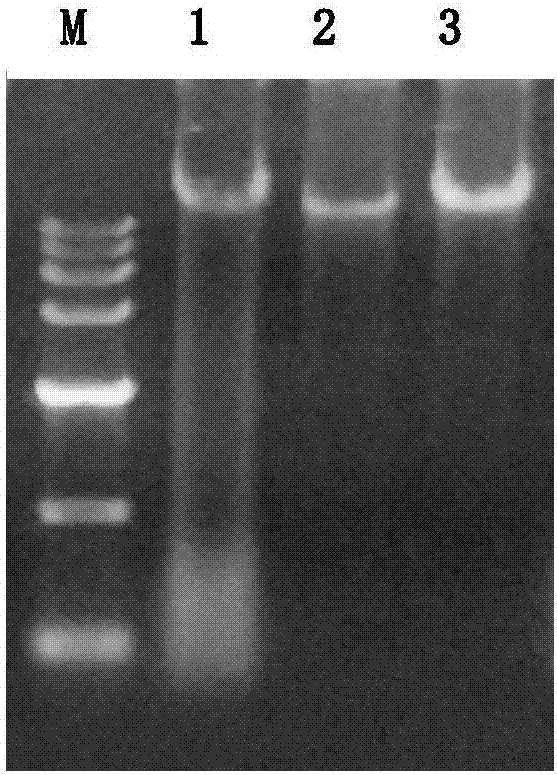
Extraction method for soil microorganism metagenome DNA and corresponding kit
A soil microorganism and metagenome technology, applied in the field of biological genomic DNA extraction, can solve problems such as low extraction efficiency and inability to meet sequencing requirements, and achieve the effects of improving extraction rate, strong practicability and good purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] 1. Reagent configuration
[0037] (1) Buffer A: 0.1M EDTA, 0.1M Tris-HCl, 1.5M NaCl, 0.1M NaH 2 PO 4 and 0.1MNa 2 HPO 4 , the pH was adjusted to 8.0, the volume was adjusted to 1L with deionized water, and sterilized under high temperature and high pressure.
[0038] (2) 50 mg / ml lysozyme: 20 mM Tris-HCl, 2 mM EDTA, Triton x-100 with a volume fraction of 1.2%.
[0039] (3) 10wt% CTAB: weigh 1g of CTAB powder, and dilute to 100ml with sterile water (requires heating in a 65°C water bath to dissolve).
[0040] (4) Buffer B: Weigh 0.5g of CTAB, and dilute to 50ml with buffer A (heating in a water bath at 65°C is required to dissolve).
[0041] (5) 20% SDS: Weigh 10 g of SDS, and dilute to 50 ml with sterile water (heating in a water bath at 68° C. to dissolve, and adjust the pH to 7.2).
[0042] (6) Phenol: chloroform: isoamyl alcohol (25:24:1).
[0043] (7) Chloroform: isoamyl alcohol (24:1).
[0044] (8) Precooling with isopropanol (4°C), 70% ethanol.
[0045] (...
Embodiment 2
[0067] Example 2 Using the SDS method to extract the sandy soil metagenome
[0068] (1) Take 15g of sand in a 50ml centrifuge tube, add 30ml of buffer A, vortex and mix, and bathe in 55°C water for 20min;
[0069] (2) 9000g, centrifuge for 10min, discard the supernatant;
[0070] (3) Add 10ml of buffer A to the centrifuge tube, and use the freeze-thaw method to disrupt the cells: -80°C for 15 minutes—65°C for 5 minutes (repeat 3 times, and do not need to invert and mix during the 65°C warm bath).
[0071] (4) Cool at room temperature (~10min is enough), add 1ml lysozyme (10mg / ml) to the centrifuge tube (final concentration is about 0.5mg / ml), and bathe in 37℃ water for 1h (during this period, gently invert and mix well every 10min) ).
[0072] (5) Add 0.5ml 10% SDS (final concentration is about 0.5% SDS) and 20μl proteinase K (final concentration is about 100μg / ml), bathe in 58℃ water for 1h (mix up and down every 10min, and pre-cool at 4℃ for the last 20min centrifuge).
...
Embodiment 3
[0086] Example 3 Using centrifugal collection method to extract sandy soil metagenome
[0087] (1) Weigh 15g of sandy soil sample into a grinding bowl pre-cooled by liquid nitrogen.
[0088] (2) Add liquid nitrogen to grind, continue to add liquid nitrogen to grind before the sample is dissolved, repeat adding three times and grind three times
[0089] (3) Transfer all samples to 50-ml Falcon tubes and add 16.5ml DNA extraction solution. Mix well (measure the pH of the supernatant if it is not 8, adjust to 8.
[0090] (4) Add 40 μl proteinase K (20mg / ml), mix gently by inversion, 37°C, 30min.
[0091] (5) 65°C, +1.83ml 20% SDS, mix well, and incubate for 1-2 hours. Mix every 20 minutes.
[0092] (6) 7000rpm, 15min, collect the supernatant into a new Fecon tube, and store it at 4°C.
[0093] (7) Add 6ml of extract solution, add 0.67ml of 20% SDS, mix well, 65°C, 15min (extract the remaining precipitated sample again).
[0094] (8) 7000rpm for 15min, and mix the supernatan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com