Method for improving cellulase expression capability of trichoderma reesei by regulating and controlling cell metabolism
A technology of Trichoderma reesei and cellulase, applied in the field of genetic engineering, to achieve the effect of improving cellulase enzyme activity and protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
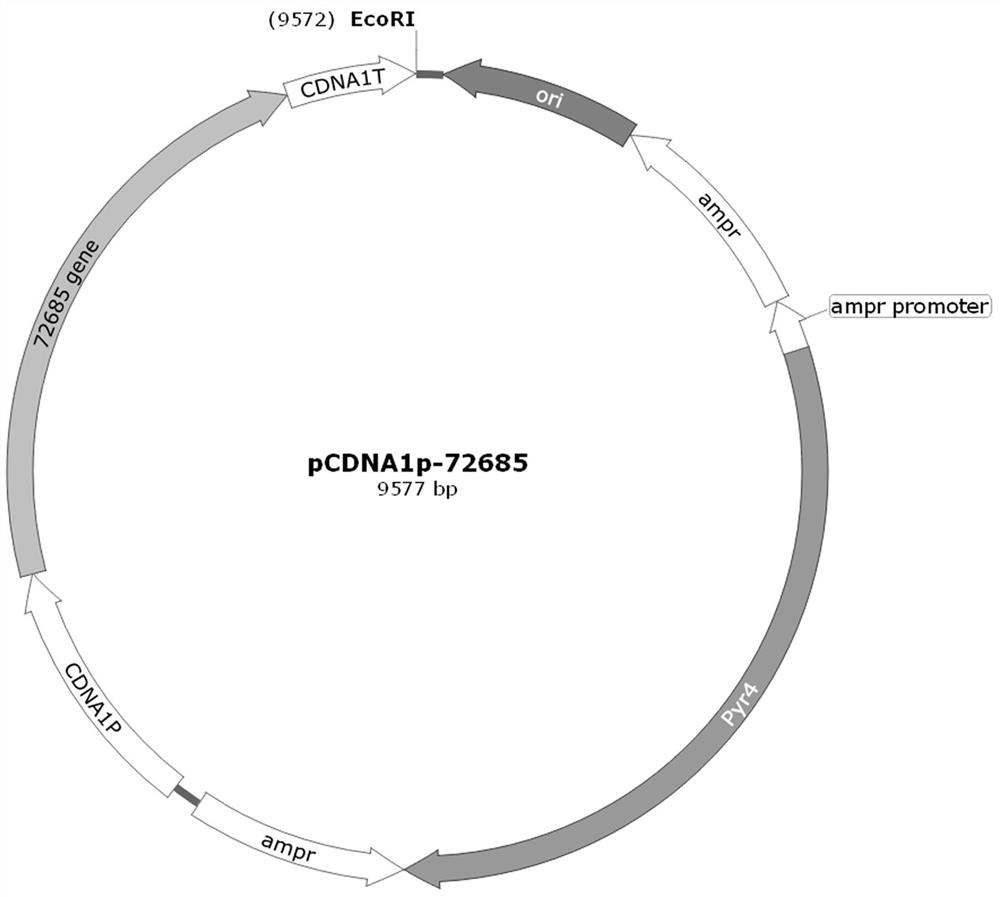
[0015] Example 1. Overexpression of regulatory genes 72685 Plasmid construction
[0016] Build overexpression 72685 Plasmid: including cDNA1 promoter, 72685 gene, cDNA1 terminator and pyr4 Expression cassette plasmid. Using the APA-GOD plasmid as a template, the APA part was amplified ( AmpR-pyr4-AmpR ), the APA sequence ( AmpR-pyr4-AmpR ) contains the Trichoderma reesei auxotrophic selection marker pyr4 An expression cassette that can be used for lack of pyr4 Genetic transformation screening of Trichoderma reesei strains. exist pyr4 There are two tandem repeated ampicillin genes at both ends of the expression cassette, and the homologous recombination of this direct tandem sequence can be used to convert the Trichoderma reesei transformant pyr4 The gene is knocked out more rapidly, so that pyr4 Filter markers can be recycled. Using the Trichoderma reesei genome as a template to amplify the cDNA1 promoter, 72685 The gene (named as cDNA) and cDNA1 terminator, elec...
Embodiment 2
[0017] Example 2. Overexpression 72685 Plasmid introduction
[0018] (1) Overexpression 72685 Plasmid transformation of Trichoderma reesei SUS4-2
[0019] Trichoderma reesei SUS4-2 was inoculated on a potato medium (PDA) plate, and cultured statically at 28 ºC for 7 days until it produced spores, and the spores were scraped off and inoculated in 100 ml of PDB medium containing uracil, at 28 ºC , 160 rpm shaking culture overnight. Collect the germinated mycelium by filtering through a 200-mesh sieve, add 10 mg / ml cellulase and digest at 30°C for 2-3 hours. After collecting the protoplasts, the pCDNAP-72685 plasmid was cleaned with E wxya I digestion, transformation of Trichoderma reesei host cells.
[0020] (2) PCR verification of the introduction of the pCDNAP-72685 plasmid into the Trichoderma reesei genome
[0021] A single transformant was picked, inoculated on a 24-well plate containing MM-glucose medium, and cultured at 28°C for 5-7 days. Extract genomic DNA and v...
Embodiment 3
[0022] Example 3. Overexpression 72685 Effects on Secretion and Expression of Trichoderma reesei Cellulase
[0023] (1) Overexpression 72685 Shake flask induction of genetic transformants
[0024] Overexpression 72685 Gene transformants and starting strains were inoculated with 2×10 7 The spores were cultured in 50 ml MM-glucose medium at 28°C and 160 rpm for 2 days. Transfer to 50 ml MM+2% Avicel medium with 10% inoculum size to induce the expression of cellulase. From the third day, samples were taken every 24 h, and the samples were taken continuously for 7 days.
[0025] (2) Overexpression 72685 Determination of the protein concentration and cellulase of the transformant of the gene
[0026] Coomassie Brilliant Blue method was used for protein quantification. After adding 250 L of 1 × dye reagent and 10 µl of protein standard, reacted at room temperature for 10 minutes, and measured the absorbance at 595 nm. The results are shown in figure 2 . visible overexpress...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com