Method for preparing bio-macromolecular monomolecular chips by virtue of high-density nano-dot arrays
A technology of biological macromolecules and nano-dot arrays is applied in the field of fabrication of high-density single-molecule chips of biological macromolecules to achieve the effect of simple fabrication process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
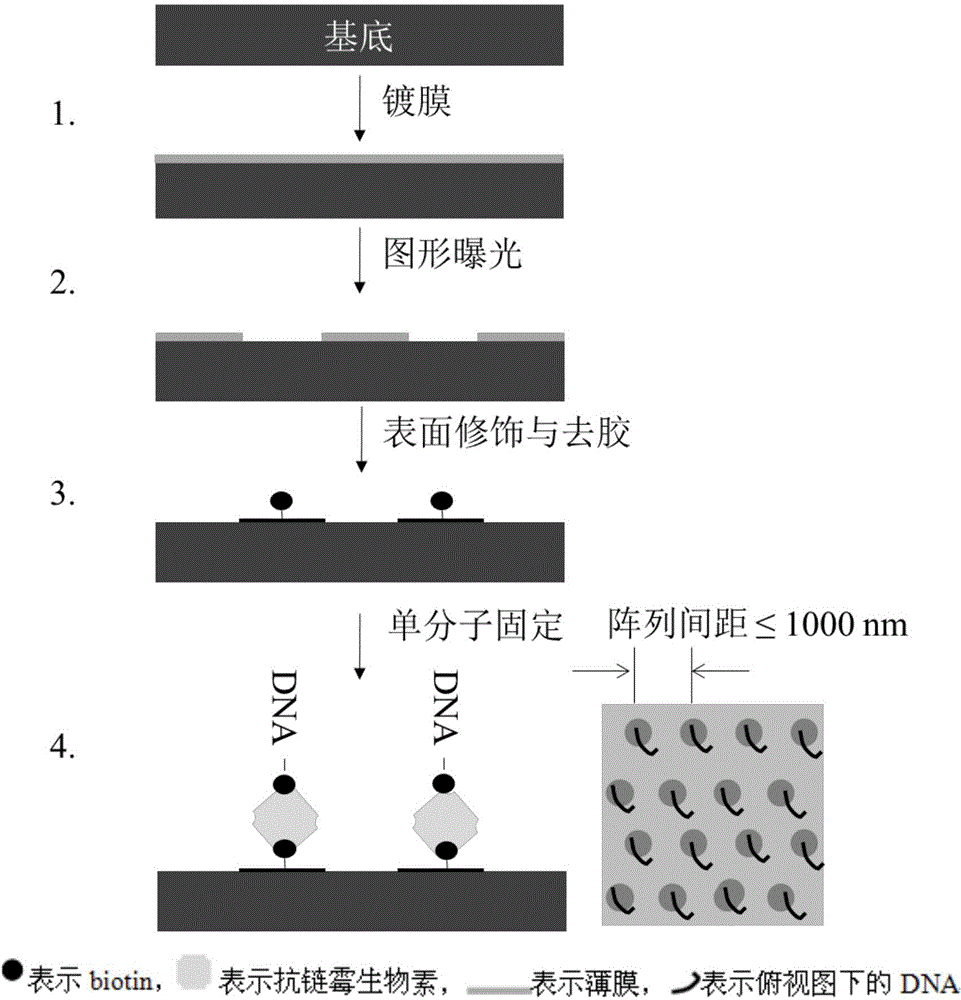
[0026] In the first step, a photoresist PMMA with a thickness of 10 nm was spin-coated on the silicon substrate after cleaning with Piranha solution and deionized water.
[0027] In the second step, a 30nm nanopit pattern was developed on the negative film by electron beam lithography (the diameter of the nanodot is controlled to accommodate only one nucleic acid loading tool), and the spacing of the nanopit is controlled at 500nm by mask design.
[0028] In the third step, the developed substrate is treated with plasma to hydroxylate the silicon surface; the substrate is washed with acetone to remove the PMMA film, and an array of hydroxylated nano-dots is obtained.
[0029] In the fourth step, the substrate is treated with an APTES solution to aminate the surface of the nano-dots, and after cleaning, it is treated with Biotin-NHS to selectively biotinylate the surface of the nano-dots.
[0030] The fifth step is to immobilize the tetramer anti-streptavidin-single DNA complex...
Embodiment 2
[0033] In the first step, the photoresist PMMA / MA with a thickness of 10nm is spin-coated on the existing glass mica substrate, so that it does not exceed the diameter of a subsequent single molecule.
[0034] In the second step, electron beam lithography is used and developed to realize a nano-dot array with a diameter of less than 20nm, and the spacing of the nano-pit is controlled at 200nm by mask plate design.
[0035] In the third step, the developed substrate is coated with O 2 The surface of the substrate was hydroxylated by plasma treatment; the substrate was washed with acetone to remove the PMMA film, and an array of hydroxylated nano-dots was obtained.
[0036] In the fourth step, the substrate is treated with an APTES solution to aminate the surface of the nano-dots, and after cleaning, it is treated with Biotin-NHS to selectively biotinylate the surface of the nano-dots.
[0037] The fifth step is to immobilize the tetrameric anti-streptavidin-single RNA complex ...
Embodiment 3
[0040] In the first step, a photoresist LIGA film with a thickness of 20 nm was spin-coated on the cleaned glass substrate.
[0041] In the second step, a pattern with a diameter of 20nm is developed on the film lithography by electron beam lithography technology, and the nanopit spacing is controlled at 500nm by mask plate design.
[0042] In the third step, the developed substrate is coated with O 2 The surface of the substrate was hydroxylated by plasma treatment, and the substrate was washed with acetone to remove the PMMA film to obtain a hydroxylated nanodot array.
[0043] In the fourth step, the substrate is treated with an APTES solution to aminate the surface of the nano-dots, and after cleaning, it is treated with Biotin-NHS to selectively biotinylate the surface of the nano-dots.
[0044] In the fifth step, the TNF-α monoclonal antibody complex bound to TNF-α (tumor necrosis factor) and labeled with biotin is immobilized on the surface of the nanodots. The diameter ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com