Method for detecting insertion flanking sequence and copying quantity of Tgf2 transposon in goldfish genome
A technology of flanking sequences and transposons, which is applied to the detection of Tgf2 transposon insertion flanking sequences and copy numbers in the goldfish genome, achieving high-efficiency and low-cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Extraction of fish genomic DNA with Tgf2 transposon insertion
[0041] Due to the insertion of Tgf2 transposon in the goldfish genome, take about 100-200 mg of caudal fin of 95% alcohol or live goldfish, cut it into pieces, and dry at 55°C for 5-6 minutes; add 410 μL of STE lysate (pH=8.10), 80 μL SDS (10%, pH=8.14) and 10 μL of proteinase K (20 μg / ml); turn over and shake well, and keep warm in a 56°C water bath for 14-15 hours; add 340 μL to 400 μL of saturated sodium chloride solution and mix well, shake gently 3min; add 340μL to 400μL of chloroform, shake well; centrifuge at 12000rpm for 20min at 4°C, transfer the supernatant to another clean centrifuge tube; add 450μL to 500μL of isopropanol (-20°C), and centrifuge at 12000rpm for 20min (4 ℃); pour off the supernatant, save the precipitate, add 800μL of 75% ethanol to wash; centrifuge at 12000rpm for 5min to 15min (4℃); to remove ethanol, dry the precipitate, add 100μL to 200μL of double distilled water...
Embodiment 2
[0043] Synthesis of embodiment 2 Splinkerette linker
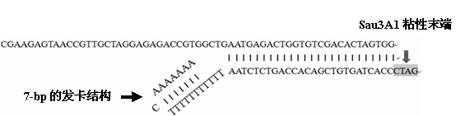
[0044] Entrusted Shanghai Shenggong Company to synthesize a long oligonucleotide sequence (SEQ ID No: 1) and a short oligonucleotide sequence (SEQ ID No: 2) required for the splinkerette adapter; The short sequence was dissolved in sterile double-distilled water to 50 μm / μl, and 50 μl of oligonucleotide long sequence and short sequence were mixed in equal volumes, incubated at 95°C for 5 minutes, and then gradually lowered to 4°C at 1°C / 15s on the PCR instrument , The long sequence and the short sequence can form a splinkerette joint during the annealing process, and store at -20°C. The synthetic splinkerette linker has the "GATC" cohesive end of restriction endonuclease Sau3A1 on one side, and a 7-bp hairpin structure formed by complementary AT base pairing in the short sequence to form a mismatch region on the other side figure 2 ).
Embodiment 3
[0045] Example 3 Digestion of Goldfish Genomic DNA and Ligation of Splinkerette Adapters
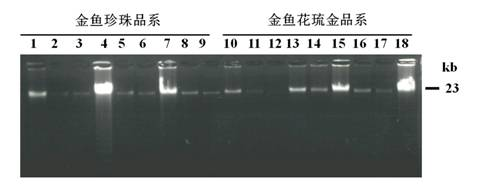
[0046] The goldfish genomic DNA containing the Tgf2 transposon extracted in Example 1 was digested with the restriction endonuclease Sau3A1 in a water bath at 37°C for 12 hours. The digestion reaction was carried out in a 30 μl system containing 10 μl Genomic DNA (2.5 μg), 3μl 10×Sau3A1 Restriction buffer, 4μl Sau3A1 enzyme (10U / μl), 0.3μl Acetylated BSA (10mg / ml), 12.7μl ddH 2 O. The digested products were subjected to 1.2% agarose gel electrophoresis, using TBE as a buffer, and electrophoresis at a voltage of 5V / cm to detect the effect of enzyme digestion ( image 3 ). The Sau3A1 digested DNA product of the complete goldfish genome was placed in a water bath at 65°C for 20 minutes to terminate the activity of the restriction endonuclease, and stored at -20°C for future use. Ligate 4.5 μl of the Sau3A1-digested DNA product and 1 μl of the splinkerette adapter with 4 μl of T4 DNA li...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com