Prostate cancer gene profiles and methods of using the same
A technology of prostate cancer and gene expression profiling, applied in biochemical equipment and methods, microbial measurement/testing, instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0151] Example 1. Comparative Genomic DNA Analysis
[0152] In a cohort of 7 AA and 7 CA CaP patients (28 samples), a comparative genome-wide analysis was performed using primary prostate tumors and corresponding normal tissues (blood). Cohorts were selected according to the following criteria: primary treatment radical prostatectomy, no neoadjuvant therapy, Gleason grades 3+3 and 3+4 (representing the majority of CaPs screened by PSA at diagnosis / primary treatment), having 80% Frozen tumor tissue with tumor cell content or higher, dissected tumor tissue with more than 2 μg of high molecular weight genomic DNA, availability of corresponding blood genomic DNA, and patient clinicopathological data.
[0153] 28 samples were sent to Illumina Inc. (UK) for sequencing. Sequences from tumor samples were mapped to a reference genome using Illumina's ELAND alignment algorithm. Sequencing reported good coverage (average 37). Apply the Strelka algorithm to simultaneously call variants...
Embodiment 2
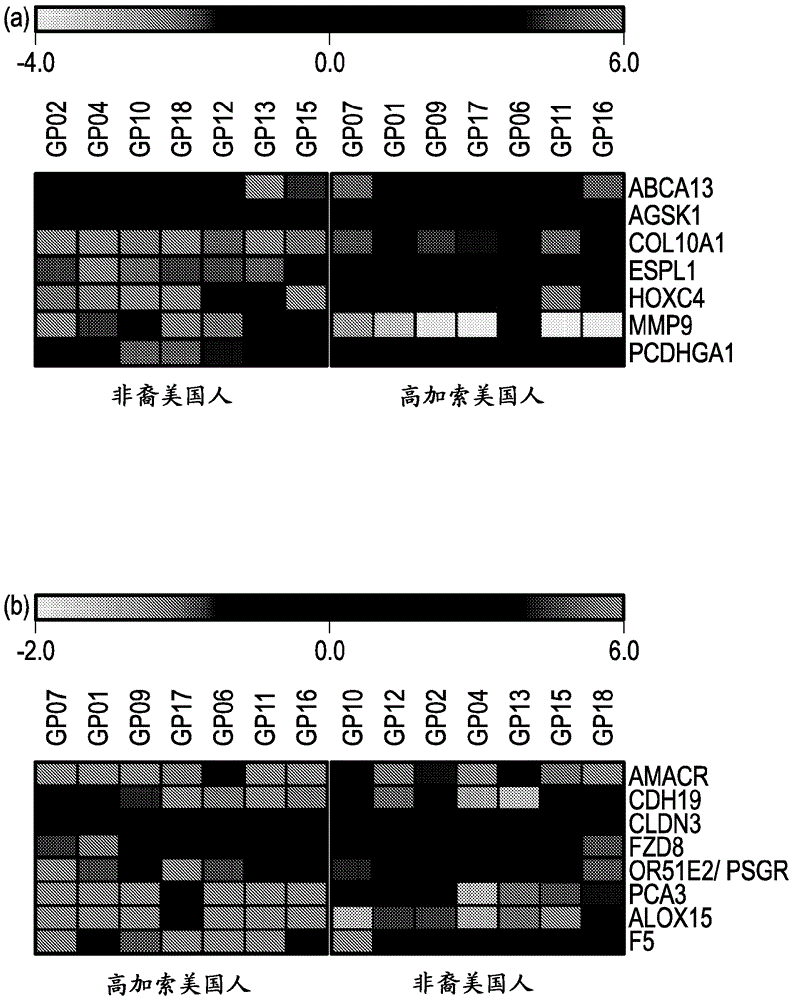
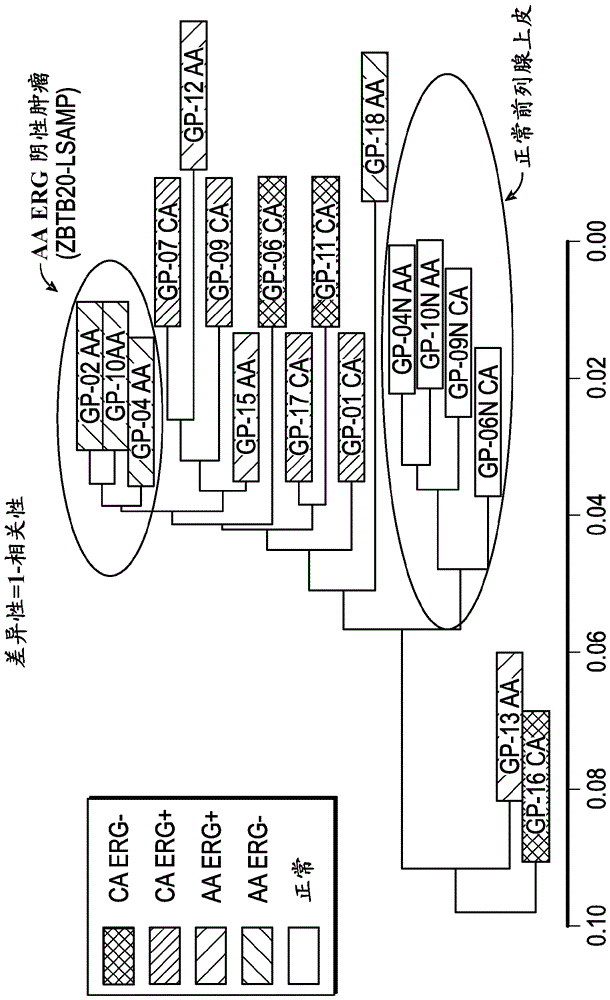
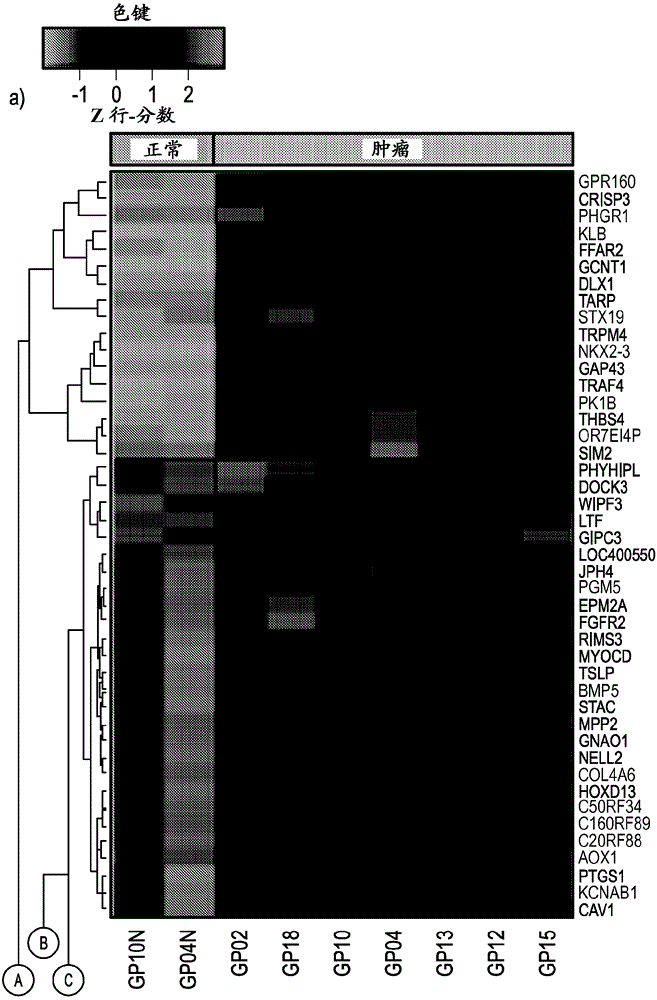
[0156] Example 2. Comparative RNA Analysis
[0157] To complement the genomic DNA analysis, RNA-Seq analysis was performed in the same cohort of prostate tumor samples. RNA-Seq technology has the ability to interrogate multiple aspects of the transcriptome including gene fusions, gene and transcript expression. Peripheral normal tissues were collected from 4 of 14 patients. 2 of the normal tissue samples were from AA males and 2 from CA males.
[0158] RNA samples were shipped to Expression Analysis (Durham, NC) for transcriptome sequencing. The details of sequencing statistics are as follows: sequencing type: paired-end, average read length for each sample: 50 nt, average read quality: 37, number of reads: approximately 31 million per sample.
[0159] Raw reads for expression analysis of each sample were obtained in fastq format. These files contain all sequences that pass Illumina's purity filters and per-base quality scores defined by the Illumina phred metric. These r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com