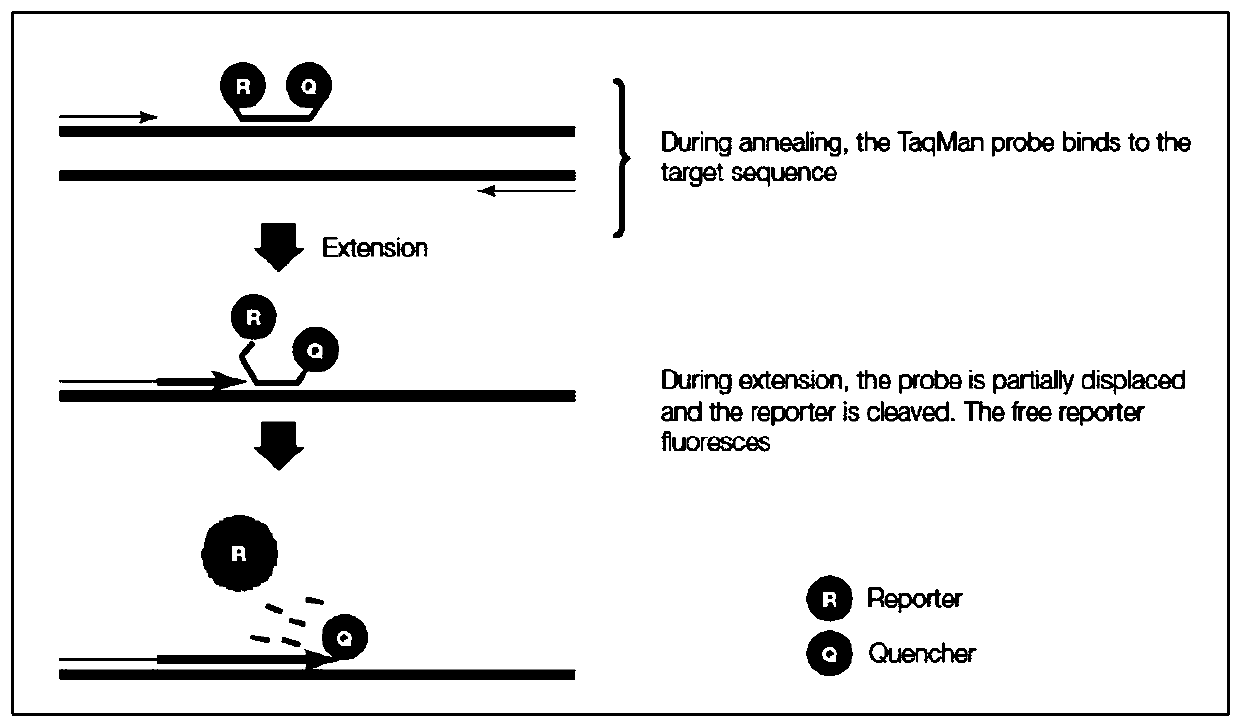
Quantitative detection method of bovine genome DNA in bovine hemoglobin product and application of quantitative detection method
A hemoglobin and detection method technology, applied in the field of molecular biology, can solve the problems of inability to realize quantitative analysis of genomic DNA, inability to detect, lack of sufficient consideration of recovery rate and linear range of detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0214] Example 1 Extraction of Bovine Genomic DNA for Control
[0215] Weigh 2g of beef, cut it into small pieces, grind it with liquid nitrogen, and weigh about 1g of the powder. The lysate (10mM Tris-Cl, 0.1M EDTA, 0.5% SDS, pH8.0) was added to the sample, the final concentration of proteinase K was 100 μg / mL, and it was lysed in a water bath at 50°C for 3 hours, with gentle shaking during the period. Add an equal volume of extract (phenol:chloroform 1:1) and mix gently, 12000rpm, 4°C, 15min. Gently transfer the upper aqueous phase to a new 50mL centrifuge tube. If the extraction effect is not good, repeat the extraction depending on the situation. Add 1 / 10 volume of 3M NaAc and an equal volume of isopropanol to the transfer water phase, mix gently and then precipitate at -20°C for more than 30min, centrifuge at 12000rpm, 4°C for 10min, and discard the supernatant. Add 20 mL of 70% ethanol to the pellet, mix gently, and wash. 12000rpm, 4°C, 10min, discard the supernatant...
Embodiment 2
[0218] Example 2 Preparation of positive control plasmid
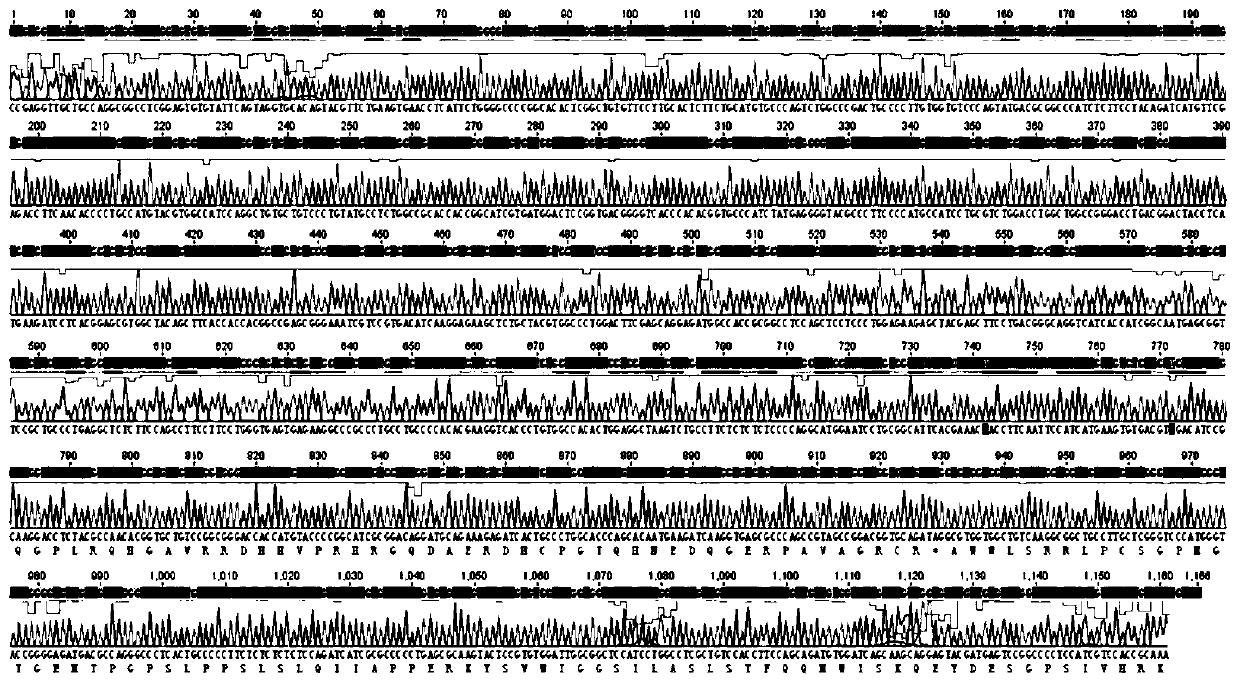
[0219] According to the literature, commonly used detection target genes include conserved single-copy genes such as ACTB and GAPDH. After sequence analysis and literature research, the ACTB gene fragment was selected as the detection object, and a pair of primers were designed to be used as detection fragments after sequence analysis. Part of the amplification product is covered, and the primers used in the amplification are Bov-ACTB-F (5'-CCGAGGTTGCTGCCAGGCGG-3') and Bov-ACTB-R (5'-AAGCATTTGCGGTGGACGAT-3') to extract bovine Genomic DNA was used as a template, PCR was carried out, and the amplified product was 1166bps. The amplified product was connected to the pUC19 plasmid vector (purchased from Beijing Huada Protein Research and Development Center Co., Ltd.), and the plasmid was sequenced and analyzed. The results are shown in image 3 , there are polymorphisms in individual sites other than the target amplificatio...
Embodiment 3
[0220] Example 3 Design and optimization of primers and probes
[0221] 1. Primer and probe design
[0222] According to the bovine genome sequence, the ACTB gene was selected as the detection target gene, and the bovine ACTB gene sequence was designed (Table 2) for the use of Taqman probe qPCR to detect DNA residues in samples. Three positions of the gene fragment and corresponding primers and probes were initially designed for detection (SEQ ID NO: 2-SEQ ID NO: 10). After all primers were designed, the primers and probes were calculated by the primer design software Primer premier 6. Annealing temperature to ensure the absence of obvious primer-dimers and complex secondary structures. After that, through the online analysis tool BLAST (Basic Local Alignment Search Tool, https: / / blast.ncbi.nlm.nih.gov / Blast.cgi) to confirm the specificity with the template sequence.
[0223] Table 2 Taqman probe qPCR primers and probe sequences
[0224]
[0225] 2. Optimization of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com