Halogenase gene of streptomyces xinghaiensis and product thereof, biosynthesis cluster of product modified by halogenase gene
A technology of marine streptomyces and halogenase, which is applied in the field of genetic engineering, can solve the problems of difficulty, many chemical synthesis steps, poor solubility of complestatin, etc., and achieve the effect of low copy number and avoiding preference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Isolation of the halogenase gene of Streptomyces marinum and the acquisition of its modified product gene cluster.
[0033] Extraction of marine Streptomyces genomic DNA:
[0034] Use TBS medium (in g / L, tryptone 17, soybean peptone 3, sodium chloride 3, glucose 2.5, dipotassium hydrogen phosphate 2.5, pH 7.5) at 28°C, 150rpm for 48 hours. Collect more than 10 ml of actinomycete cells, resuspend the cells in 5 ml of SET buffer, add an appropriate amount of 0.1 mm glass beads for vortexing. Digest with 1ml of 20mg / ml lysozyme at 37°C for more than 2h, then add 500 μl of 15mg / ml proteinase K, and react at 37°C for 30min. Then add 1 / 10 volume of 20% SDS solution and react at 55°C for 1h. Use 1 / 3 volume of 5 M NaCl solution, react at room temperature for 30 min, then add saturated phenol / chloroform at a volume ratio of 1:1, mix well, and react at room temperature for 30 min. Centrifuge at 8000 r / min at 4°C for 20 min, discard the protein precipitate, and add a...
Embodiment 2
[0058] Example 2: Analysis of sequencing results of gene clusters encoding Complestatin-like Compound in marine Streptomyces
[0059] Sequencing Results and Analysis of Halogenase from Streptomyces marinum
[0060] Halogenase gene sequence
[0061] > sin H SEQ ID NO. 1
[0062] ATGACCCGCCGGGTGACAAGGGGAGGAGGGATGGCCTTGCCGGATTCCGAGGAATTCGATGTGGTGGTCGTCGGTGGAGGGCCCGCCGGATCGACGCTGGCCGCGTTGACGGCCATGCAGGGACACCGGGTGCTGGTCCTGGAGAAGGAGTTCTTCCCCCGTCACCAGATCGGGGAGTCGCTCCTGCCGGCCACCGTGCACGGCGTGTGCCGGCTGACCGGCGTGGCGGACGAGCTCGCCGCCGCGGGCTTCCCGCGCAAGCGCGGCGGCACGTTCAAGTGGGGCGCCAACCCCGAGCCGTGGACCTTCTCCTTCTCCGTCTCCCCGCGCATGACCGGGCCGACGTCCTACGCCTACCAGGTCGAGCGGGCCAAGTTCGACGAGATCCTGCTCAACAACGCCCGCCGGGTGGGCGCCGAGGTGCGCGAGGGCTGTGCCGCCGTCGACGTCGTCGAGGACGGGGAGCGGGTCCGGGGCGTCCGGTACACCGACGCCGACGGCCGCGAGCACCGGGCGTCGGCCACGTTCGTCGTGGACGCCTCCGGCAACGGAAGCCGGCTGTACCGGCGGGTGGGCGGAACCCGGGAGTACTCGGAGTTCTTCCGCAGCCTGGCCCTGTACGGCTACTTCGAGGGCGGCAAGCGGCTGCCGGAACCGAACTCGGGCAACATCCTGTCGGTGGCGTTCGAGAGCGGCTGGTTCTGGTACA...
Embodiment 3
[0071] The gene and product thereof of embodiment 2 are detected and analyzed as follows:
[0072] (1) Sequence analysis of the halogenase-positive plasmid of the marine Streptomyces fosmid library:
[0073] Combining the halogenase-positive Fosmid plasmid sequencing results with the 454 and Solexa high-throughput sequencing of the aforementioned marine Streptomyces, the sequence was analyzed, and the DNA sequence analysis open reading frame (ORF) analysis was provided by the National Center for Bioinformatics of the United States. The ORF Finder function of the NLM (http: / / www.ncbi.nlm.nih.gov / gorf / gorf.html). The DNA homology analysis was compared with various DNA databases using the worldwide Blast engine (http: / / www.ncbi.nlm.nih.gov / BLAST / ) provided by the National Center for Bioinformatics of the United States.
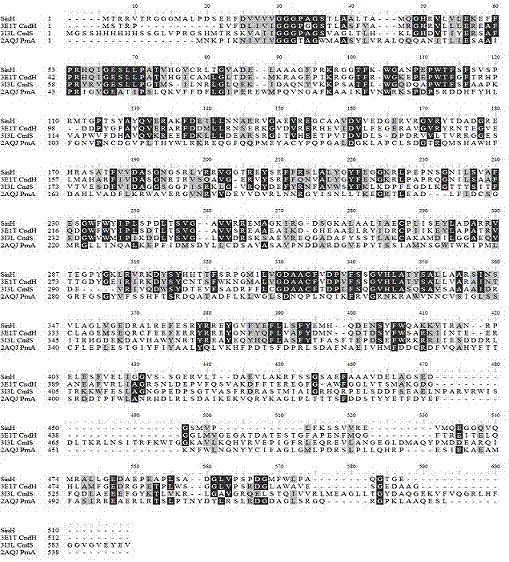
[0074] After annotating the 42.81kB sequencing information (Table 2) and S. lavendulae Complestatin gene clusters in the comparison ( image 3 ), it can be p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com