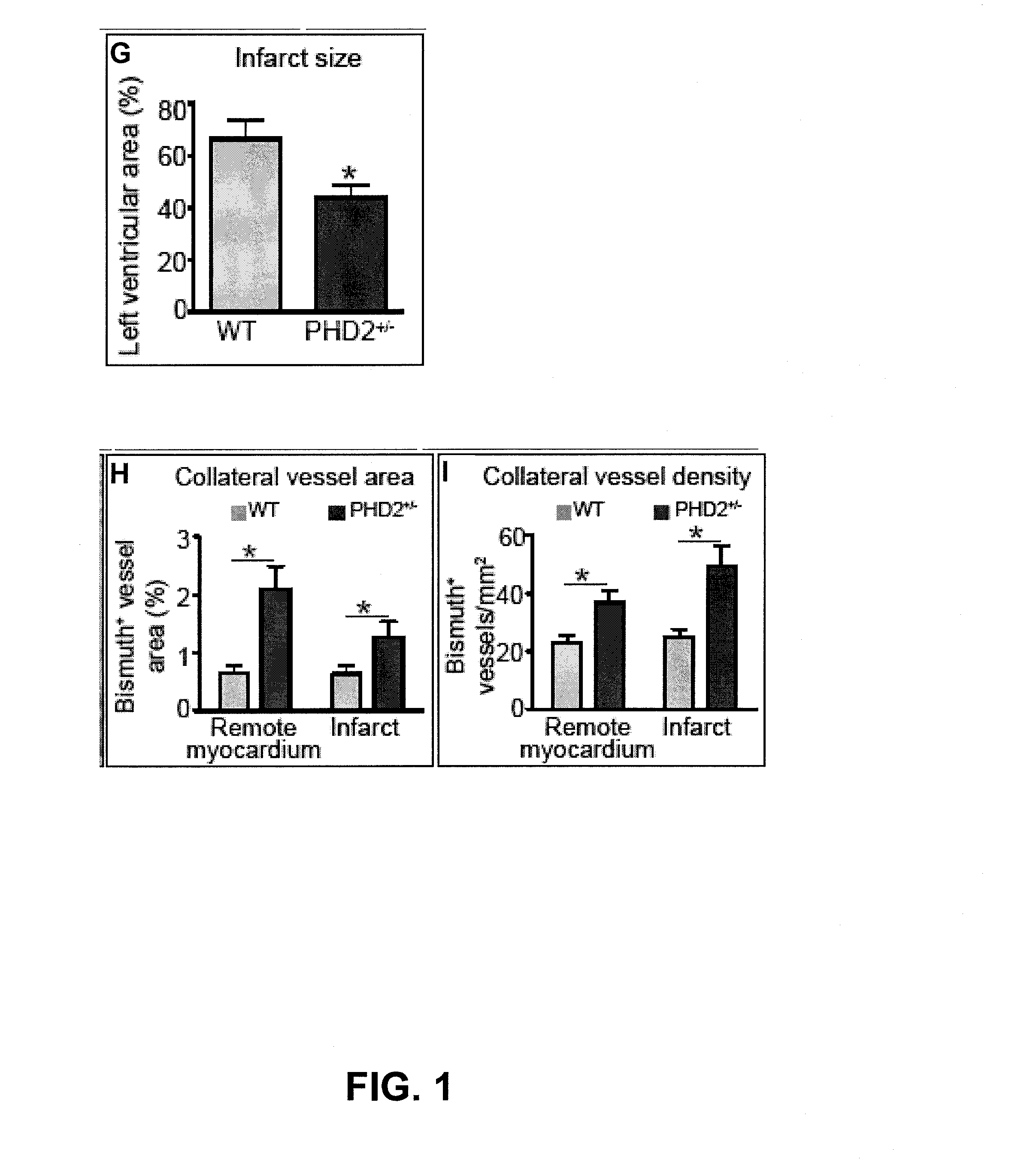
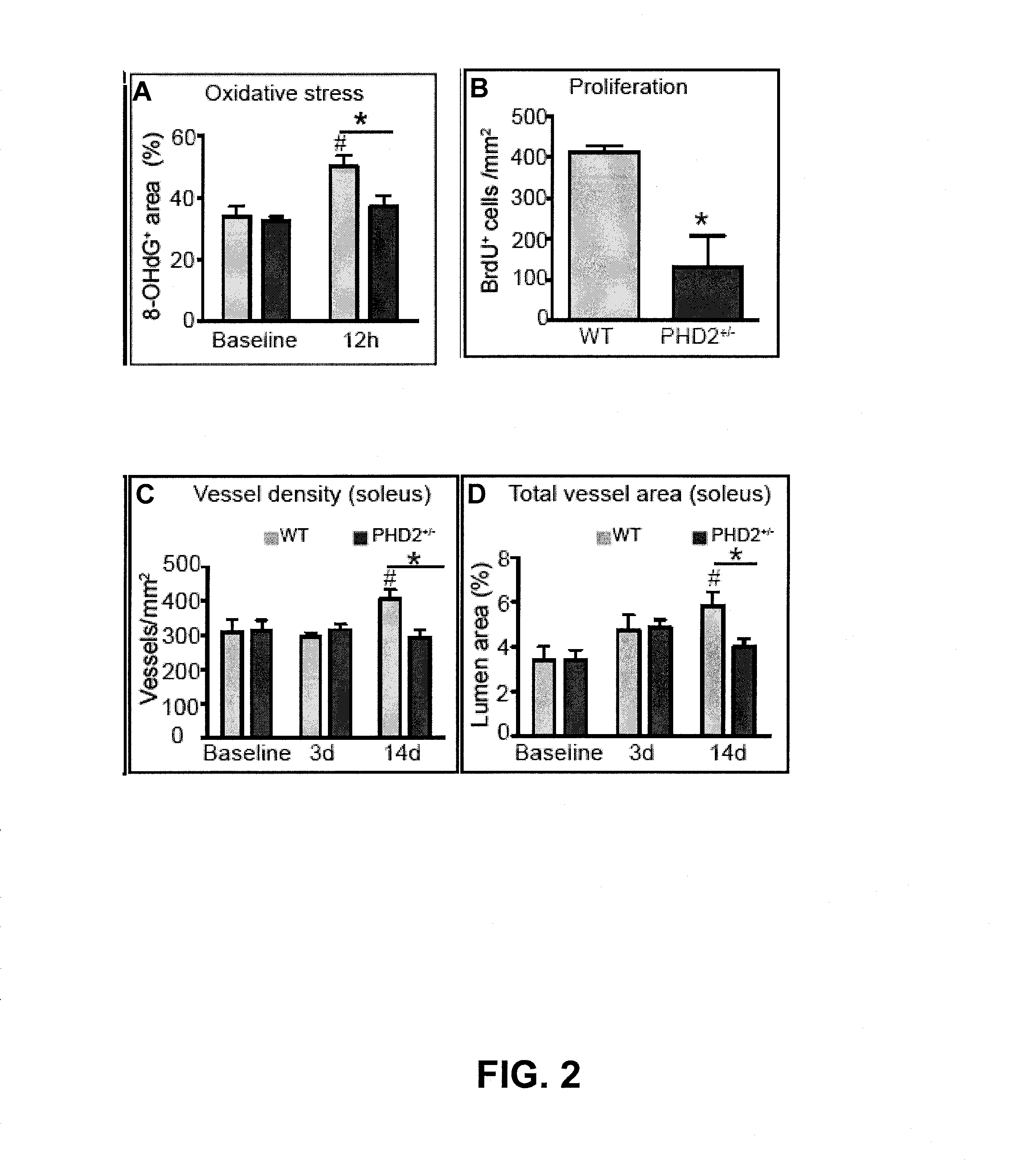
Induction/monitoring of arteriogenesis using sdf1 and pdgfb or inhibition of phd2
InactiveUS20130078224A1Smooth maturationSmooth migrationBiocidePeptide/protein ingredientsPDGFBPerfusion
The disclosure relates to the field of ischemia and how to increase tissue perfusion in ischemic tissue by cellular therapy. Specifically, the beneficial effects of myeloid (bone marrow-derived) cells with a particular arteriogenic gene expression profile are shown, and it is shown that increased arteriogenesis and perfusion is specifically due to the effects of combined PDGFB and SDF-1. The arteriogenic gene profile of the myeloid cells used for therapy can, for instance, be obtained by inhibition of PHD2.
Owner:VLAAMS INTERUNIVERSITAIR INST VOOR BIOTECHNOLOGIE VZW +1
Cell sensor having multifunctional reactions for the definition of quality criteria during the production of materials
InactiveUS20090118138A1Microbiological testing/measurementLibrary screeningCell type specificTest group
Method for producing a cell sensor system for the definition of quality criteria during the production of materials, characterised by the following method steps:a) cultivation of first cells of a specific type under standardised culture conditions (control group),b) cultivation of second cells of the specific type on / in / between different materials to be tested (test group),c) harvesting of the cells,d) determination of the gene activities of the cells of the control group and of the cells of the test group,e) comparison of the gene activities of the test group with the control group,f) identification of the genes for which there is a difference in the gene activities between the control group and the test group,g) construction of a microarray using the identified genes with different gene activity as the gene profile, this created microarray being defined as the standard for the specific cell type, andh) provision of third cells of the specific cell type as cell sensor.
Owner:GERRESHEIMER WILDEN GMBH
Pro-angiogenic genes in ovarian tumor endothelial cell isolates
InactiveUS8440393B2Microbiological testing/measurementScreening processCell tumorAngiogenesis growth factor
A gene profiling signature for ovarian tumor endothelial cells is disclosed herein. The gene signature can be used to diagnosis or prognosis an ovarian tumor, identify agents to treat an ovarian tumor, to predict the metastatic potential of an ovarian tumor and to determine the effectiveness of ovarian tumor treatments. Thus, methods are provided for identifying agents that can be used to treat ovarian cancer, for determining the effectiveness of an ovarian tumor treatment, or to diagnose or prognose an ovarian tumor. Methods of treatment are also disclosed which include administering a composition that includes a specific binding agent that specifically binds to one of the disclosed ovarian endothelial cell tumor-associated molecules and inhibits ovarian tumor in the subject.
Owner:UNITED STATES OF AMERICA +1
Gene expression profile detection kit for predicting breast cancer reoccurrence of Chinese population
The invention discloses a characteristic gene expression profile applied to predicting breast cancer reoccurrence of Chinese population and also provides a recurrence index calculation formula and a recurrence prediction judgment standard. The invention further provides a fluorescent quantitative detection kit for the breast cancer characteristic gene profile. A PCR (Polymerase Chain Reaction) amplification primer disclosed by the invention is composed of eight pairs of primers, and the following eight gene correlated expressions can be subjected to combined detection: CCNA2, CCND1, SERP1NE1,SPRR1B, TOP2A, beta-ACTIN, GAPDH and HPRT1. The characteristic gene expression profile disclosed by the invention can be used for guiding individualized treatment of clinical breast cancer and preventing breast cancer reoccurrence.
Owner:JIANGSU CANCER HOSPITAL
Differential expression gene profiles and applications in molecular staging of human gastric cancer
InactiveUS20090117561A1High expressionReduce expressionMicrobiological testing/measurementMammalStomach cancer
The invention provides methods for detecting differential gene expression in intestinal gastric tissue in a mammal by comparing the expression of specific genes in an intestinal gastric tissue suspected of being cancerous with that of the corresponding adjacent intestinal gastric tissue or a normal gastric mucosa tissue. The methods can be used in diagnosing or monitoring the progression of intestinal gastric cancer and determining the levels of differentiation of intestinal gastric cancer Systems and kits for methods of the invention are also provided.
Owner:LU YOUYONG +6
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com