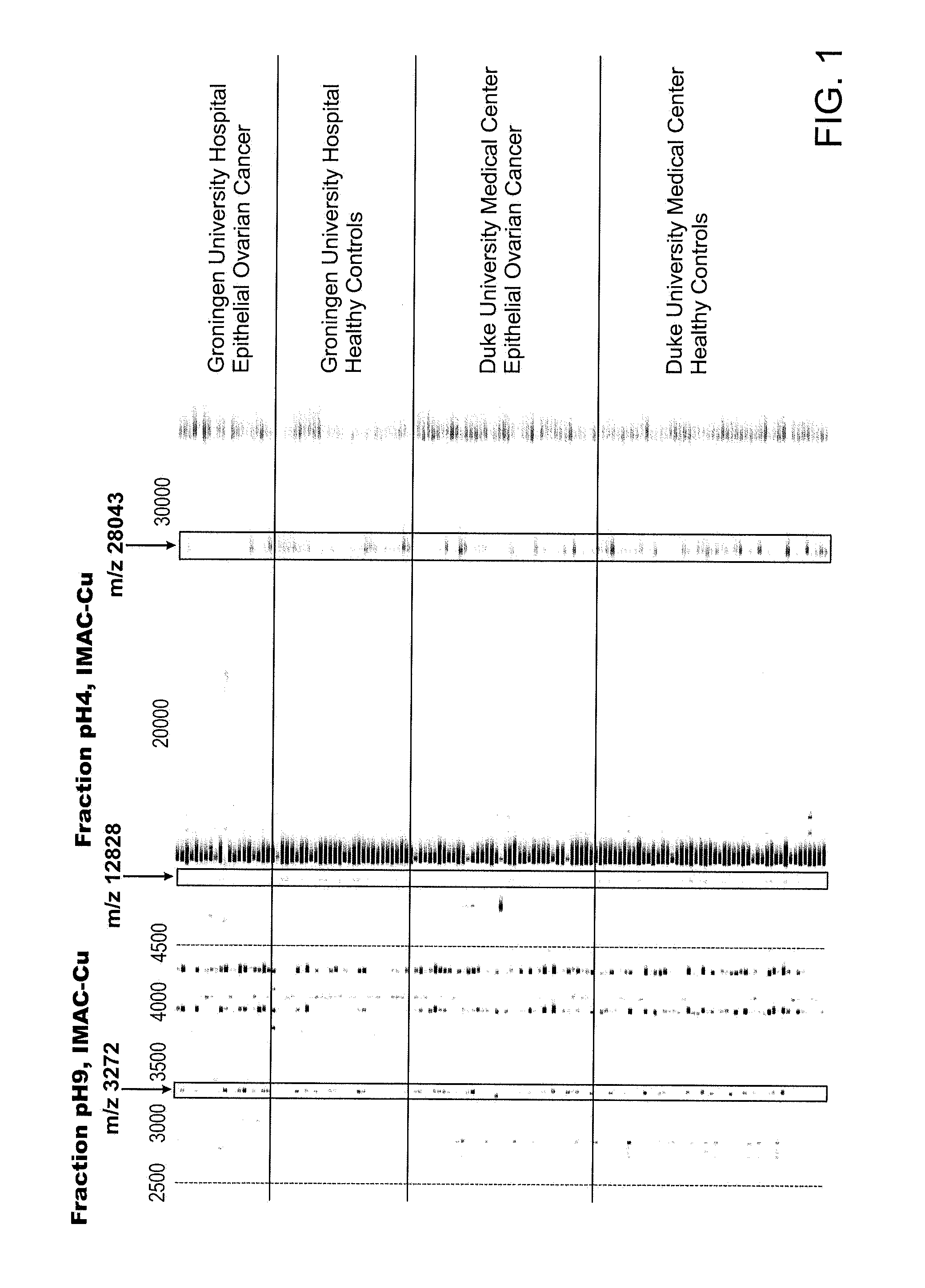
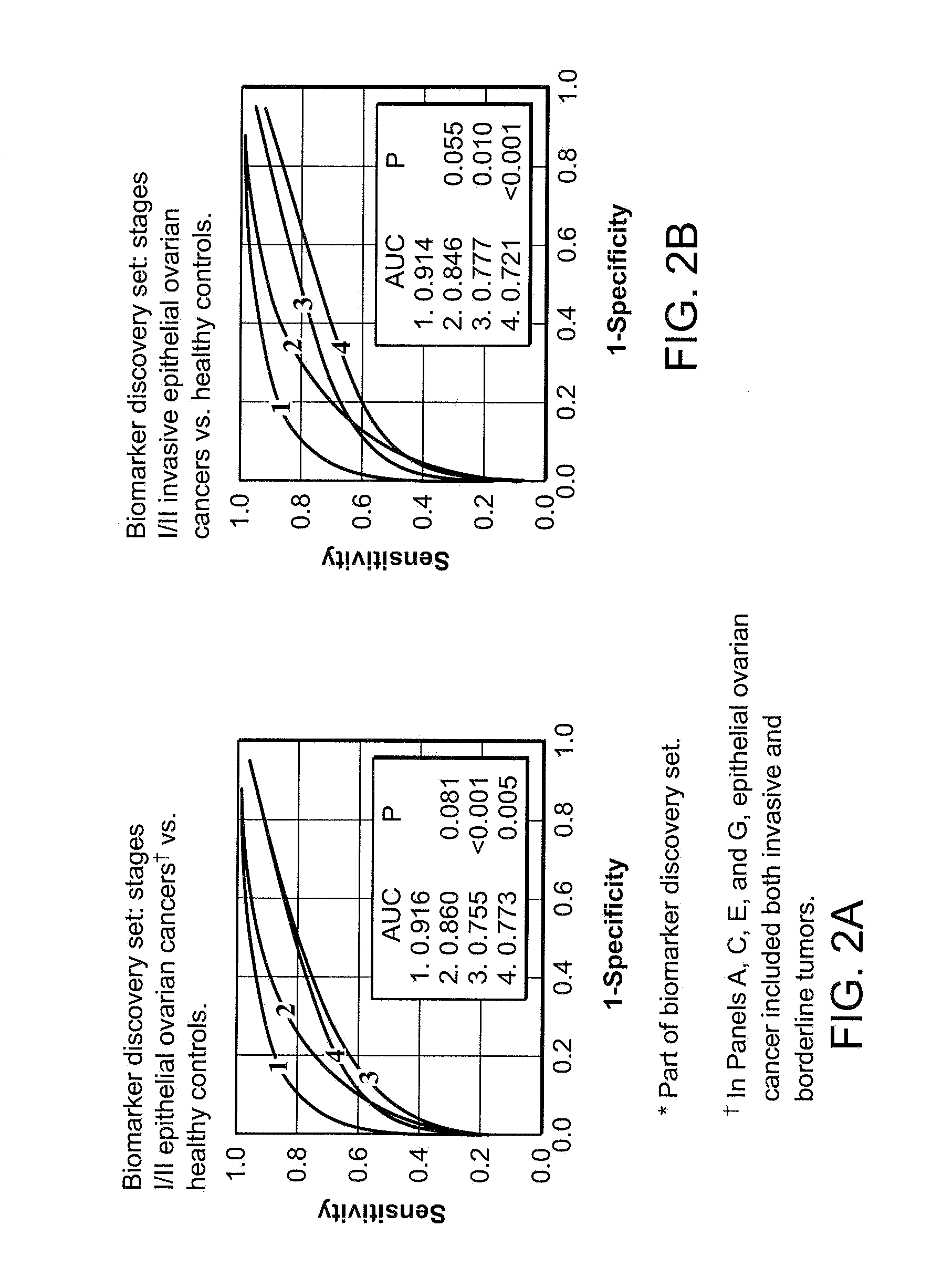
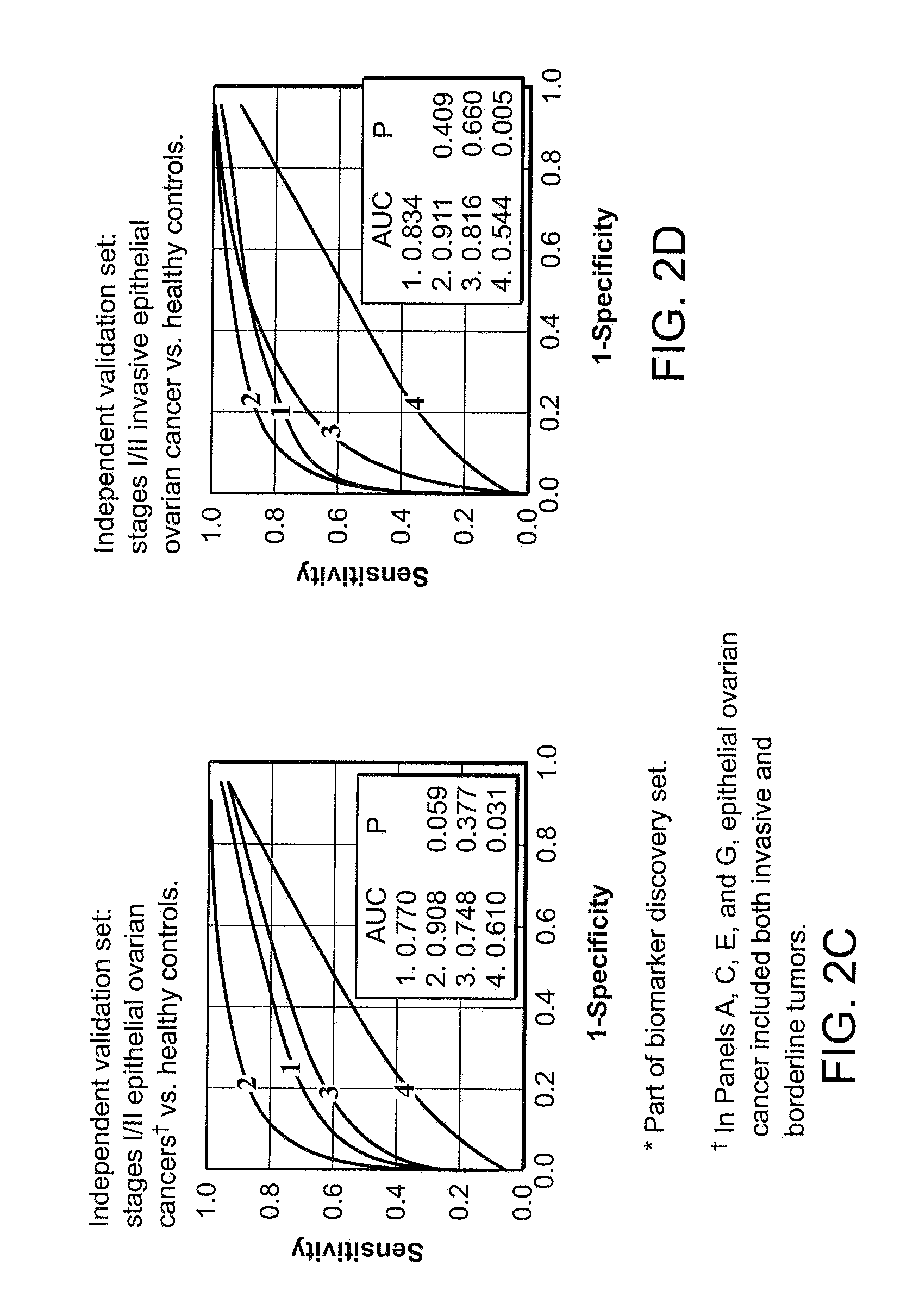
Use of biomarkers for detecting ovarian cancer
a biomarker and ovarian cancer technology, applied in the direction of enzymology, peptides, drug compositions, etc., can solve the problems of low sensitivity and specificity of population-based screening tools for early detection and diagnosis of ovarian cancer, and neither technique has sufficient sensitivity and specificity to be applied to the general population, so as to limit the number of markers detected
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Protein Expression Profiling
[0192]Serum fractionation: Serum samples were thawed on ice and then centrifuged at 20000 g for 10 minutes to remove precipitate. 20 μl of serum were mixed with 30 μl of a denaturing buffer (U9: 9 M urea, 2% CHAPS, 50 mM Tris pH 9.0) and vortexed for twenty minutes at 4 degrees. For each sample, 180 μl of Hyper Q DF anion exchange resin was equilibrated in 200 μl of U1 buffer (U9 that was diluted 1:9 in 50 mM Tris pH 9.0) three times. The denatured serum was applied to the resin and allowed to bind for thirty minutes. The unbound material was collected and then 100 μl of 50 mM Tris 9.0 containing 0.1% OGP was added to the resin. This wash was collected and combined with the unbound material (flow through; fraction 1). Fractions were then collected in a stepwise pH gradient using two times 100 ul each aliquots of wash buffers at pH 7, 5, 4, 3, and organic solvent). This led to the collection of a total of six fractions. Fractionation was performed on a Bio...
example 2
[0203]Biomarker discovery: Qualified mass peaks (S / N>5, cluster mass window at 0.3%) within the mass range of M / Z 2 kD-50 kD were selected from the SELDI spectra. In order to obtain a more consistent level of data variance across the range of spectrum of interest, logarithmic transformation was applied to the peak intensity prior to further analysis. The peak intensity data of early stage epithelial ovarian cancer patients and healthy controls from Duke University Medical Center (Ca n=36, HC n=47) and Groningen University Hospital (Ca n=20, HC n=30) were analyzed using the Unified Maximum Separability Analysis (UMSA) algorithm that was first used for microarray data analysis and subsequently for protein expression data (ProPeak, 3Z Informatics). ((Li J, et al., Clin Chem 2002; 48:1296-304; Rai A J, et al., Zhang Z, et al. Arch Pathol Lab Med 2002; 126:1518-26; Zhang Z, et al., Applying classification separability analysis to microarray data. In: Lin S M, Johnson ...
example 3
Purification of Biomarkers
[0205]For all markers, serum was initially fractionated using the anion exchange protocol used for the protein expression profiling. For each purification step, fractions were monitored either on NP20 or IMAC-copper ProteinChip arrays.
Purification of the 28 kD marker: 1 ml of the pH 4 fraction from the anion exchange separation was added to 500 ul of RPC PolyBio 10-15 (Biosepra) and incubated at 40 C for 1 hour. Fractions containing increasing amounts of acetonitrile with 0.1% trifluoroacetic acid were collected. The 75% acetonitrile / 0.1% trifluoracetic acid fraction was dried down by speed-vac and rehydrated in 100 ul SDS-tricine sample loading buffer without DTT. 40 ul sample was loaded onto 16% tricine gel and run at 100 mV for 4 hrs. The gel was destained with colloid blue kit (Pierce) and the 28 kDa was excised.
Purification of the 12.8 kDa marker: 10 ml of the pH 4 fraction from the anion exchange separation was adjusted to pH 7.5 with 1 M Tris HCl, pH...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com