Pseudomonas. chlororaphis subsp. Aurantiaca Pa40 and application thereof
A technology of orange subspecies and green needle false single, applied in the direction of application, chemicals for biological control, biocides, etc., to achieve the effect of high research and application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
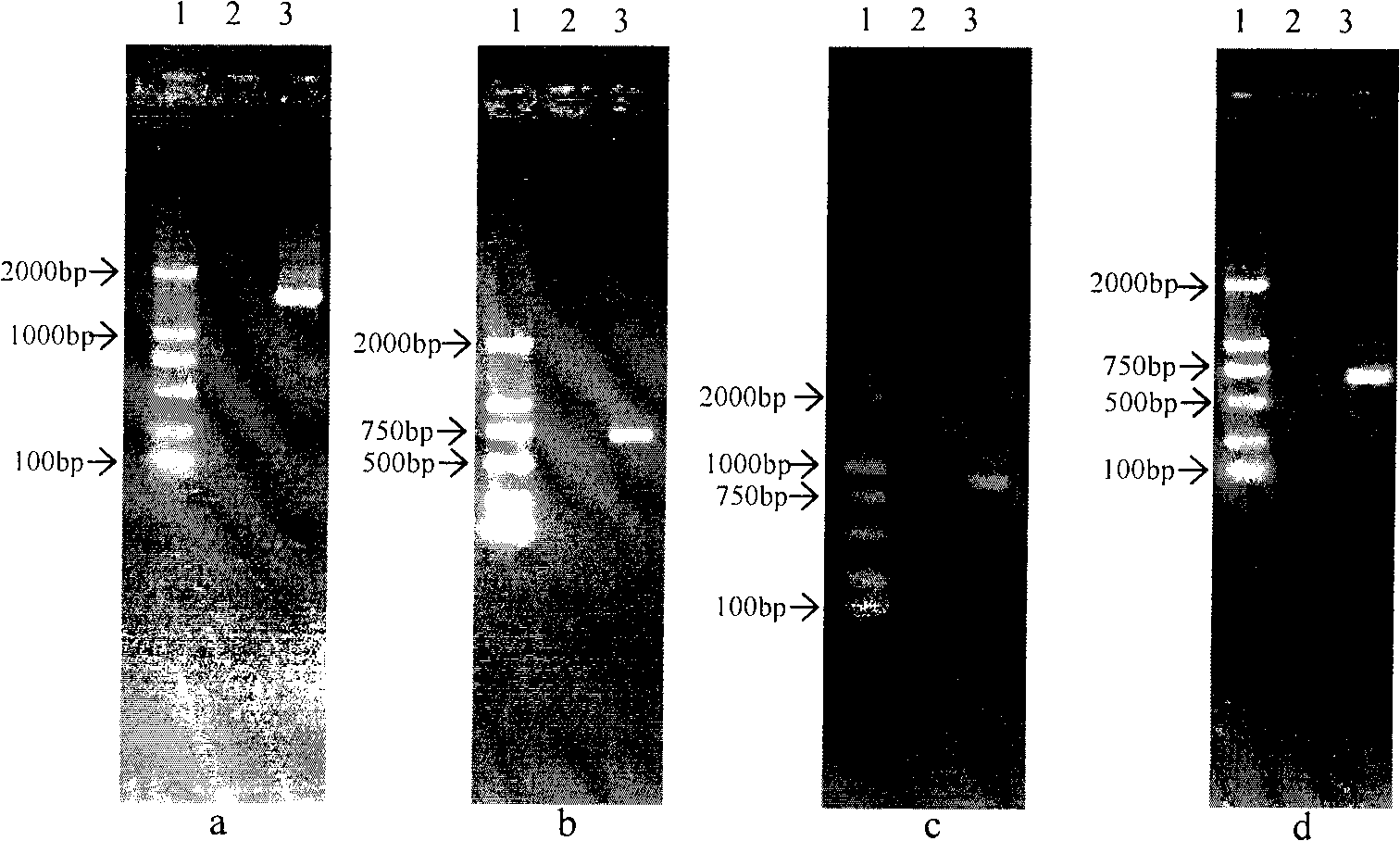
[0027] The identification of embodiment 1 Pa40
[0028] Based on the morphological characteristics, physiological and biochemical characteristics, 16S rDNA sequence, atpD gene sequence, recA gene sequence, carA gene sequence and BIOLOG carbon source metabolic profile of Pseudomonas, it was identified as Pseudomonas chloropinus subsp. Pseudomonas. chlorophis subsp. aurantiaca. The specific identification results are as follows:
[0029] 1. Morphological characteristics of bacteria
[0030] Gram staining was negative, electron microscope observation, bacteria oblong.
[0031] 2. Physiological and biochemical characteristics
[0032] The physiological and biochemical characteristics of Pseudomonas.chlorophis subsp.Aurantiaca Pa40 CGMCCNo.2764 are shown in Table 1: Gram-negative bacteria, strict aerobic, nitrate reduction, starch hydrolysis, esculin utilization, Negative α-galactase reaction, oxidase, arginine double hydrolysis, gelatin hydrolysis, lecithinase, lipase, glucose...
Embodiment 2
[0040] Embodiment 2 antibacterial spectrum analysis
[0041] Plate inhibition test method for pathogenic fungi. Take a 5mm-diameter target pathogenic fungus and inoculate it in the center of the PDA plate. Pseudomonas and the target fungus are inoculated at 2.5cm from both sides of the target pathogenic fungus at an angle of 180°. After culturing at 25°C for 4 to 6 days, measure the colony diameter of the pathogenic fungus . Plates inoculated with target fungi but not pseudomonas were used as controls. The growth inhibition rate was calculated according to the following formula:
[0042] Growth inhibition rate=(C-T) / C×100%
[0043] C: Control fungal growth diameter
[0044] T: diameter of fungal growth after inoculation with pseudomonas
[0045] Plate inhibition detection method for pathogenic bacteria. The inhibition of Pseudomonas to pathogenic bacteria was detected by double-layer culture method. Use King`B (peptone 20g, glycerin 10ml, K 2 HPO 4 1.5g, MgSO 4 ·7H ...
Embodiment 3
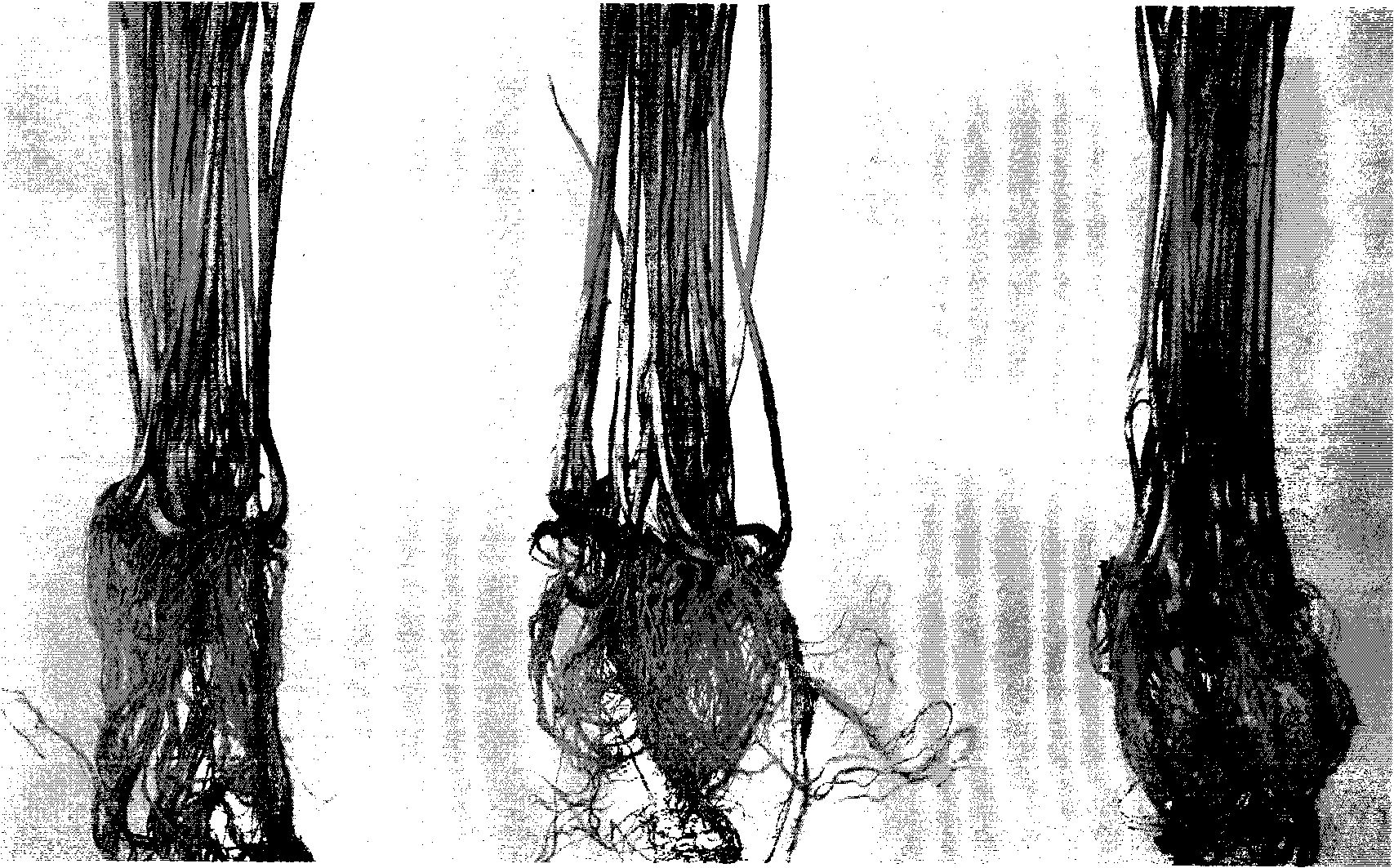
[0055] Example 3 Detection of biocontrol-related traits
[0056] Protease detection: use skim milk plate (tryptone 5g, yeast extract 2.5g, glucose 1g, 7% skim milk 250ml, agar 15g, add 1000ml of water) to detect protease. Inoculate Pa40 into the center of the plate and incubate at 28°C for 72 hours. There will be a transparent circle around the colony, such as image 3 As shown, Pseudomonas chlororaphis subsp.aurantiaca Pa40 CGMCCNo.2764 produces protease.
[0057] HCN detection: HCN test paper preparation, 10mg copper (II) ethyl acetoacetate (copper(II) ethyl acetoacetate) and 10mg 4,4'-methylbis(xylaniline) [4,4'-methylenebis-(N,N-dimethylaniline) ] dissolved in 4ml of chloroform, cut Whatman filter paper strips, soaked in the prepared solution, after the chloroform volatilized completely, HCN detection test paper was prepared. Puncture and inoculate pseudomonas in a centrifuge tube containing 1ml of PDA medium, suspend the HCN detection test paper in the centrifuge tube, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com