Construction method of single-cell or trace sample full-length transcriptome library for nanopore sequencing
A technology for nanopore sequencing and library construction, applied in chemical libraries, biochemical equipment and methods, combinatorial chemistry, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
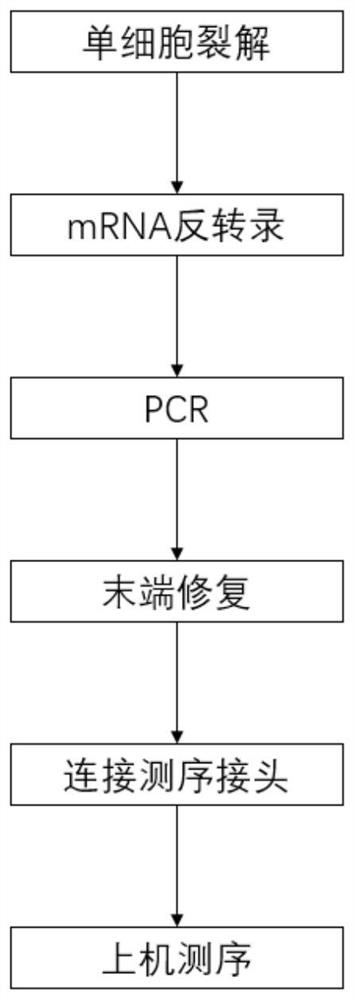
[0046] This embodiment provides a method for constructing a full-length transcriptome library of a single cell or micro sample for nanopore sequencing (the basic process is as follows: figure 1 shown), including the following steps:
[0047] 1. Single cell lysis
[0048] Prepare 10×Reaction Buffer according to Table 1, add it to a single-cell tube, flick to mix, and centrifuge immediately (the lysis buffer contains detergent, bubbles must be avoided when flicking and mixing), and lyse at room temperature for 5 minutes.
[0049] Table 1 Recipe of single cell lysis reaction buffer
[0050] components Dosage 10× Lysis Buffer 19μL RNase inhibitor 1μL total capacity 20 μL
[0051] 2. Reverse transcription
[0052] (1) Take a 0.2mL centrifuge tube and prepare reverse transcription reagents according to Table 2.
[0053] Table 2 RT Reaction System Recipe (1)
[0054]
[0055]
[0056] (2) Put the sample on ice, and add 2 μL of 3’SMART-S...
Embodiment 2
[0116] This embodiment provides a method for constructing a full-length transcriptome library of a single cell or micro sample for nanopore sequencing (the basic process is as follows: figure 1 shown), the difference from the method in Example 1 is that the end repair reaction condition in step 5 is to first add 2 μL of FFPE Master Mix and 3.5 μL of FFPE buffer to the PCR purified product, react at 20°C for 15min, and then Add 2 μL of Endprep Master Mix, 3.5 μL of Endprep buffer and water to the reaction system, react at 20°C for 30 minutes, and at 65°C for 10 minutes. The linker ligation reaction condition in step 7 is 22°C for 1.5 hours.
experiment example
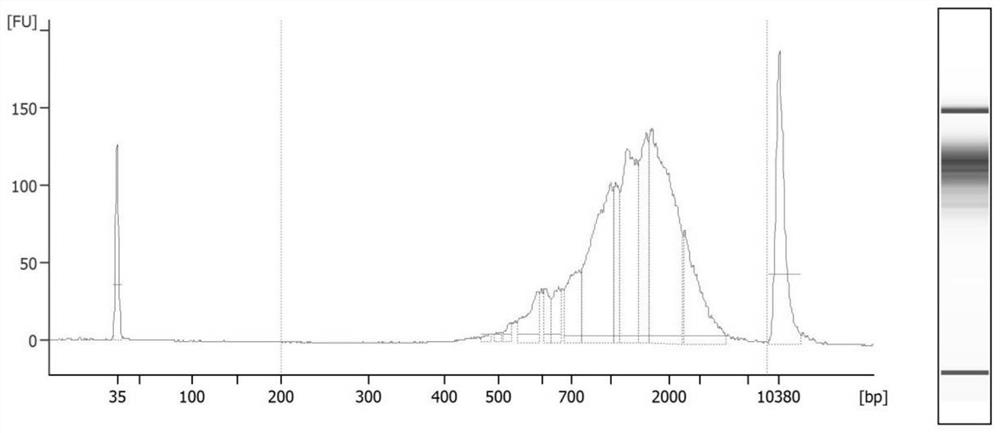
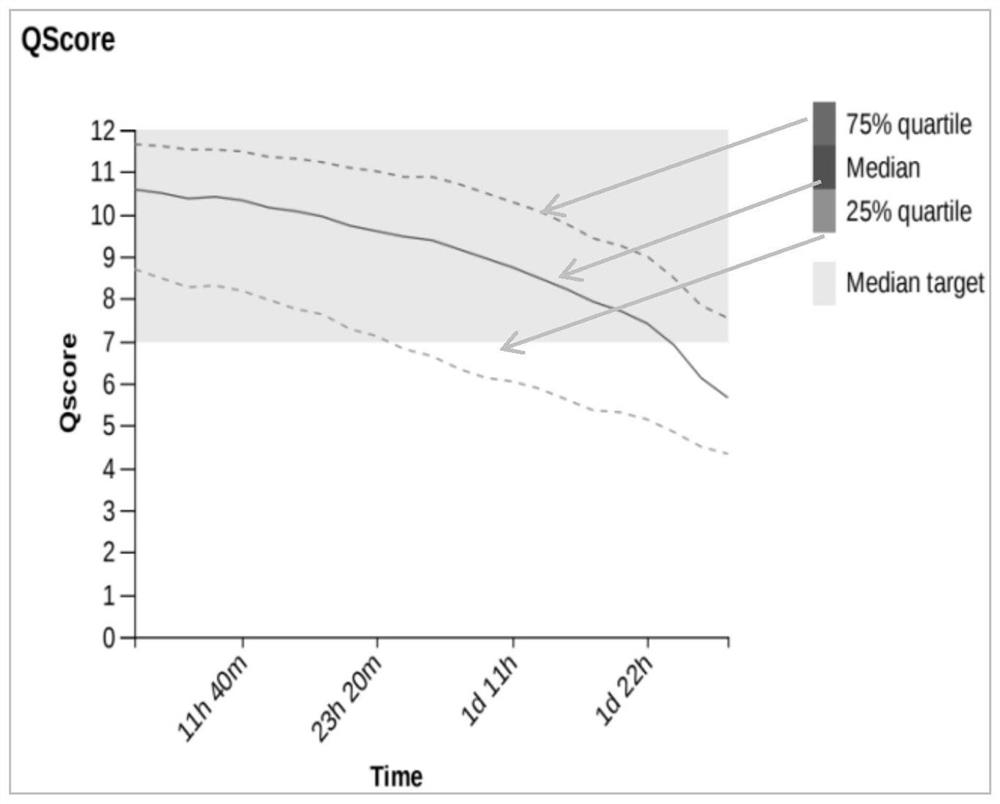
[0120] The full-length transcriptome library construction method for single-cell or micro-sample for nanopore sequencing in Examples 1 and 2 and the comparative example is used to construct a full-length transcriptome library for a single-cell sample, wherein the library sequencing quality and library sequencing pass the pore The speed test results are as follows image 3 , Figure 4 , Figure 5 , Figure 6 , Figure 7 and Figure 8 As shown, the off-machine results of the measured data obtained from the two samples are shown in Table 9, and the statistical results of the full-length sequence of the measured transcripts obtained are shown in Table 10. The results show that the sequencing effects of Example 1 (N01, N02) and Example 2 (N03, N04) are equivalent, and compared with the comparative example (N05, N06), the library quality of Example 1 and 2 is significantly improved ( image 3 , Figure 5 and Figure 7 ), the hole speed is relatively stable in a short time ( ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com