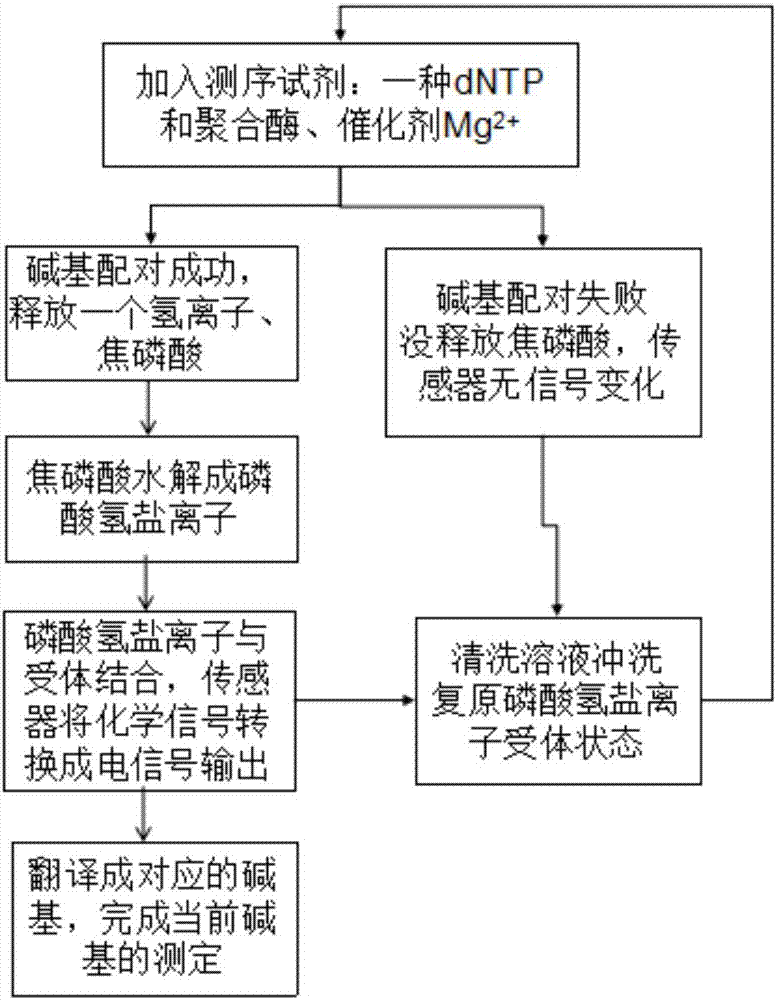
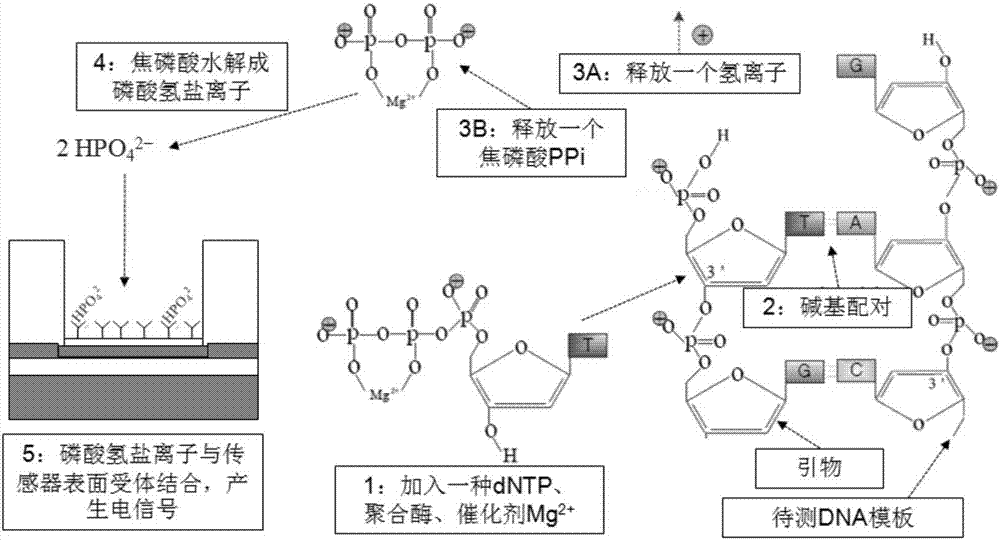
Gene sequencing method realized by detecting pyrophosphoric acid charges
A technology of gene sequencing and pyrophosphoric acid, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of heavy workload, large amount of template consumption, and long sequencing time, so as to improve the accuracy rate and improve the sequencing throughput. Quantity, the effect of reducing the cost of sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0032] Such as figure 2 As shown, a traditional metal gate ion-sensitive field effect transistor is used to realize the gene sequencing method of the present invention; the transistor is arranged on the base, including a sensor gate plate, and the sensor gate plate passes through a metal layer and a gate dielectric material The silicon channel is connected under the gate dielectric material, and the two ends of the silicon channel are respectively connected to the source and the drain. The sensor grid plate is also provided with a sensing dielectric layer; the sensing dielectric layer is fixed with a PPi receiver. A body, a solution cavity is arranged above the dielectric layer, and the dielectric layer and the PPi receptors on the dielectric layer are exposed to the solution in the cavity; the solution in the cavity contains a DNA template to be tested.
[0033] The base can be a silicon wafer, and the transistor is arranged on the upper layer of the base.
[0034] Further,...
example 2
[0044] Such as image 3 As shown, a silicon nanowire ion-sensitive field effect transistor is used to realize the gene sequencing method of the present invention; the transistor is arranged on the base, including a silicon nanowire, the surface of the nanowire is a dielectric layer, and the two ends of the nanowire are connected to the source The pole and the drain are connected; PPi receptors are fixed on the dielectric layer, a solution cavity is arranged above the nanoscale channel, and the channel and the PPi receptors on the channel are exposed to the solution in the cavity. The solution in the chamber contains the DNA template to be tested.
[0045] The base may include an upper layer and a lower layer, the upper layer is silicon dioxide, the lower layer is a silicon wafer, and the transistor is arranged on the upper layer of the base.
[0046] Further, one option is that a layer of gold film can be provided on the dielectric layer of the nanoscale channel, which can re...
example 3
[0050] Such as Figure 5 As shown, an interleaved double-electrode sensor is used to realize the gene sequencing method of the present invention. The preparation and sequencing steps of the DNA template sample were the same as in Example 1. The difference is that the electrical signal is sensed in a different way. The two electrodes on the chip form a staggered structure to maximize the capacitance between the electrodes. The capacitive surface can be directly immobilized with PPi receptors.
[0051] Further, a dielectric layer may be provided on the surface of the electrode, and a PPi acceptor may be provided on the surface of the dielectric layer.
[0052] When the PPi released when the receptor binds to DNA elongation is converted into hydrogen phosphate ions, the interelectrode capacitance changes and is detected.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com