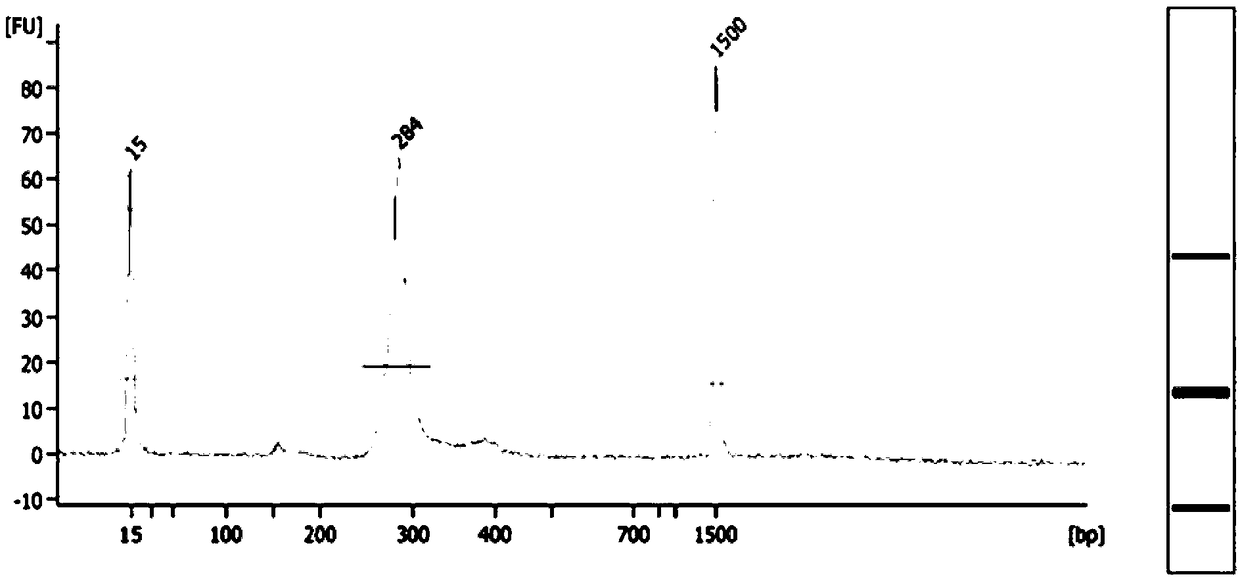
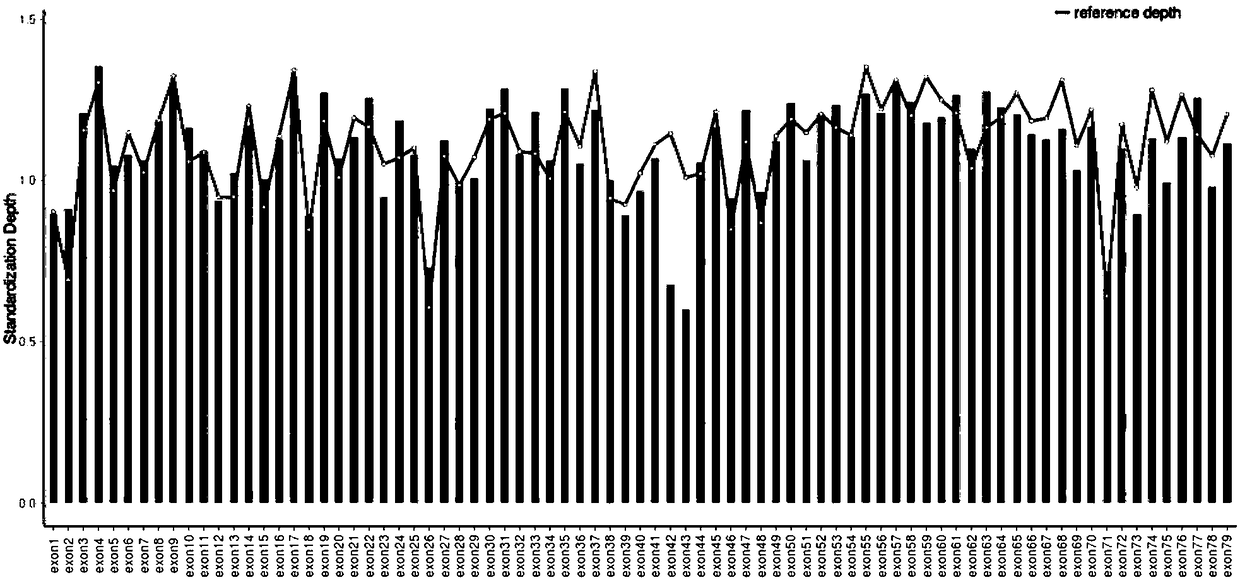
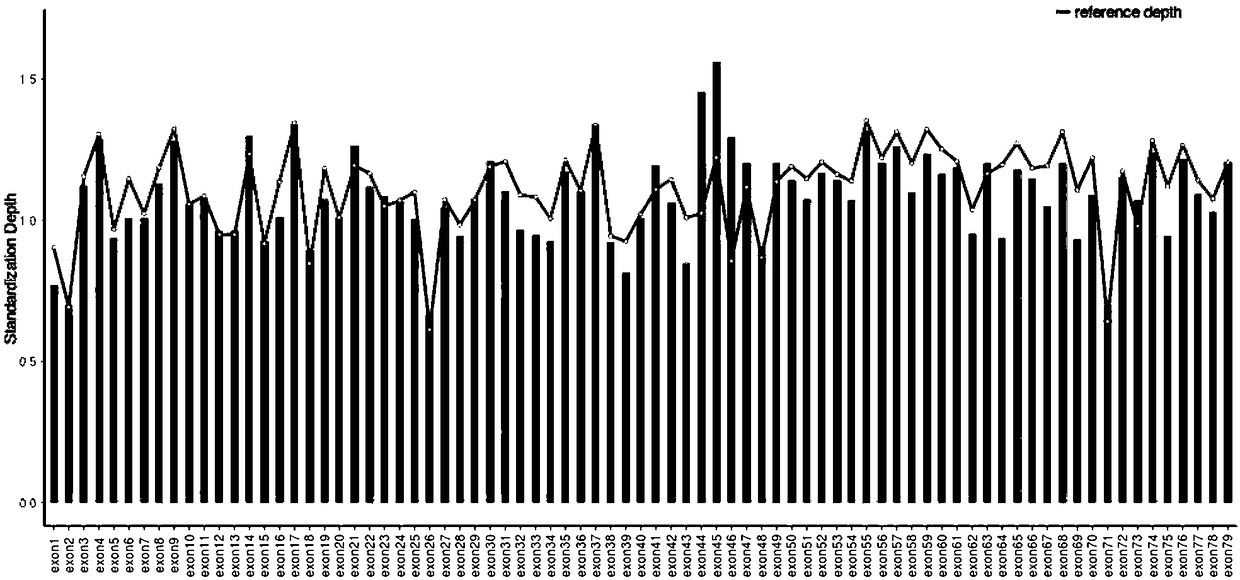
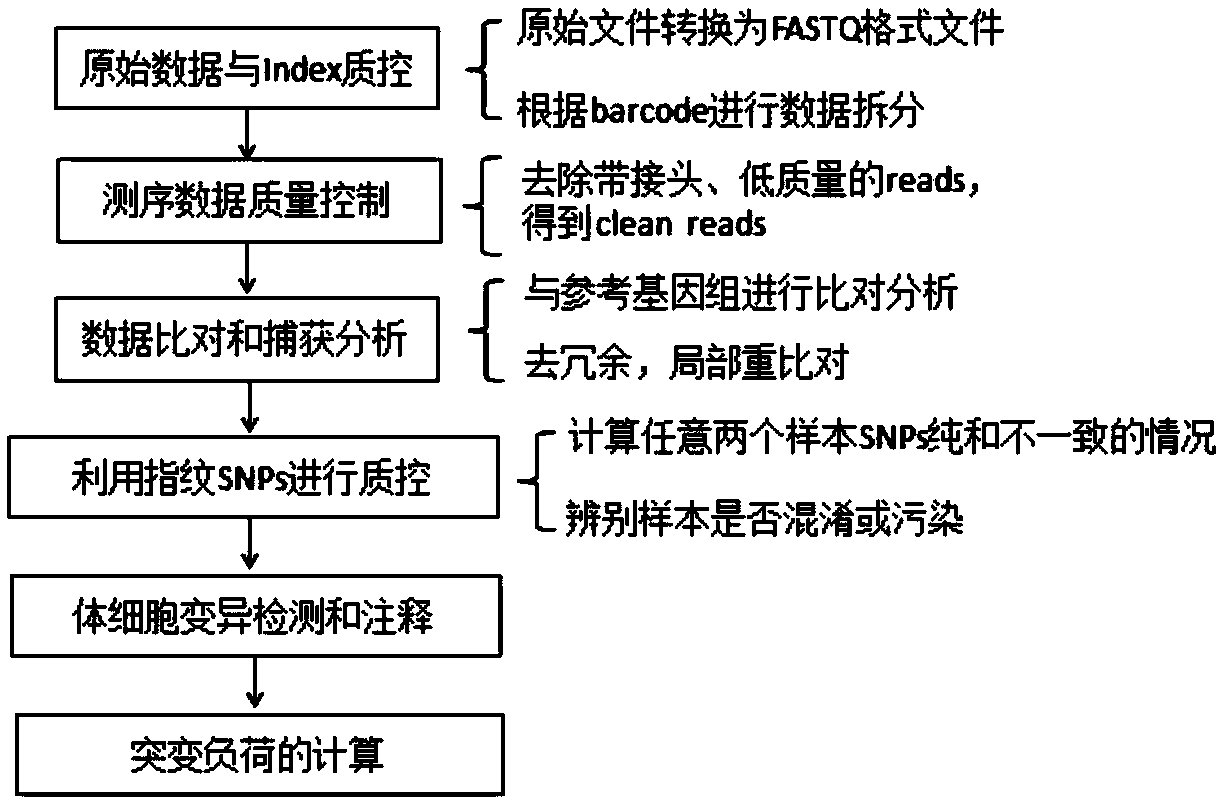
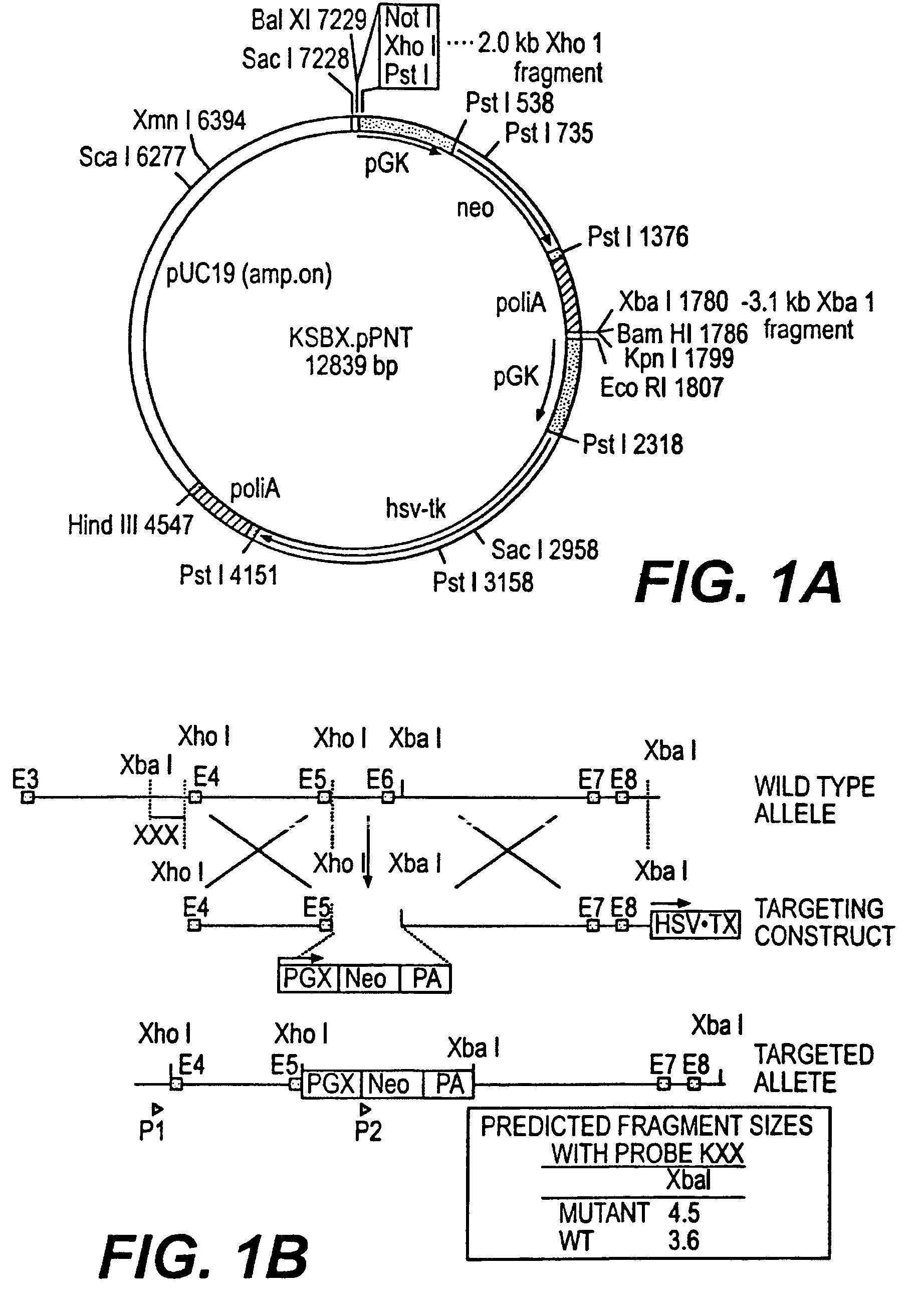
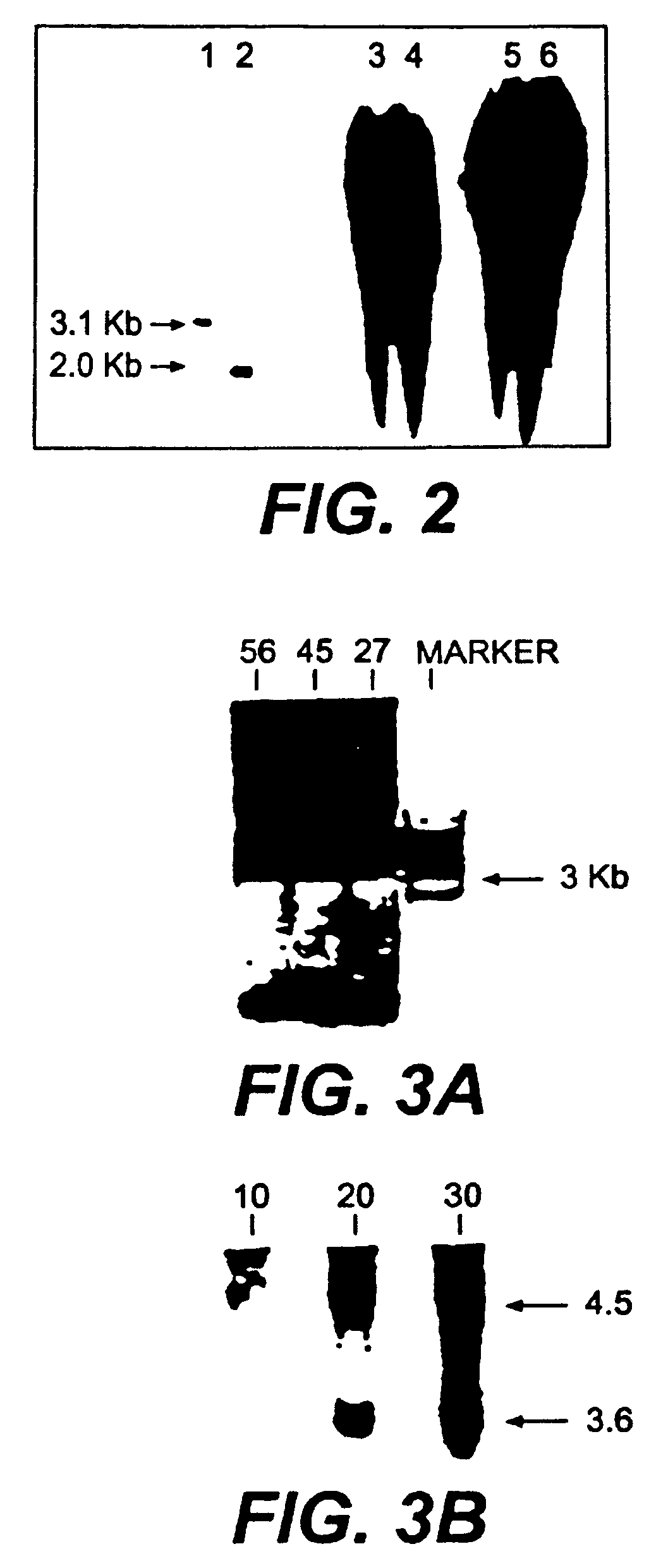
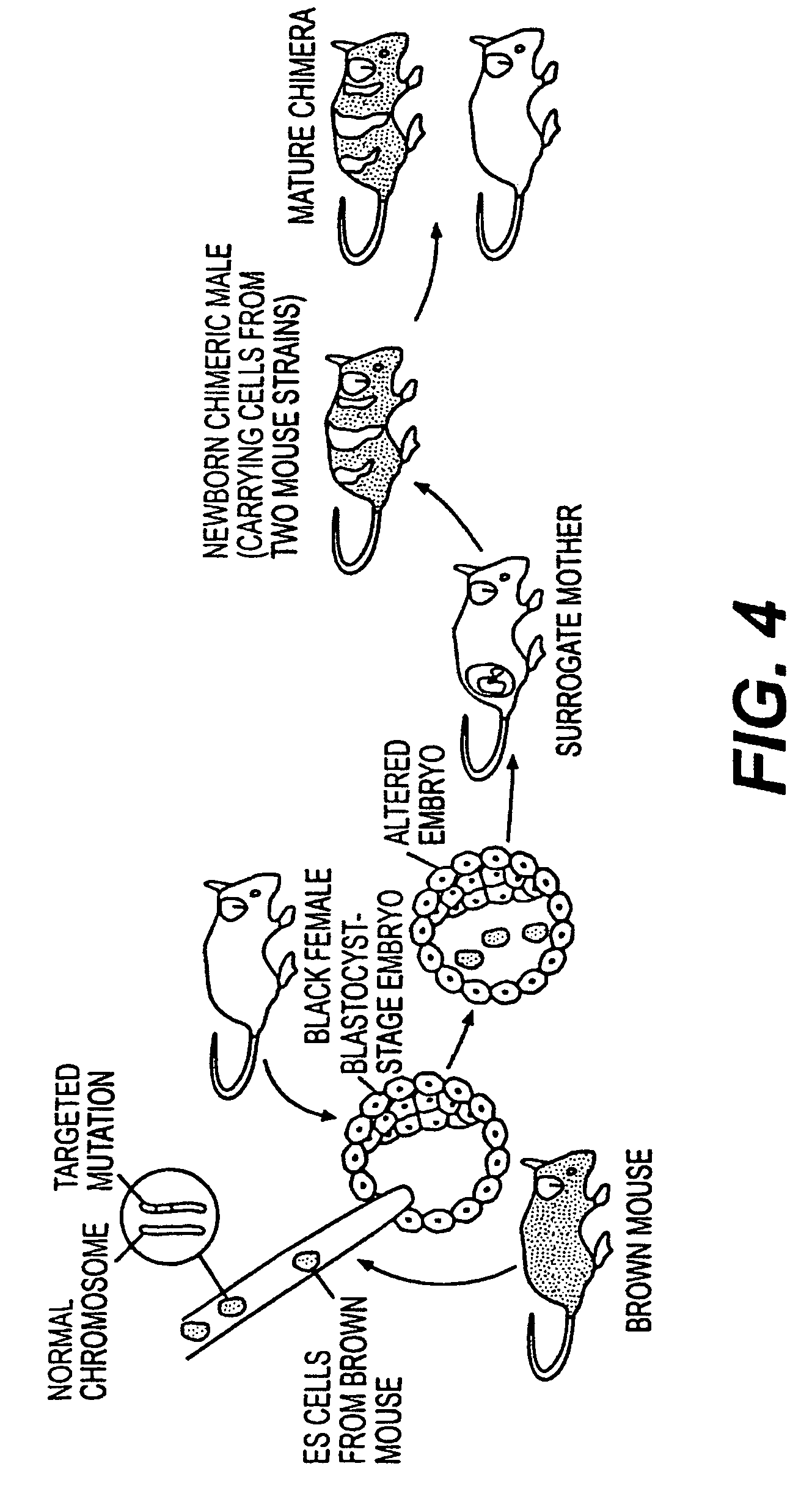
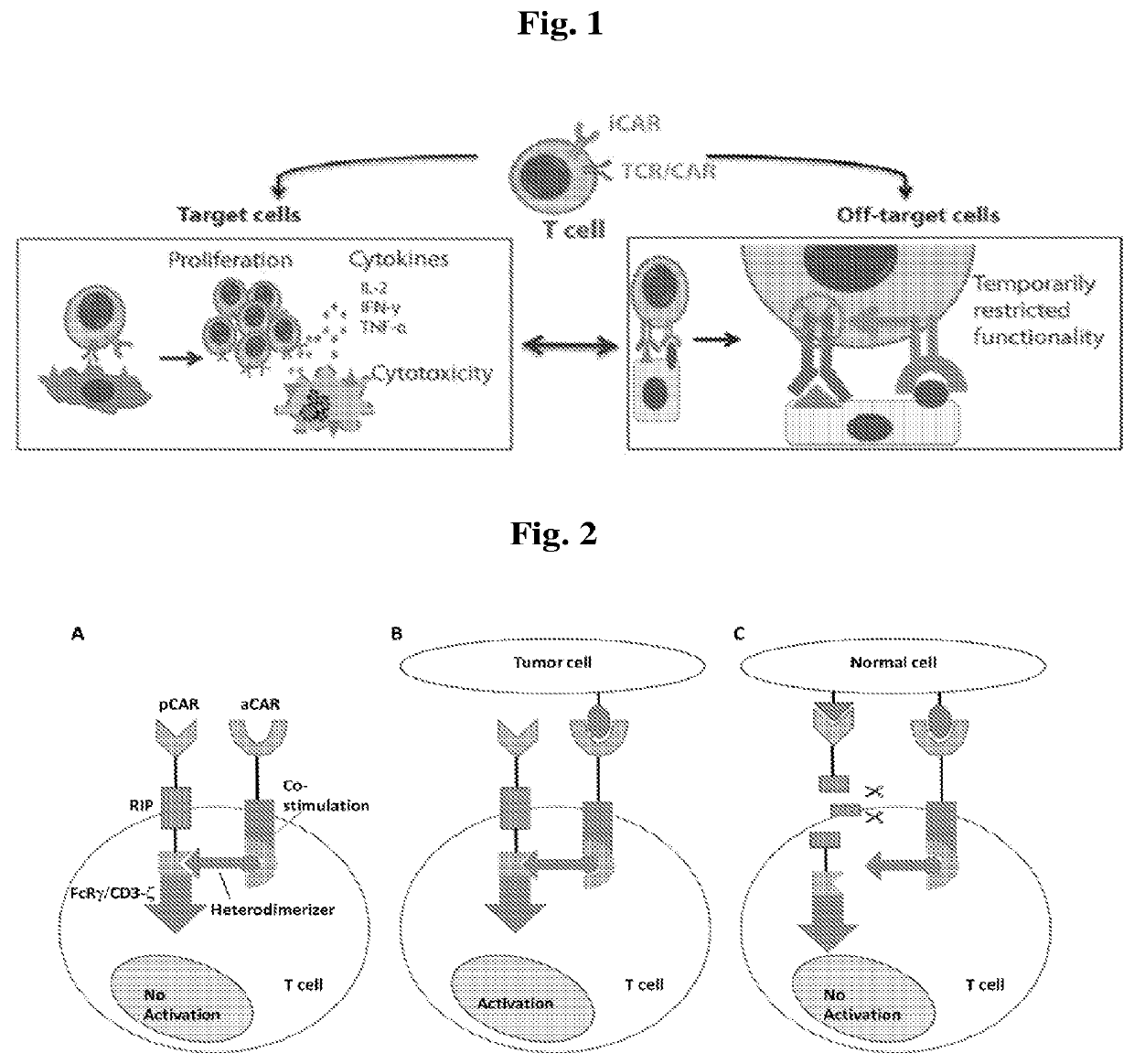
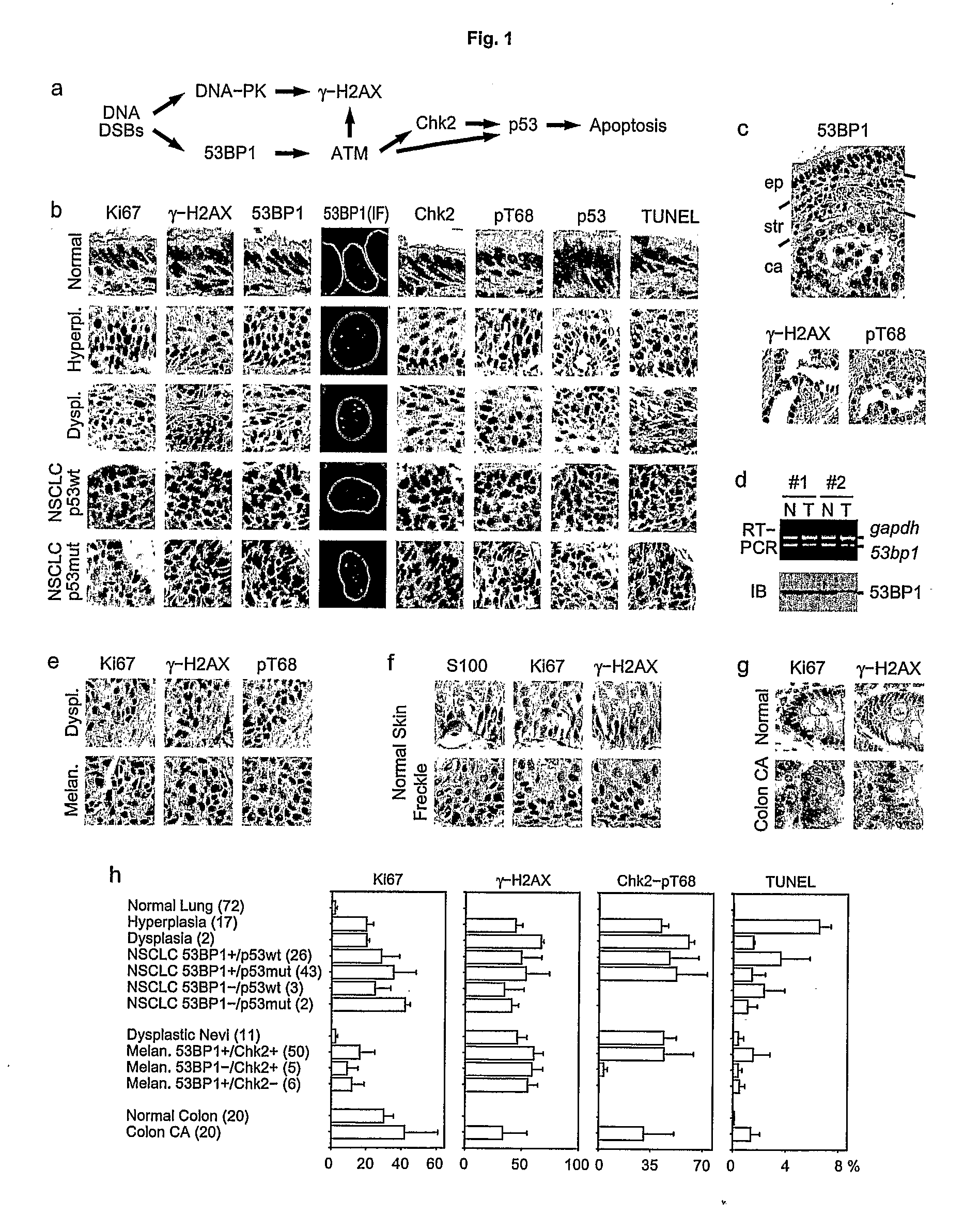
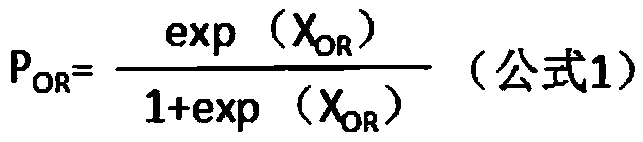Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
59 results about "Heterozygosity Loss" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
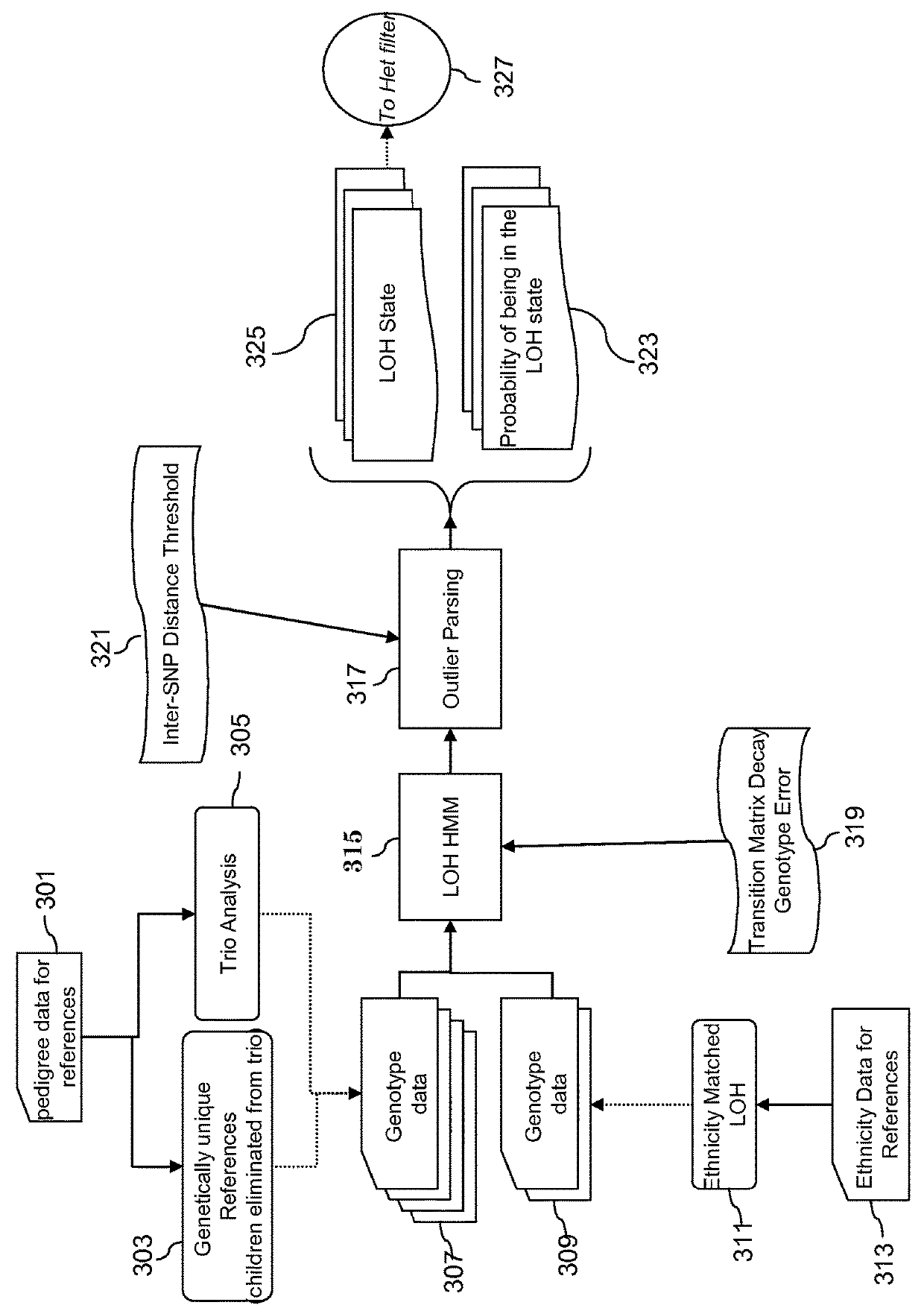
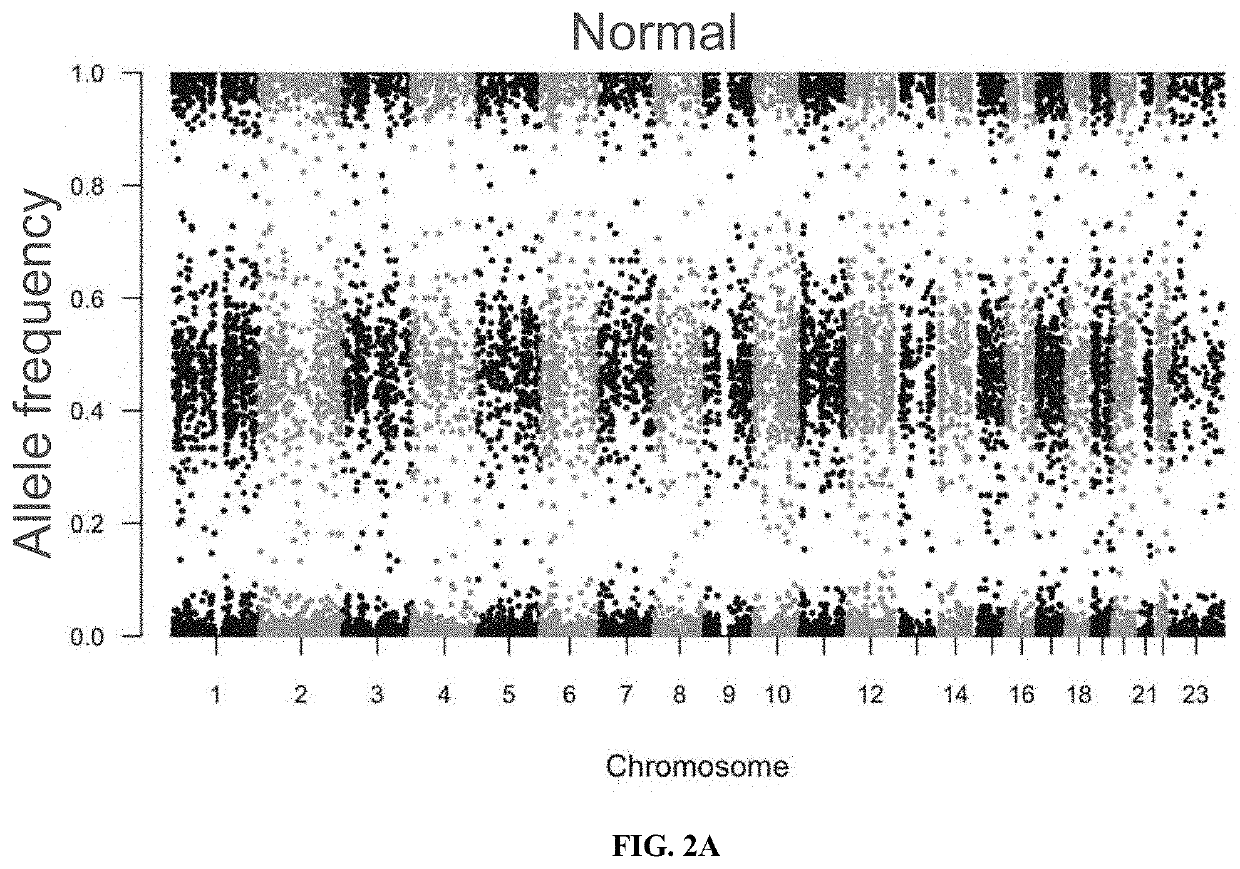
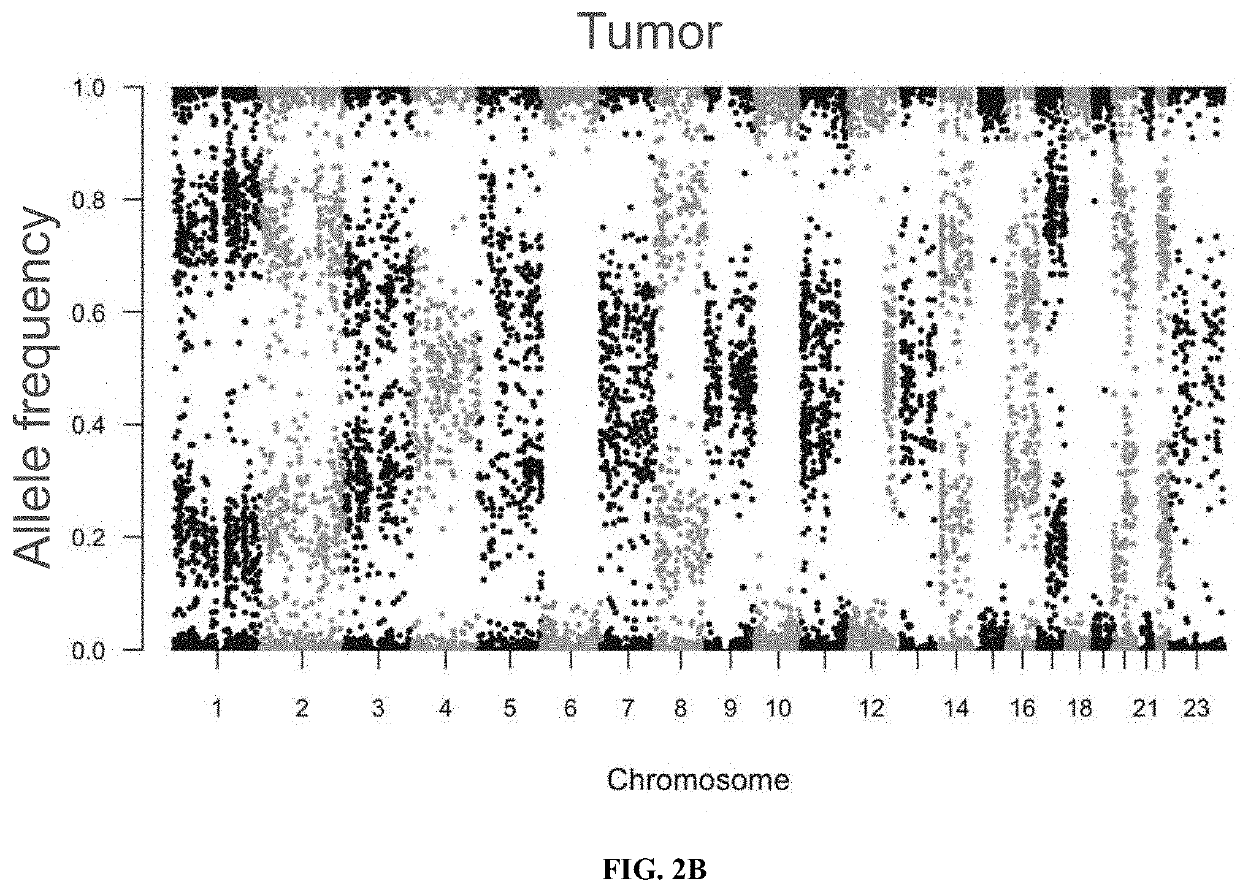
Loss of heterozygosity (LOH) is a cross chromosomal event that results in loss of the entire gene and the surrounding chromosomal region.
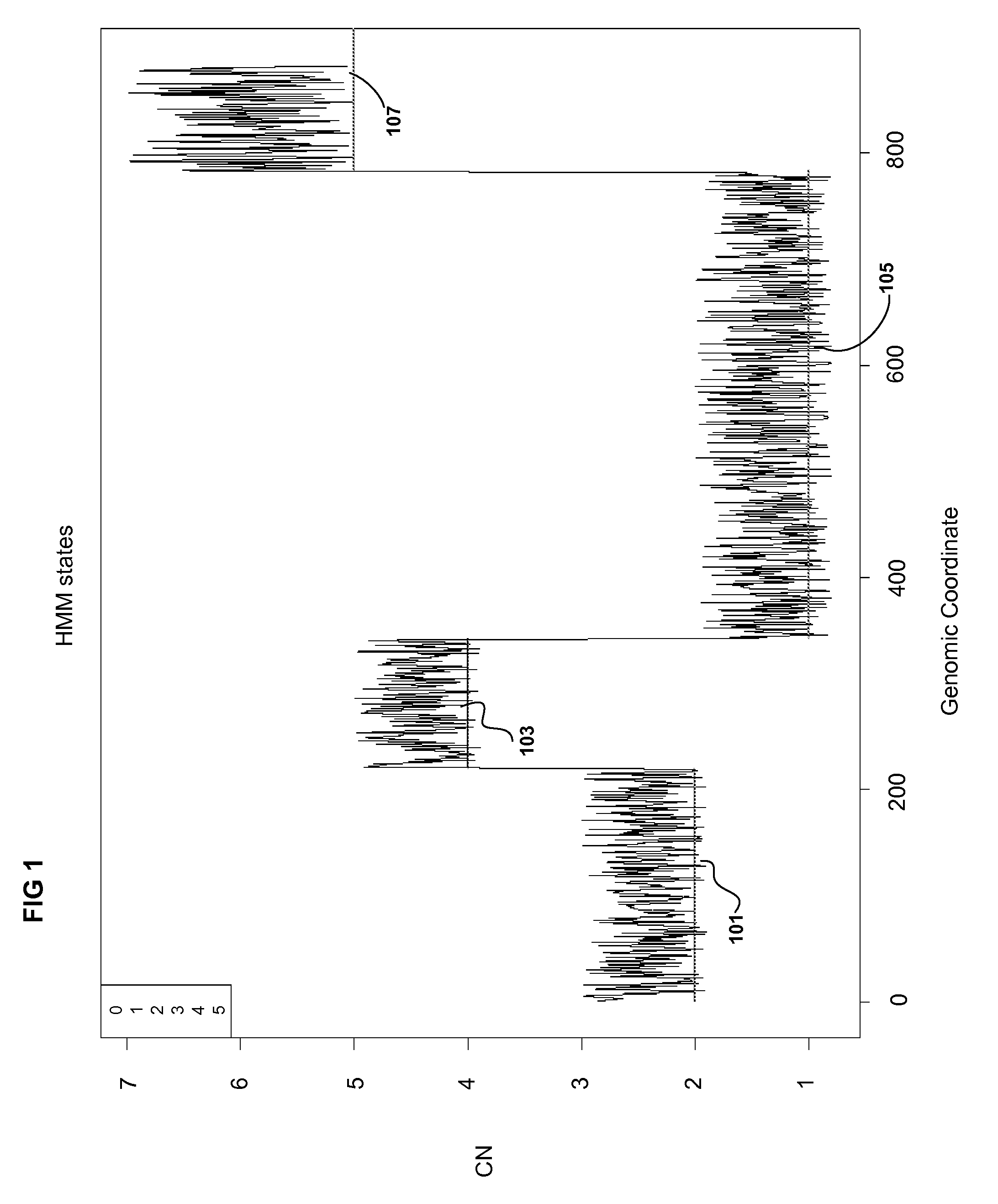
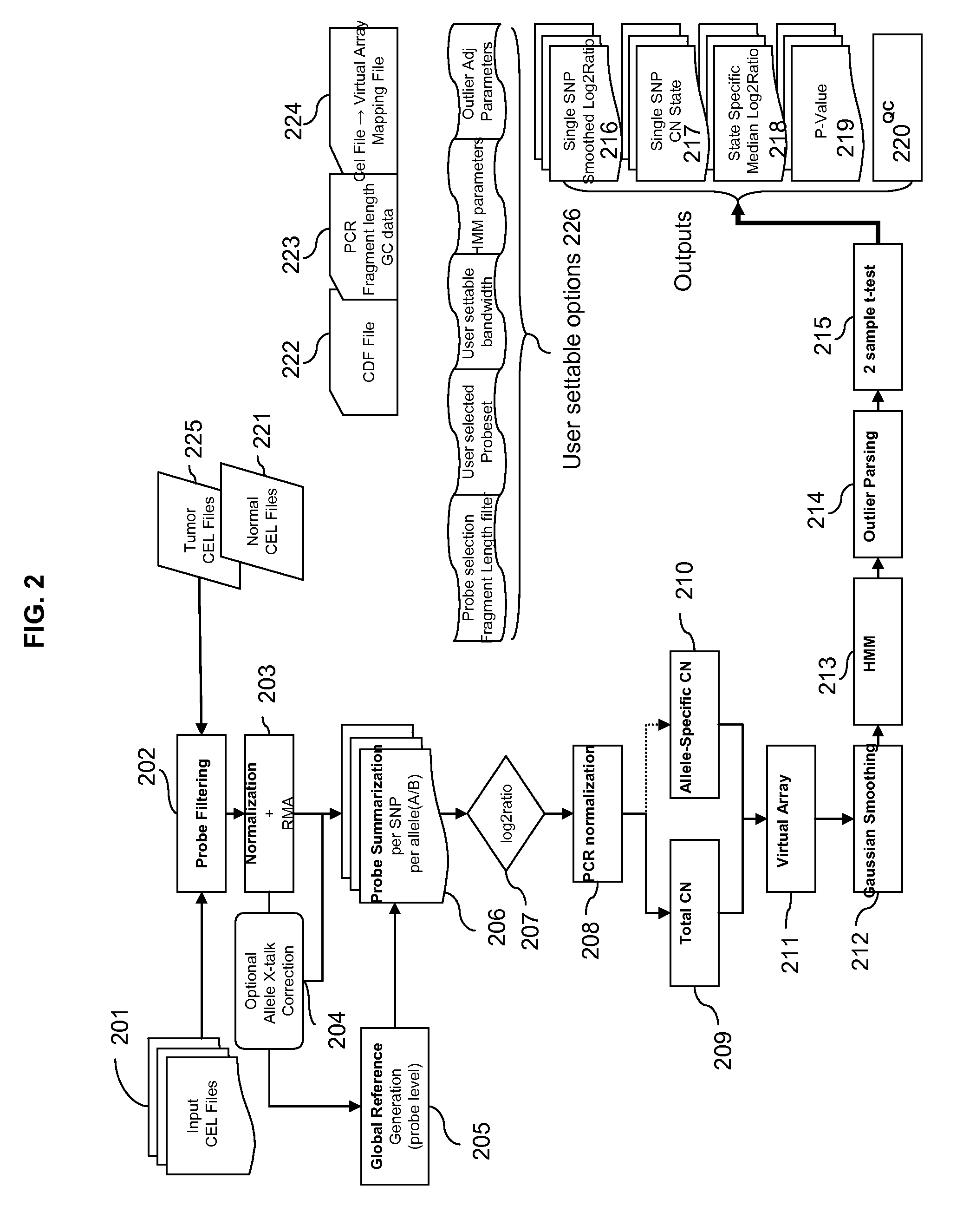
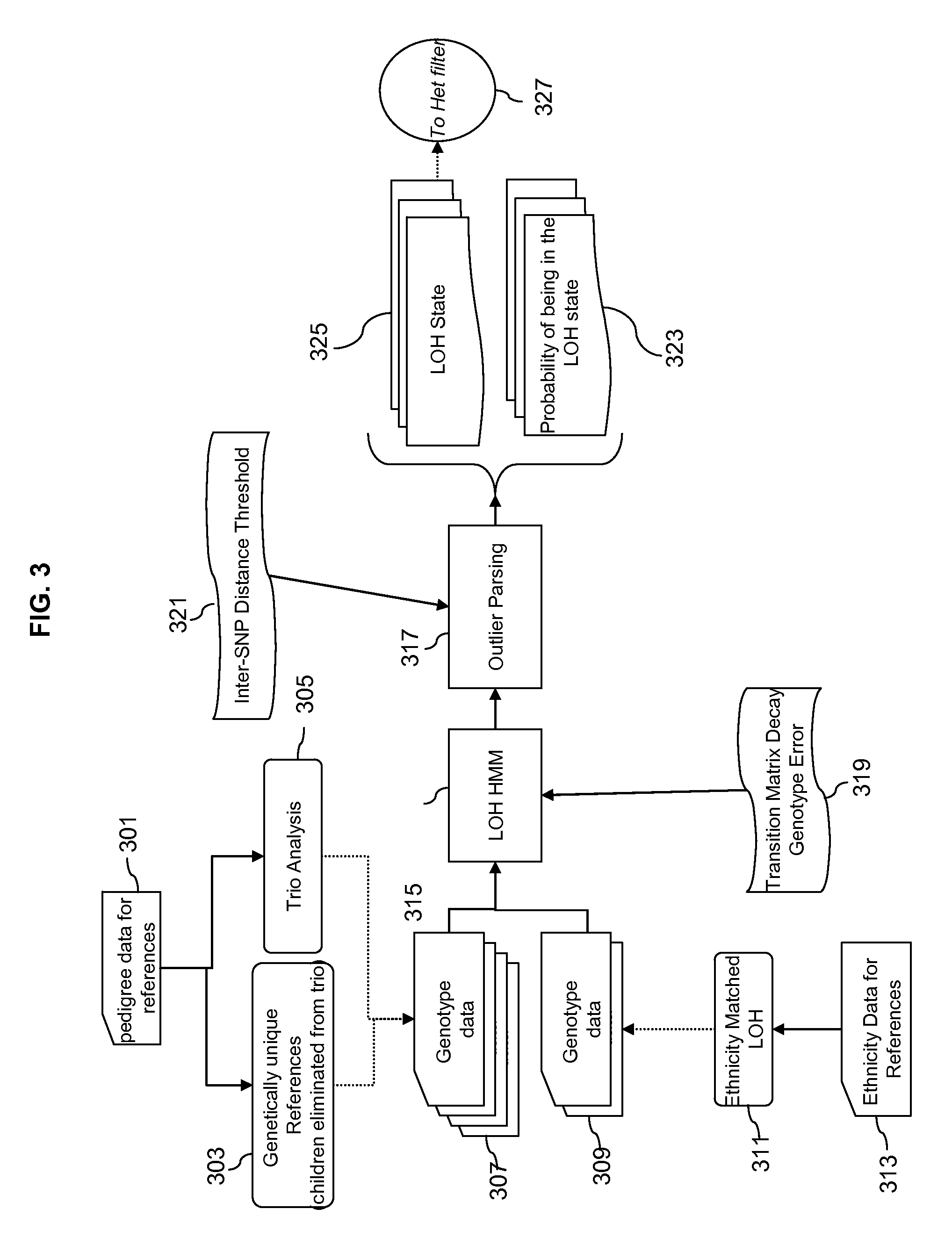
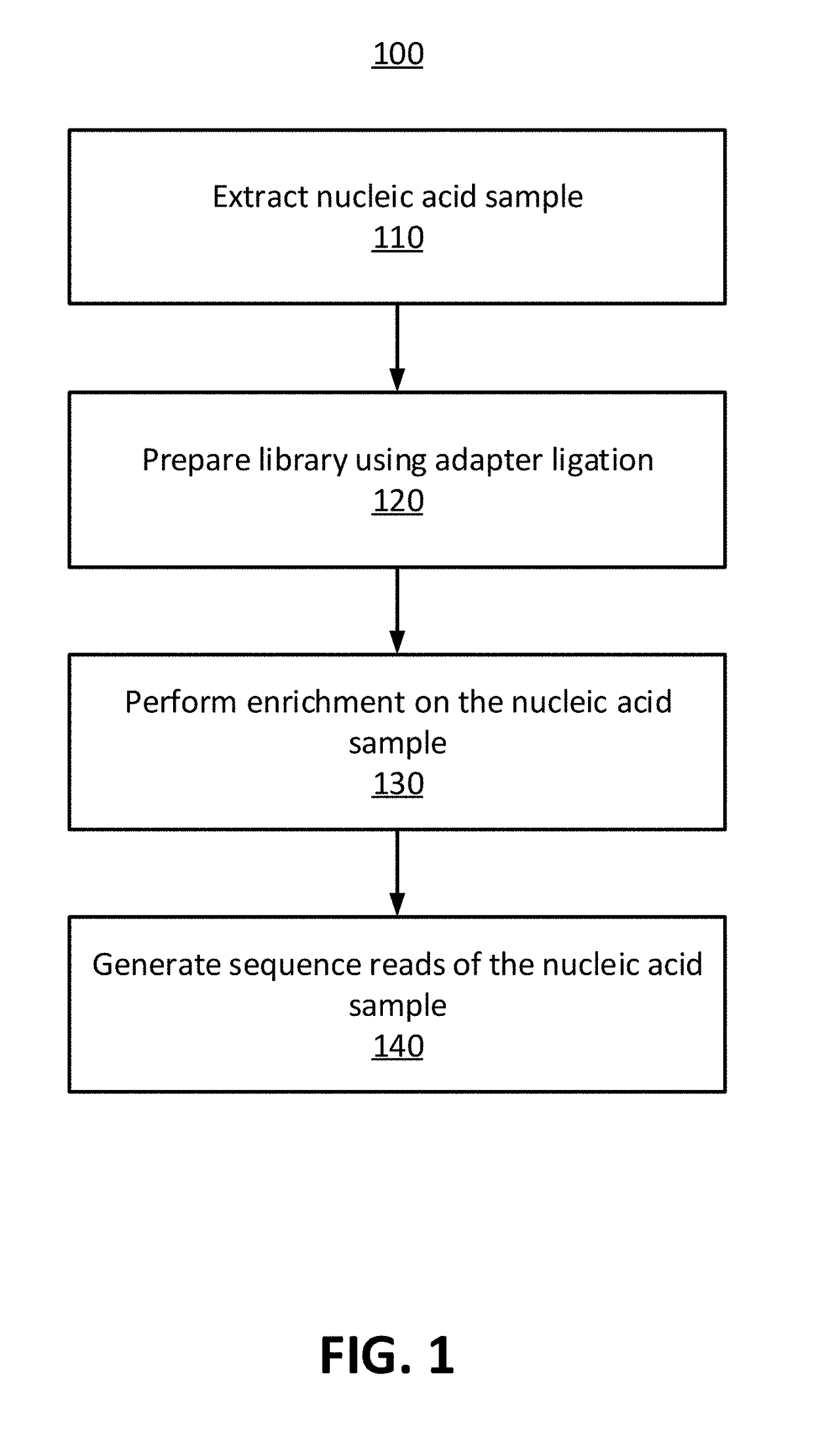
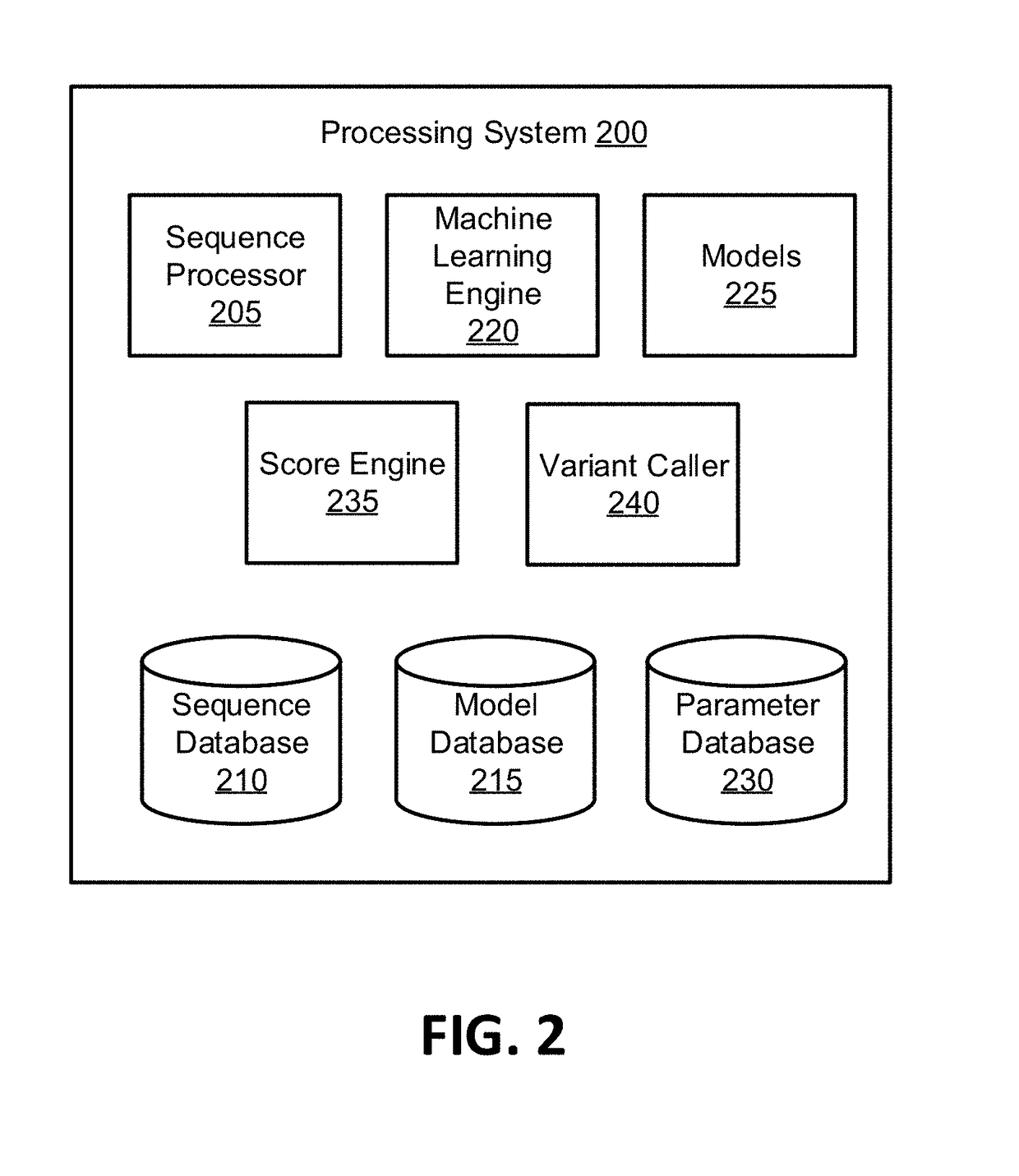
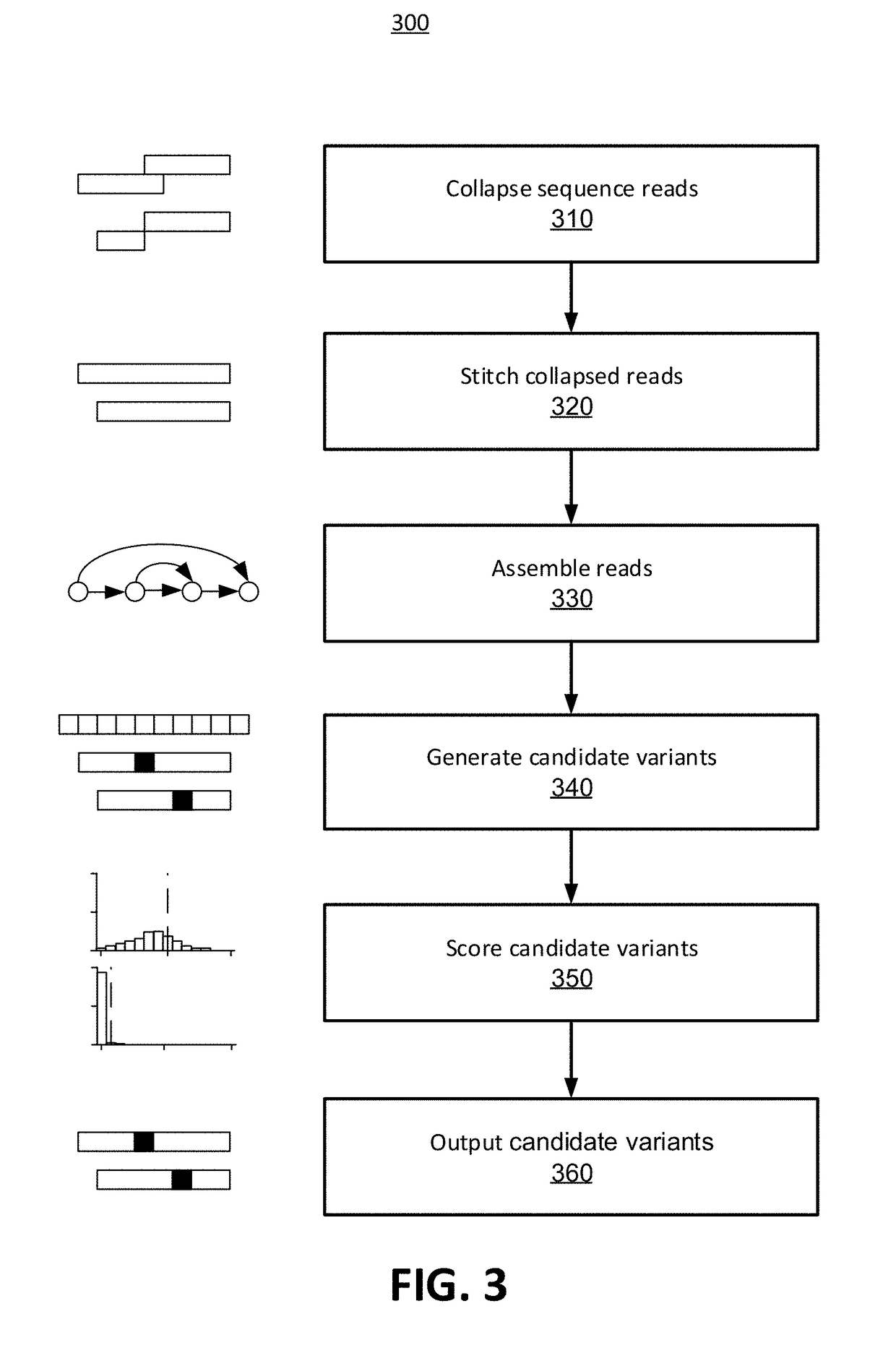
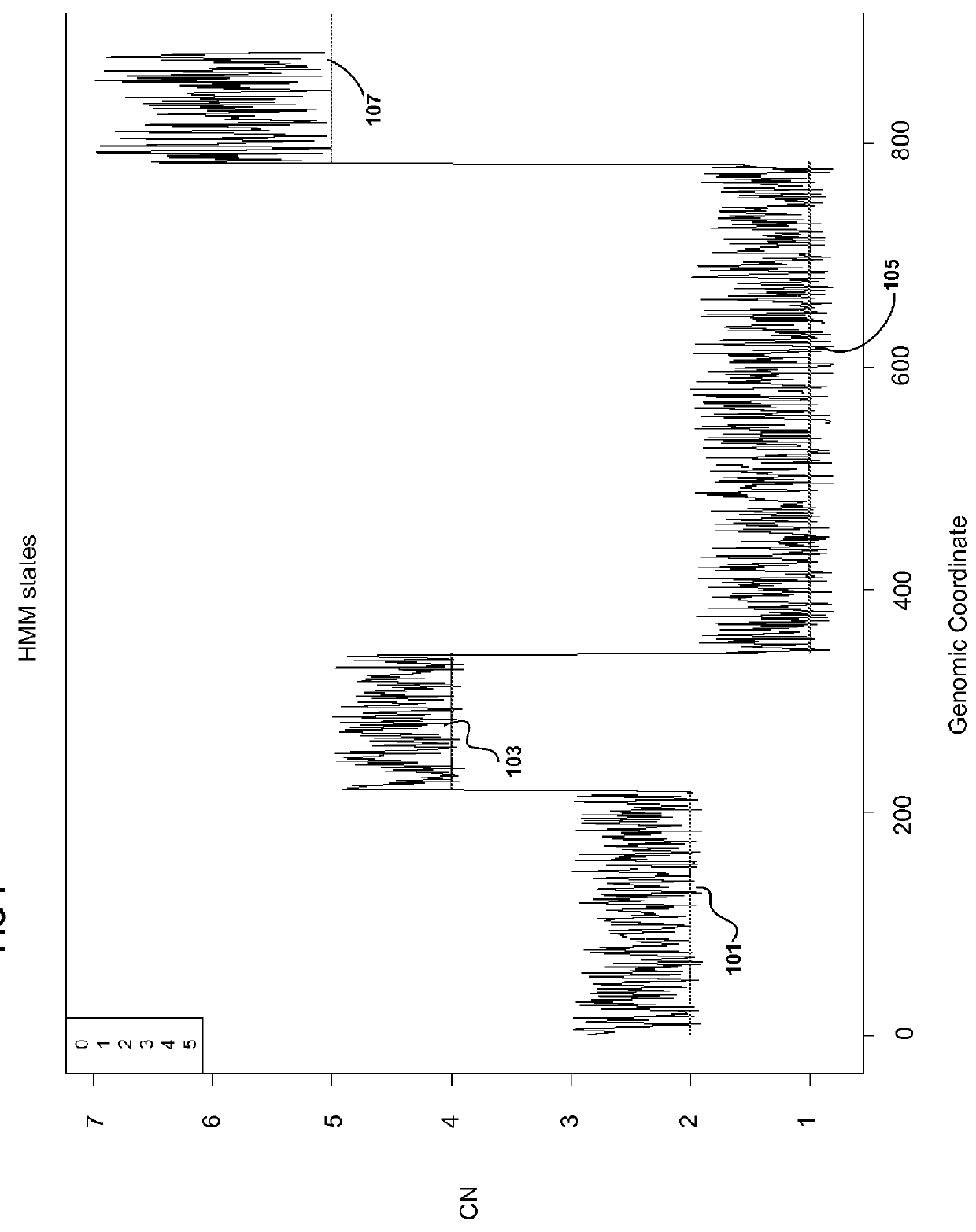
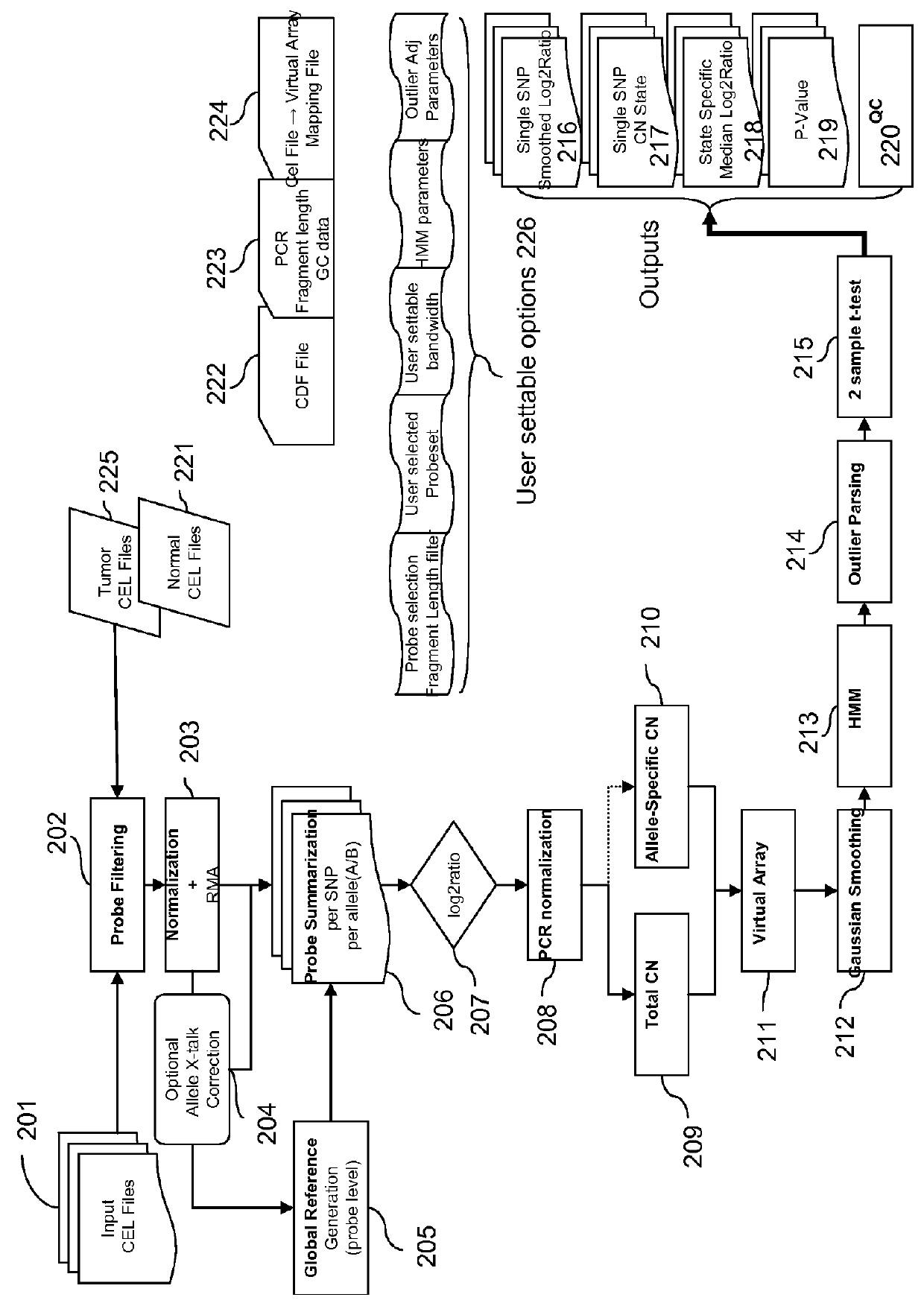
Methods for Identifying DNA Copy Number Changes Using Hidden Markov Model Based Estimations
Methods for estimating genomic copy number and loss of heterozygosity using Hidden Markov Model based estimation are disclosed.
Owner:AFFYMETRIX INC
DNA amplification of a single cell
InactiveUS6673541B1Improve accuracyImprove comprehensive applicabilitySugar derivativesMicrobiological testing/measurementSingle-strand conformation polymorphismComparative genomic hybridization
The present invention relates to a novel method for the amplification of DNA, this method being particularly useful for the amplification of the DNA or the whole genome of a single cell, chromosomes or fragments thereof. Described is also the use of the method in DNA analysis for medical, forensic, diagnostic or scientific purposes, like comparative genomic hybridization (CGH)-, fluorescence in situ hybridization (FISH)-, polymerase chain reaction (PCR)-, single strand conformation polymorphism (SSCP)-, DNA sequence-, "loss of heterozygosity" (LOH)-, fingerprint- and / or restriction fragment length polymorphism (RFLP)-analysis.
Owner:AMGEN RES (MUNICH) GMBH
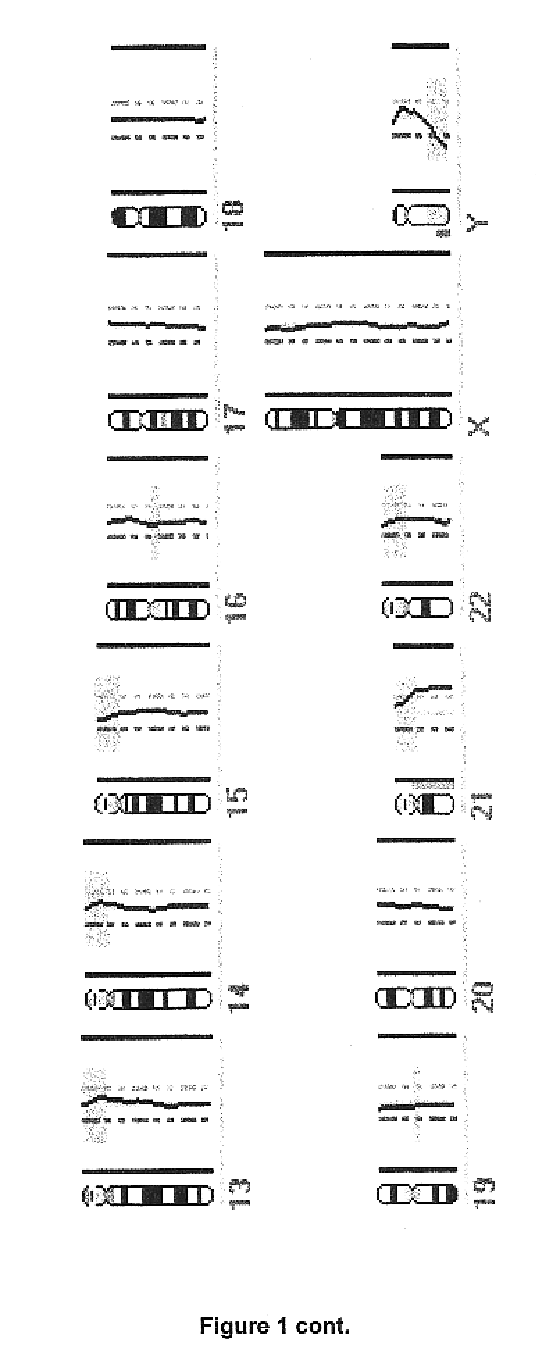
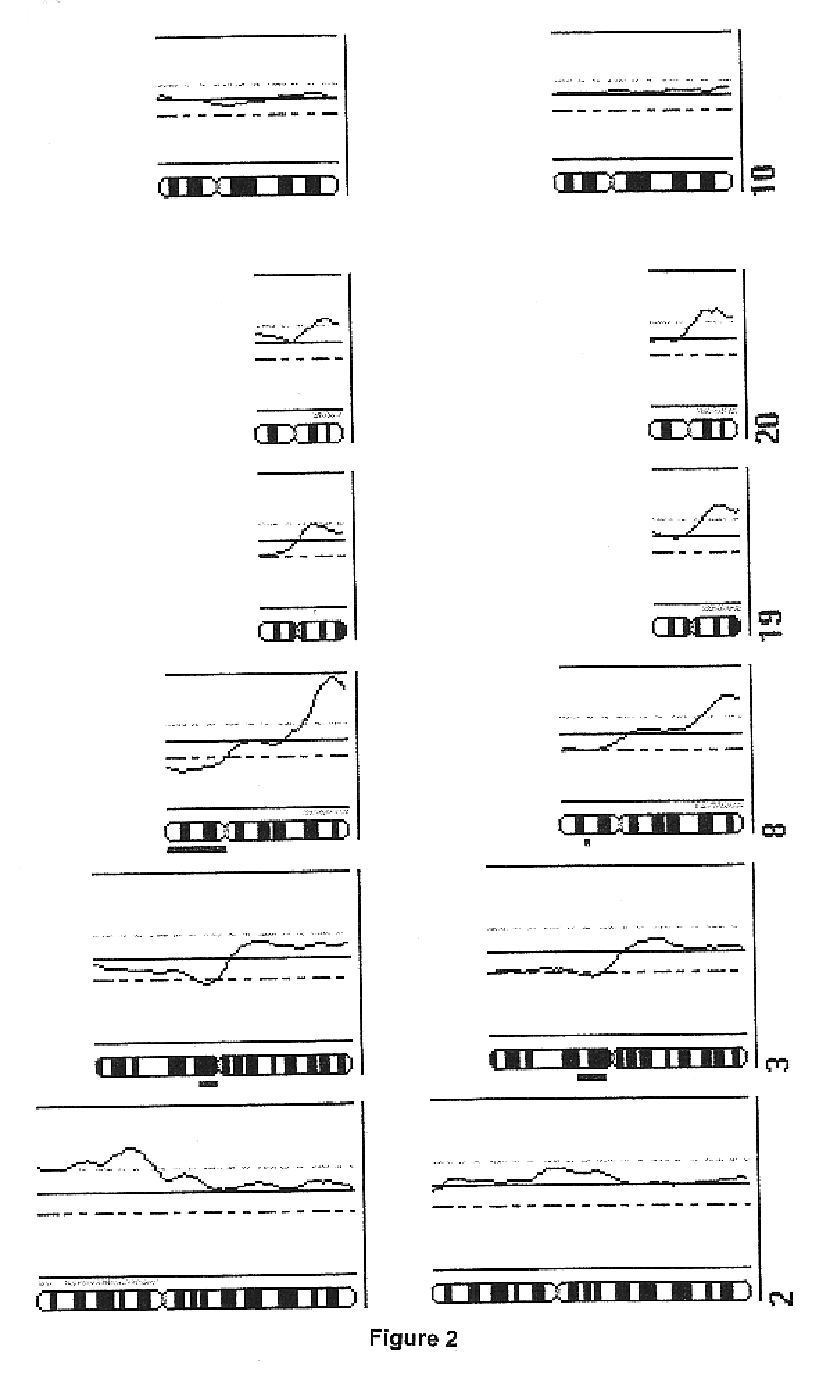
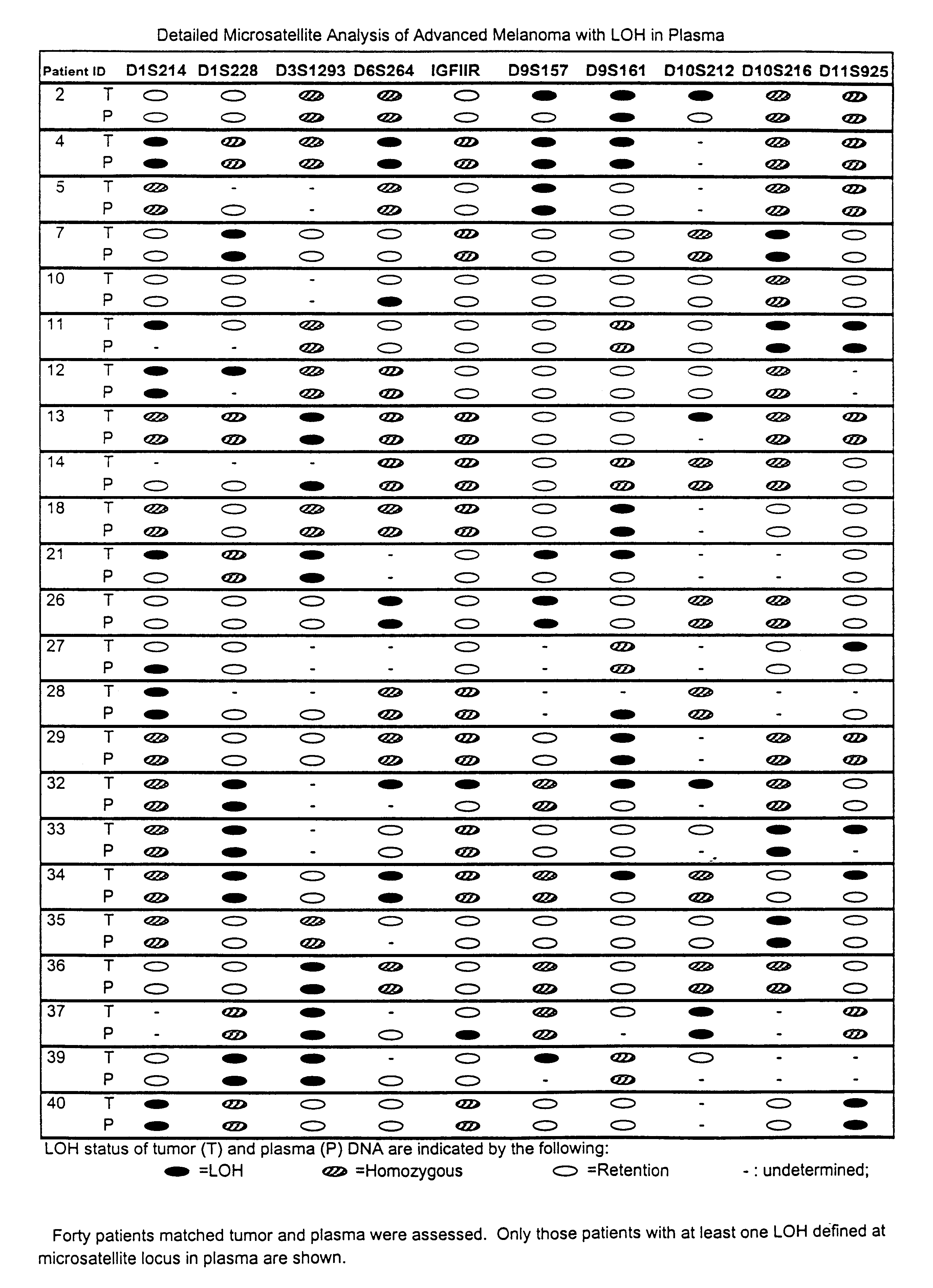
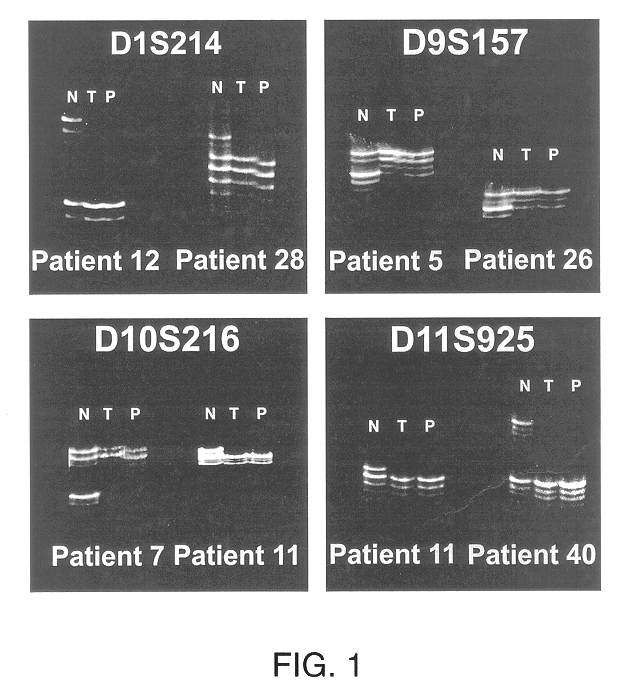
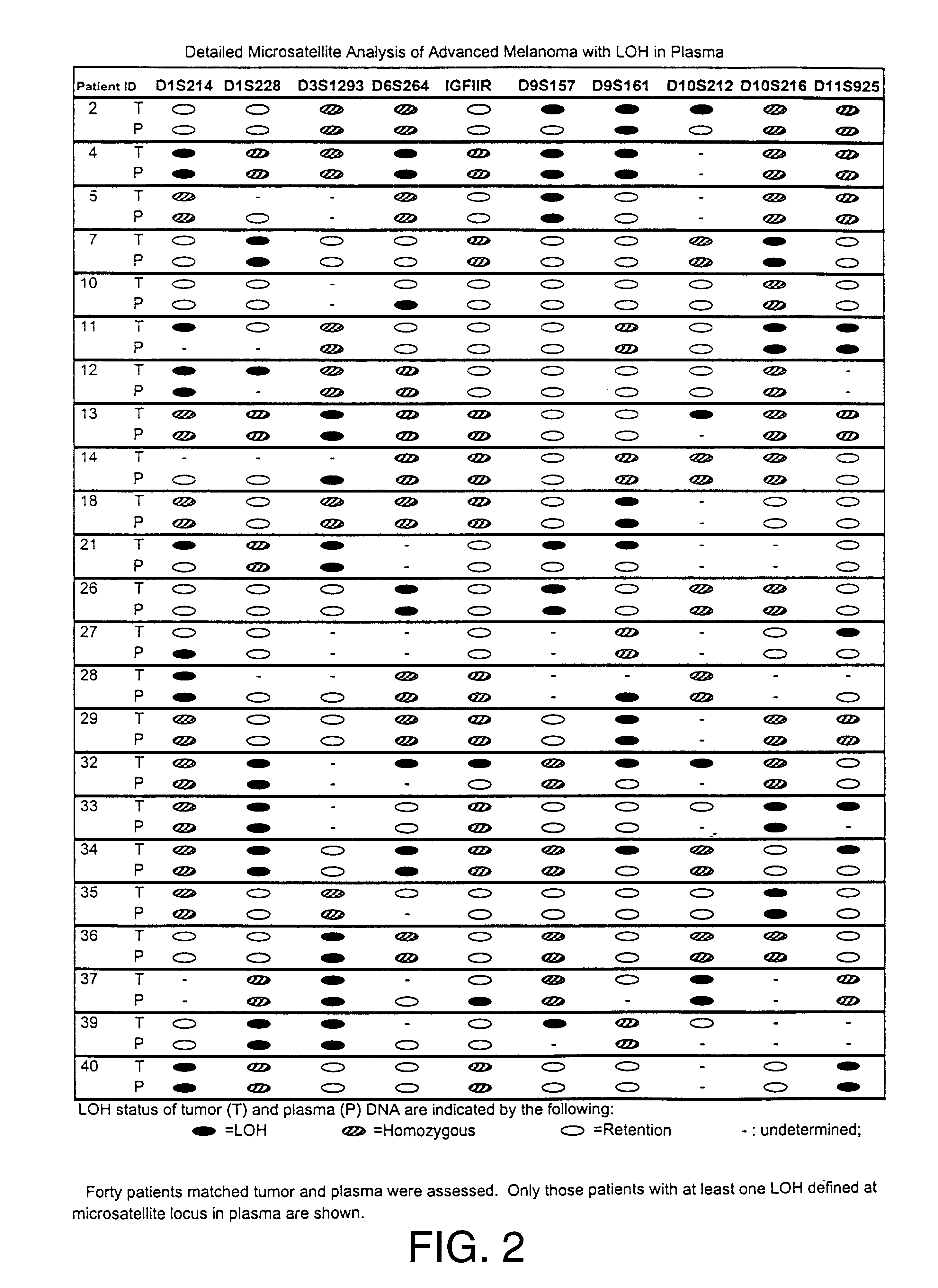
Detection of loss of heterozygosity in tumor and serum of melanoma patients
A method is provided for assessing allelic losses on specific chromosomal regions in melanoma patents. The method relies on the evidence that free DNA may be released in the plasma / serum of cancer patients allowing the detection of DNA with LOH in the plasma / serum of cancer patients by analysis for microsatellite markers. The amount of and specific allelic loss allows a prognosis to be made regarding tumor diagnosis and progression, tumor metastasis, tumor recurrence, and mortality.
Owner:JOHN WAYNE CANCER INST
Probe biochips and methods for use thereof
InactiveUS20060199183A1Bioreactor/fermenter combinationsBiological substance pretreatmentsPolynucleotideAlternative splicing
The invention relates to fields of use of unlabelled polynucleotide probes able to form hairpins, the biochips comprising such probes and methods allowing use thereof. The present invention also concerns methods for designing such probes and biochips. More particularly, the invention concerns the use of such unlabelled probes and biochips for manipulating and analysing polynucleotide sequences and optionally molecules which are associated therewith. This invention further concerns methods for preparing and use such probes and biochips for analysing mutations, sequencing, detection of alternative splicing variants, gene expression analysis, analysis of allelic imbalances and loss of heterozygosity and the detection of any nucleic acid present in organisms or residues from said organisms.
Owner:GENEWAVE SAS
Methods and materials for assessing loss of heterozygosity
InactiveUS20120015050A1Raise the possibilityReduce in quantityHeavy metal active ingredientsBiocideCancer cellRegimen
This document provides methods and materials involved in assessing samples (e.g., cancer cells) for the presence of a loss of heterozygosity (LOH) signature. For example, methods and materials for determining whether or not a cell (e.g., a cancer cell) contains an LOH signature are provided. Materials and methods for identifying cells (e.g., cancer cells) having a deficiency in homology directed repair (HDR) as well as materials and methods for identifying cancer patients likely to respond to a particular cancer treatment regimen also are provided.
Owner:MYRIAD GENETICS +1
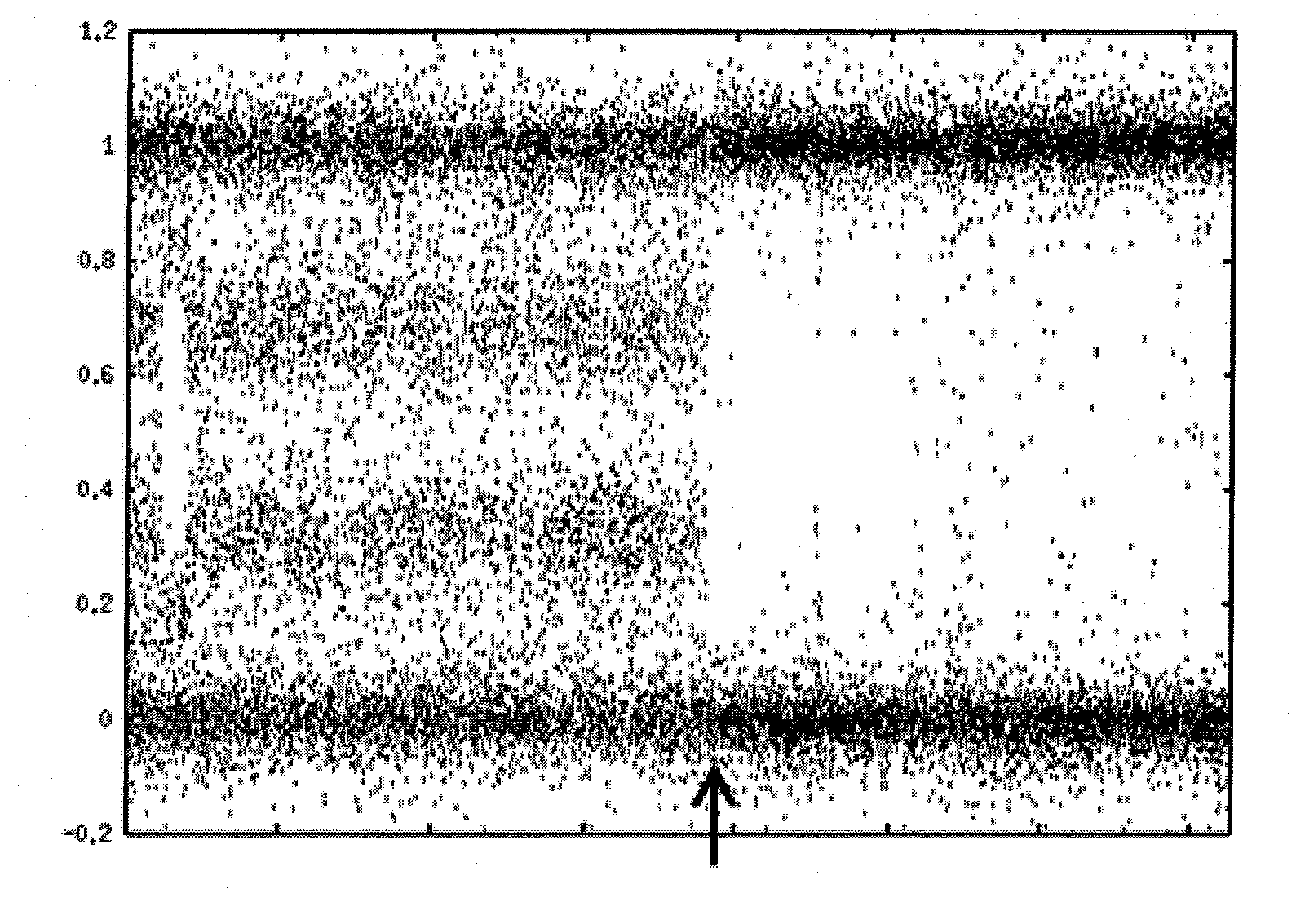
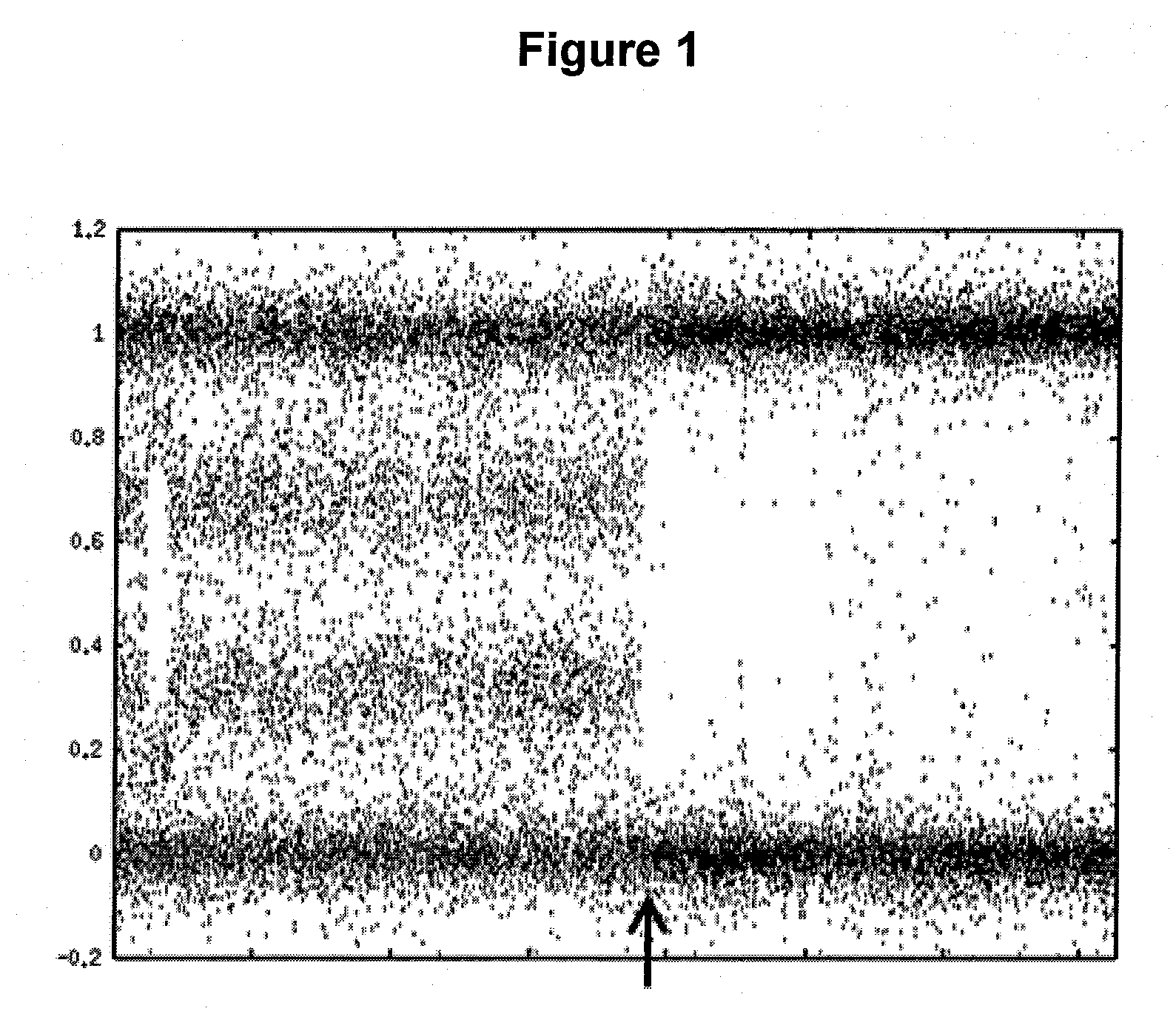
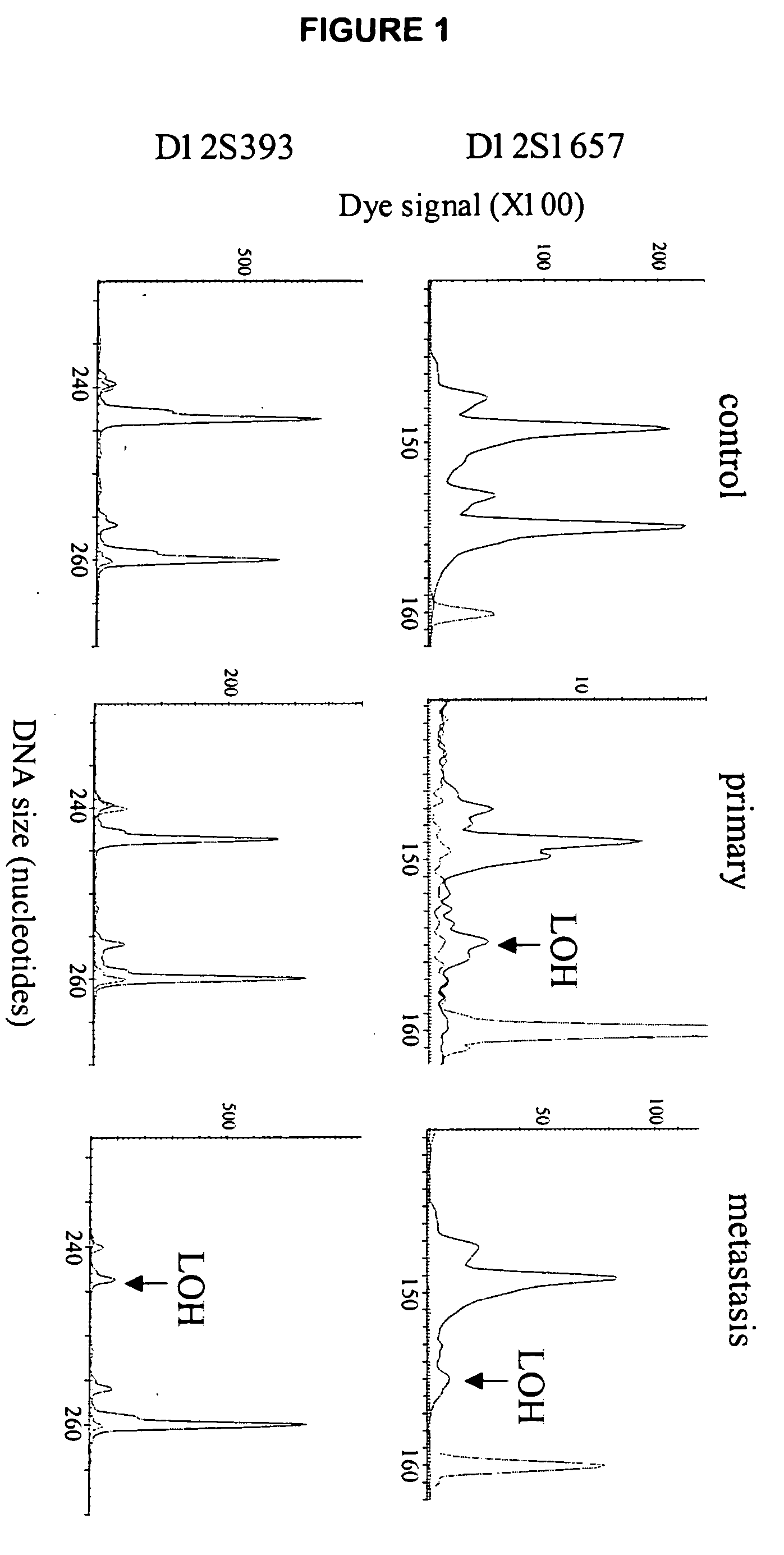
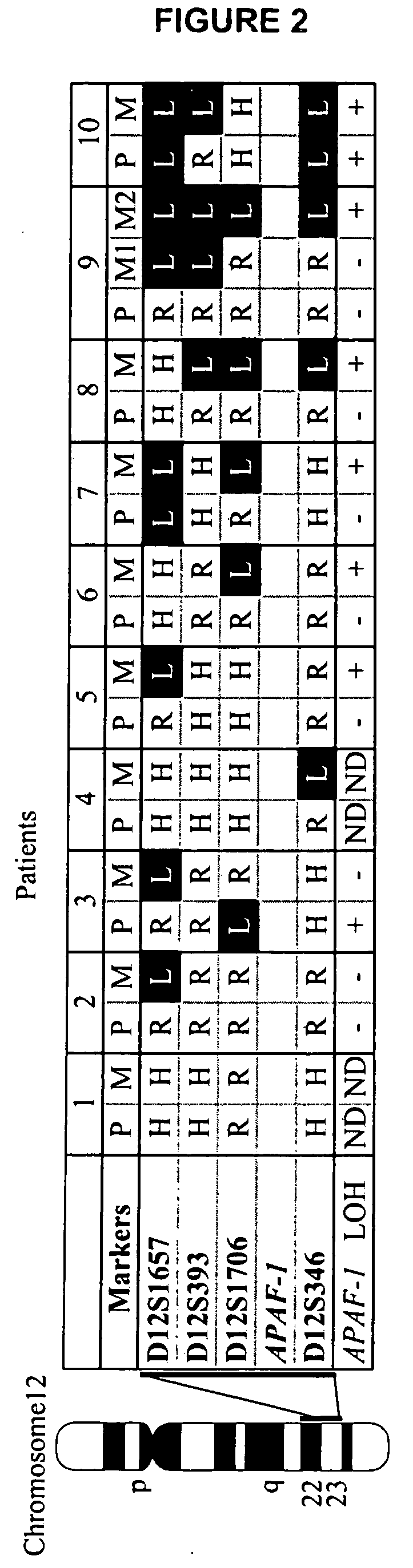
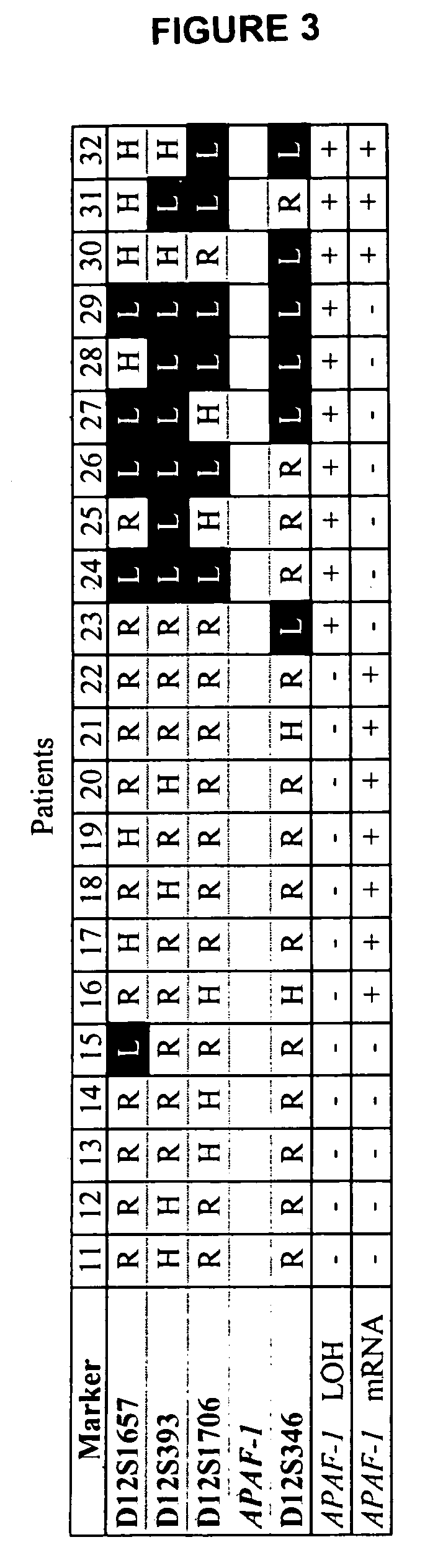
Loss of heterozygosity of the DNA markers in the 12q22-23 region
InactiveUS20050112604A1Increase probabilityPoor efficacySugar derivativesMicrobiological testing/measurementCell freeProbability of survival
A method of detecting DNA markers in the 12q22-23 region. The method comprises providing a sample containing acellular DNA from a subject and detecting one or more DNA markers in the 12q22-23 region in the sample. Also disclosed are methods of diagnosing and monitoring cancer; methods of determining the efficacy of a therapy, and the probabilities of survival and responsiveness to a therapy; and packaged products for using these methods.
Owner:JOHN WAYNE CANCER INST
Method for predicating homologous recombination deficiency mechanism and method for predicating response of patients to cancer therapy
InactiveCN107287285AInnovativeOvercoming the pitfalls of inaccurate forecastsMicrobiological testing/measurementSequence analysisAbnormal tissue growthPolymerase L
The invention discloses a method for predicating a homologous recombination deficiency (HRD) mechanism and a method for predicating response of patients to cancer therapy and relates to the field of biological information predication. The method comprises the step of judging whether a tumor sample has homologous recombination deficiency or not according to one or more comprehensive values in a large-segment INDEL (Insertion / Deletion) fraction, a copy number variation fraction and a tumor mutation load fraction, wherein the comprehensive values can also comprise a loss of heterozygosity variation fraction. By adopting the method disclosed by the invention, predication of a chromosome large-segment structure, a chromosome gene type number, a chromosome gene copy number, a chromosome variation interval and abnormal loss of heterozygosity and chromosome telomeric imbalance is realized, so that an evaluation range is more complete and HRD can be accurately predicated; the comprehensive values also can be used for determining whether the patients have response to a therapeutic regimen containing one or more of a PARP (Poly Adenosine Diphosphate Ribose Polymerase) inhibitor, an DNA (Deoxyribonucleic Acid) injury inhibitor, a topoisomerase II / II+inhibitor, a topoisomerase I inhibitor and radiotherapy; the method is simple and has wide general applicability.
Owner:SHANGHAI ORIGIMED CO LTD
Methods and materials for assessing loss of heterozygosity
InactiveUS20150080260A1Raise the possibilityReduce in quantityTherapiesMicrobiological testing/measurementCancer cellRegimen
This document provides methods and materials involved in assessing samples (e.g., cancer cells) for the presence of a loss of heterozygosity (LOH) signature. For example, methods and materials for determining whether or not a cell (e.g., a cancer cell) contains an LOH signature are provided. Materials and methods for identifying cells (e.g., cancer cells) having a deficiency in homology directed repair (HDR) as well as materials and methods for identifying cancer patients likely to respond to a particular cancer treatment regimen also are provided.
Owner:MYRIAD GENETICS +1
Detecting cross-contamination in sequencing data
Detecting cross-contamination between test samples used for determining cancer in a subject is beneficial. To detect cross-contamination, test sequences including at least one single nucleotide polymorphism are prepared using genome sequencing techniques. Some of the test sequences can be filtered to improve accuracy and precision. A prior contamination probability for each test sequence is determined based on a minor allele frequency. A contamination model including a likelihood test is applied to a test sequence. The likelihood test obtains a current contamination probability representing the likelihood that the test sample is contaminated. The contamination model can also determine a likelihood that the sample includes loss of heterozygosity representing the likelihood that the test sequence is contaminated. Test samples that are contaminated are removed. A source for the contaminated test sample can be found by comparing contaminated test sequences to other test sequences.
Owner:GRAIL LLC
Methods for identifying DNA copy number changes using hidden markov model based estimations
Methods for estimating genomic copy number and loss of heterozygosity using Hidden Markov Model based estimation are disclosed.
Owner:AFFYMETRIX INC
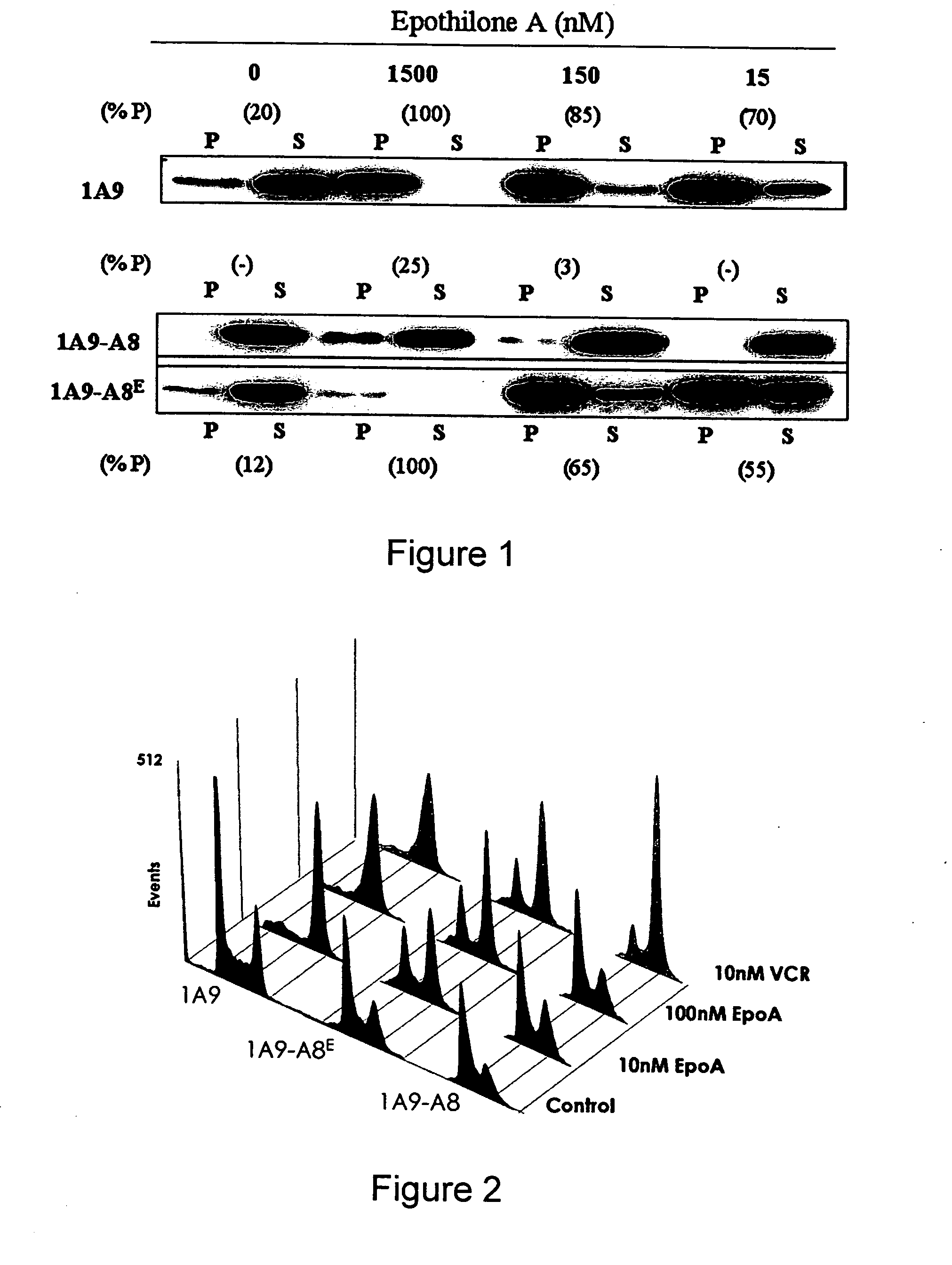
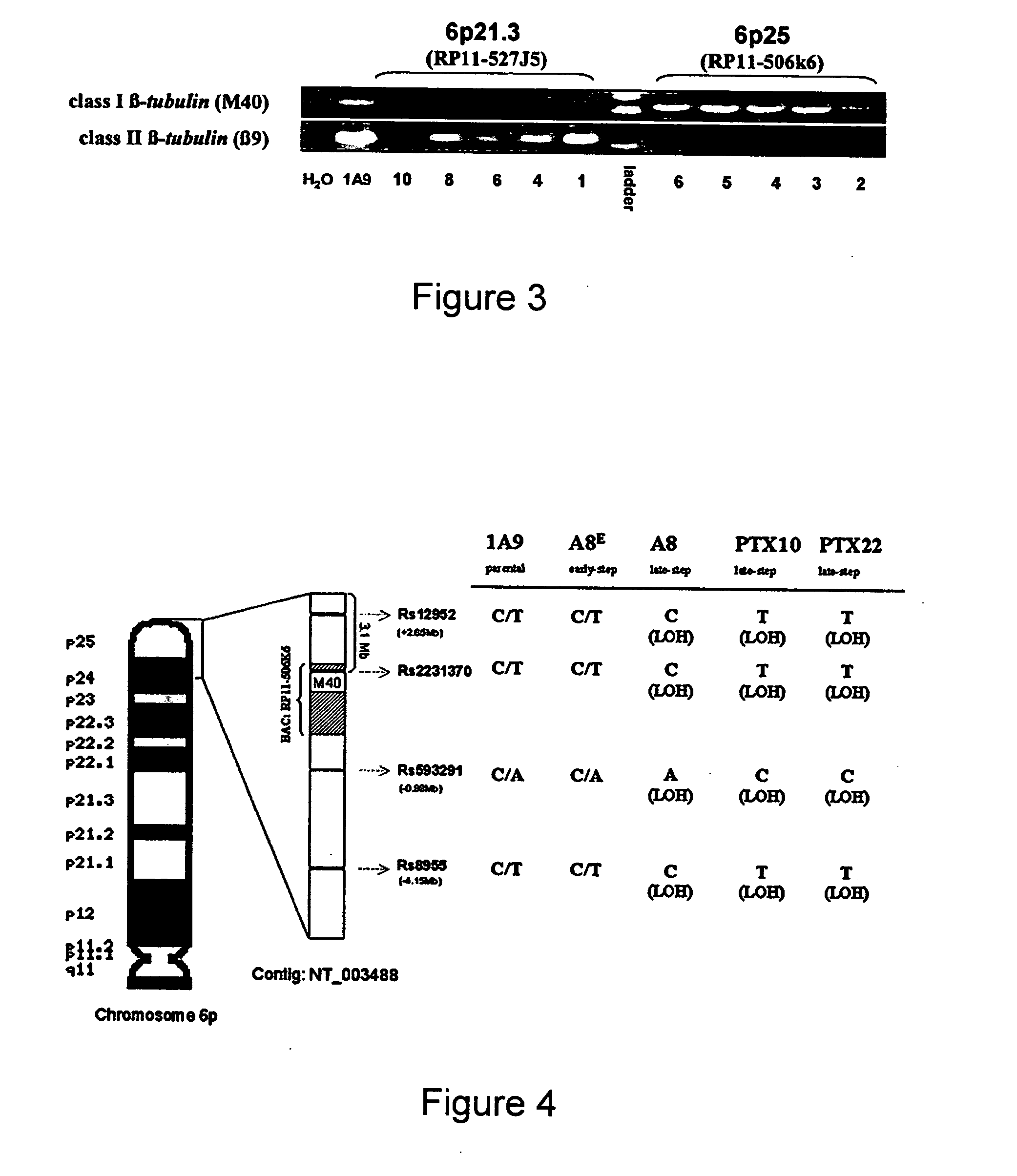
Methods of screening for resistance to microtuble-targeting drugs
InactiveUS20070190544A1Reduce the possibilityMicrobiological testing/measurementDrug treatmentΒ tubulin gene
The invention relates to methods for determining resistance or responsivity to microtubule-targeting drug treatment in cancer patients. The methods comprise obtaining a tumor cell sample from a cancer patient and analyzing DNA in the tumor cell sample to determine the presence or absence of a loss of heterozygosity (LOH) at the M40 β-tubulin gene locus within chromosomal locus 6p25, where determining LOH comprises screening for at least one mutation in the M40 β-tubulin gene that affects the binding of a microtubule-targeting drug to β-tubulin. In such methods, the presence of LOH is indicative of microtubule-targeting drug resistance in the cancer patient or of a decreased likelihood that the cancer patient will respond to therapy with a microtubule-targeting drug.
Owner:EMORY UNIVERSITY
Prognostic significance of molecular genetic aberrations on chromosome segment 11p15.5 in non-small-cell lung cancer
The present invention provides a method for detection of loss of heterozygosity (LOH) at human chromosome segment 11p15.5 in a tumor sample from a patient with non-small-cell lung cancer (NSCLC), and a method for prognosis of the patient's disease progression. The prognosis is useful for determining the course of treatment following surgical resection of tumor.
Owner:HEALTH RES INC
Method for detecting mutation and HRD scoring of tumor patient to guide medication
PendingCN112820351ACalculation speedEasy to detectDrug and medicationsBiostatisticsAllele ImbalanceWhite blood cell
The invention relates to a method for for detecting mutation and HRD scoring of tumor patient to guide medication, which comprises the following steps: detecting a tumor tissue sample and a control leukocyte sample thereof on the basis of taking a BAM file as an input file, and detecting mutation information of an HRR pathway in panels of different sample types and large-fragment chromosome abnormality HRD caused by gene recombination repair defects, the method for detecting the tumor tissue sample comprises the following steps: 1) detecting mononucleotide mutation of an HRR pathway; 2) detecting the loss of heterozygosity (LOH), telomere allele imbalance (TAI) and large fragment state transition (LST) of the HRR region, wherein the HRD score is equal to the weighted sum of LOH, TAI and LST scores; the invention provides a mutation and HRD scoring medication guidance method for tumor patients. According to the method, a traditional method that the HRD value score is calculated through whole genome detection and is not beneficial to clinical actual effect is optimized, the calculation speed is increased, the detection effect is good, the detection result is accurate, and the requirements of actual application can be well met.
Owner:江苏医联生物科技有限公司
Detection method and kit for detecting 1p/19q combination loss of heterozygosity of chromosome
PendingCN109666745AHigh sensitivityImprove featuresMicrobiological testing/measurementType frequencyAP site
The invention discloses a detection method for detecting 1p / 19q combination loss of heterozygosity of chromosome, which comprises the following steps: 1) selecting SNP sites with the same number on 1pand 19q; 2) designing an original primer, modifying a T of each original primer, closest to the 3'end but not at the tail end, into U, if the tail end of the 3'end is T, modifying a second T towardsthe 5'direction into U to obtain a multiplex amplification primer sequence; 3) synthesizing a multiplex amplification primer and dissolving and mixing the multiplex amplification primer; 4) carrying out multiplex PCR amplification; 5) processing the amplification product to enable a single chain of U to form an AP site, and then exposing the 5'phosphoric acid tail end and 3'phosphoric acid tail end of the AP site; 6) sequencing, and judging whether 1p / 19q combination loss of heterozygosity occurs on the chromosome according to SNP allelic type frequency. According to the invention, the NGS method based on multiplex amplification is used for detecting Chr1p / 19qco-LOH, not only is the detection sensitivity high, and are the specificity and the efficiency high, but also the limitation on samples is small.
Owner:CARRIER GENE TECH SUZHOU CO LTD +1
A universal platform for car therapy targeting a novel antigenic signature of cancer
PendingUS20190248869A1Peptide/protein ingredientsAntibody mimetics/scaffoldsEffector Immune CellNucleotide
A nucleic acid molecule comprising a nucleotide sequence encoding an inhibitory chimeric antigen receptor (i CAR) capable of preventing or attenuating undesired activation of an effector immune cell, wherein the i CAR comprises an extracellular domain that specifically binds to a single allelic variant of a polymorphic cell surface epitope absent from mammalian tumor cells due to loss of heterozygosity (LOH) but present at least on all cells of related mammalian normal tissue; and an intracellular domain comprising at least one signal transduction element that inhibits an effector immune cell is provided. Vectors and transduced effector immune cells comprising the nucleic acid molecule and methods for treatment of cancer comprising administering the transduced effector immune cells are further provided.
Owner:GAVISH GALILEE BIO APPL +1
Detection kit and method for Duchenne/Becker muscular dystrophy based on multiplex PCR (Polymerase Chain Reaction) trapping technique
ActiveCN108220418ASample quality requirements are lowHigh detection sensitivityMicrobiological testing/measurementDNA/RNA fragmentationDmd geneIntein
The invention discloses a multiplex PCR (Polymerase Chain Reaction) trapping primer group for a DMD (Duchenne Muscular Dystrophy) gene, a mutation detection kit for the DMD gene and a mutation detection method for the DMD gene. The inventors provide the multiplex PCR trapping primer aiming at the sequences of 79 exons and bilateral 50bp introns and 23 known intron pathogenic mutations of the DMD gene; moreover, the mutation detection method of the DMD gene is provided based on the primer group; the mutation detection method is high in detection sensitivity, good in accuracy, highly consistentin amplification efficiency and low in requirement on quality of a DNA (Deoxyribonucleic Acid) sample, and is used for solving the problem that the complete detection on the mutation information, particularly on loss of heterozygosity / repetition, of the DMD gene is difficultly carried out in one time by adopting a multiplex PCR trapping technique in the prior art.
Owner:CAPITALBIO GENOMICS
Method and detection kit to judge whether solid tumors are suited for immunotherapy
ActiveCN108624650AIncreased objective response rateMicrobiological testing/measurementMaterial analysisPeripheral blood mononuclear cellCD16
The invention provides a detection kit to judge whether solid tumors are suited for immunotherapy. The detection kit includes detection reagents for detecting the indexes: tumor mutation load of peripheral blood circulating tumor DNA, HLA typing, HLA state of loss of heterozygosity, PD-L1 membrane expression positive tumor cell percentage, infiltration level of CD8+ immune cells, infiltration level of FOXP3+ immune cells, mRNA expression score of T-cell inflammation related genes, percentage of CD14+CD16-HLA-DR+ monocytes in peripheral blood mononuclear cells, and micro-satellite instability.The invention also provides, correspondingly, a method to judge whether solid tumors are suited for immunotherapy. The kit and method herein can be used to screen patients with solid tumors so as to provide significantly increased objective response rate and have a promising application prospect in terms of clinical screening of patients with solid tumors suited for immunotherapy.
Owner:LEPU MEDICAL TECH (BEIJING) CO LTD
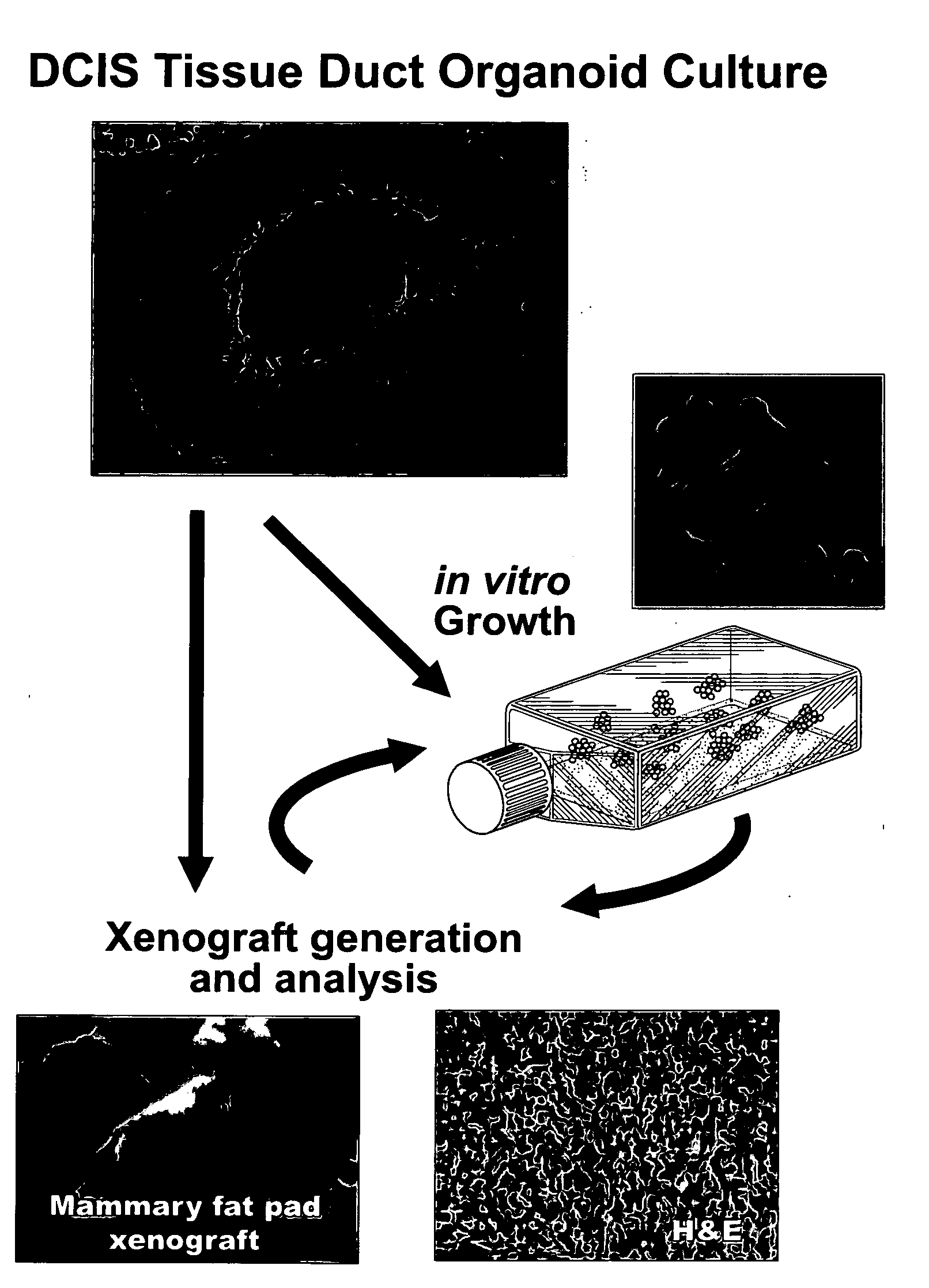
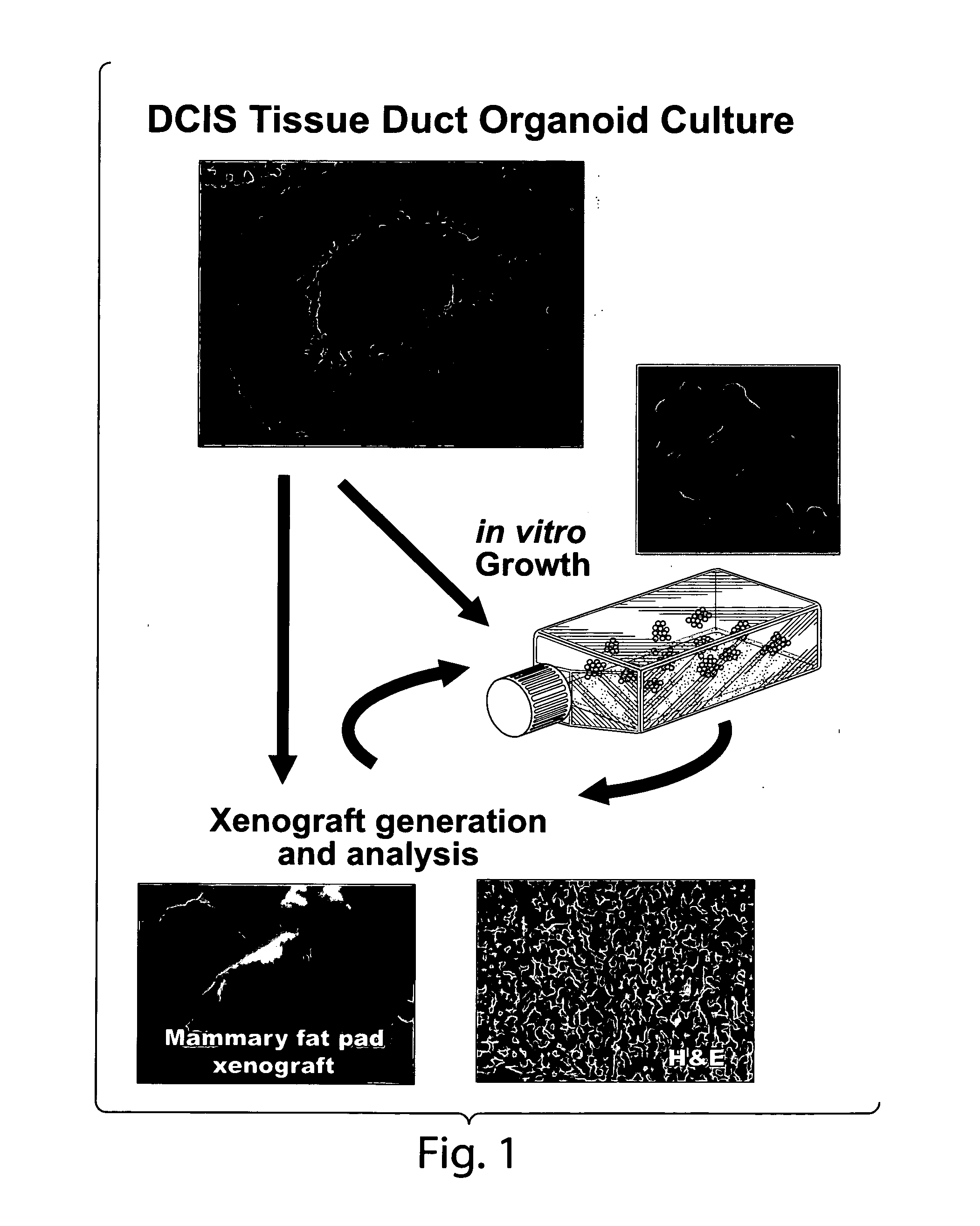
Genetic alteration associated with pre-malignant cancer
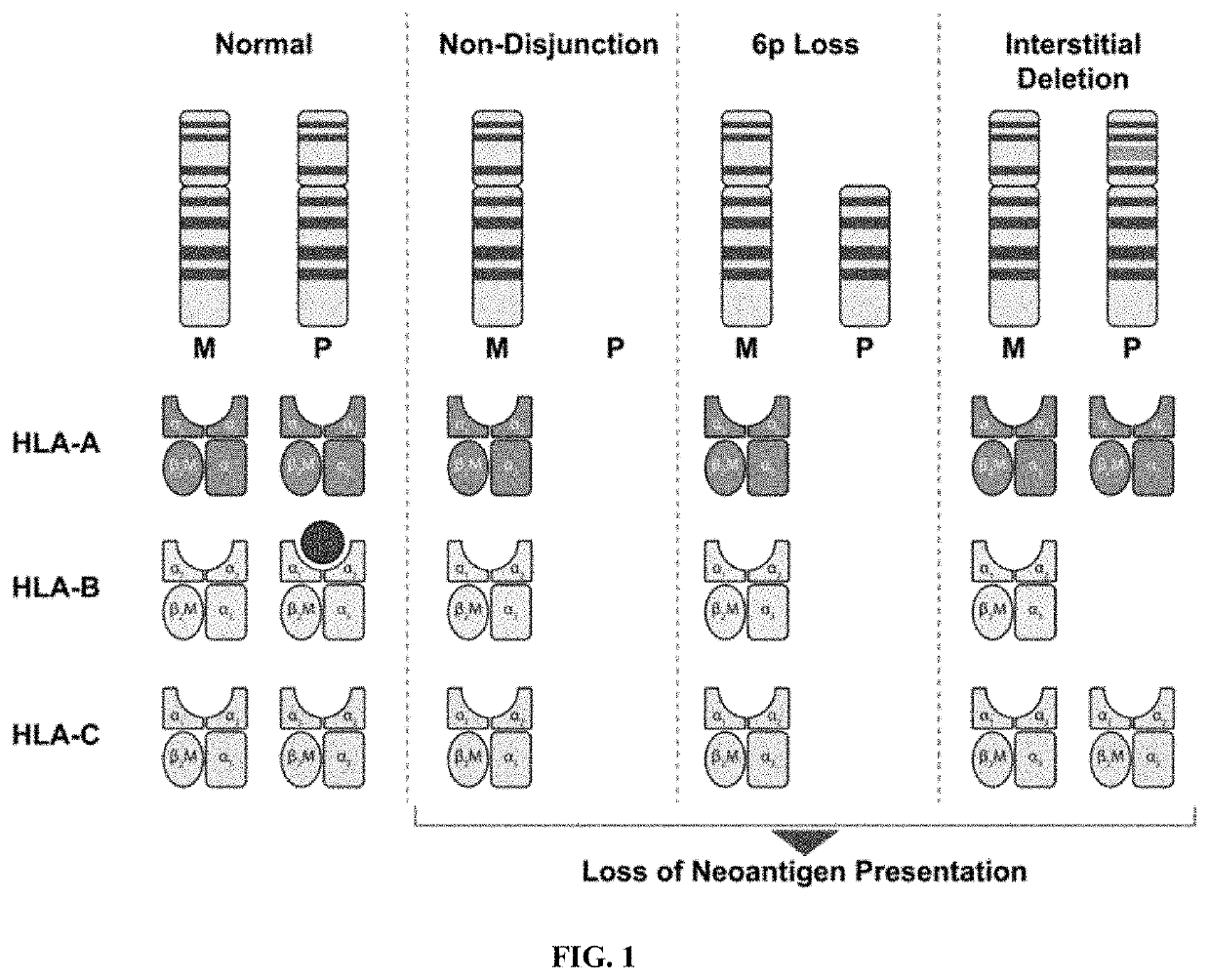
Described herein are progenitor cancer cells and cell lines isolated from human breast ductal carcinoma in situ (DCIS) lesions and the uses of these cells or cell lines in drug design, drug screening, and monitoring in vivo therapy. The DCIS malignant precursor cells or cell lines are epithelial in origin, are positive for markers of autophagy, show at least one genetic difference from normal cells of said fragment, form 3-D tube-like structures or ball aggregates, or are inhibited in formation of 3-D structures and migration by treatment with chloroquine. In one embodiment, there is a loss of heterozygosity (LOH) that is narrowly confined to a region of chromosome 6p (6p21.1-6p12.3) that contains the SUPT3H gene.
Owner:GEORGE MASON UNIVERSITY
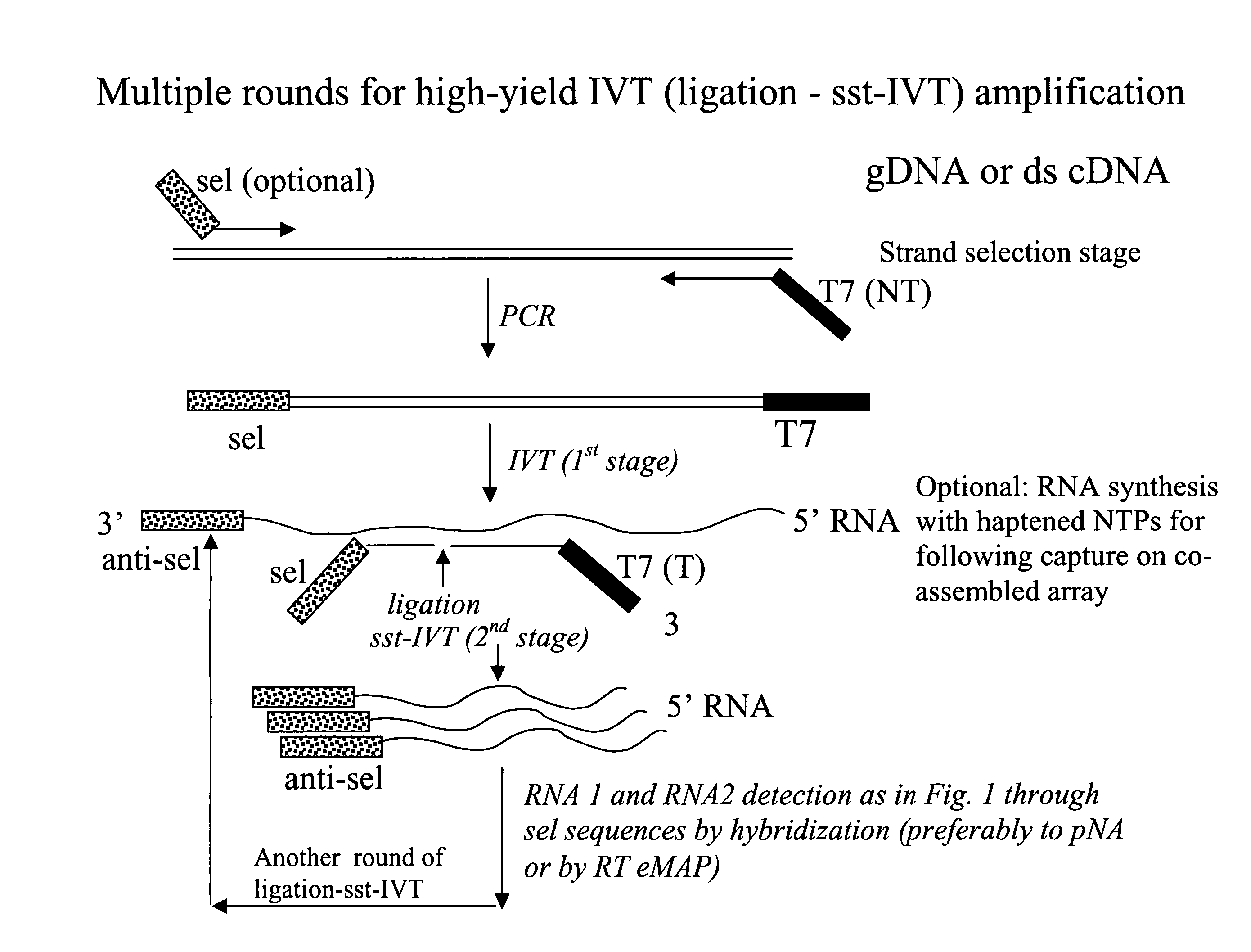
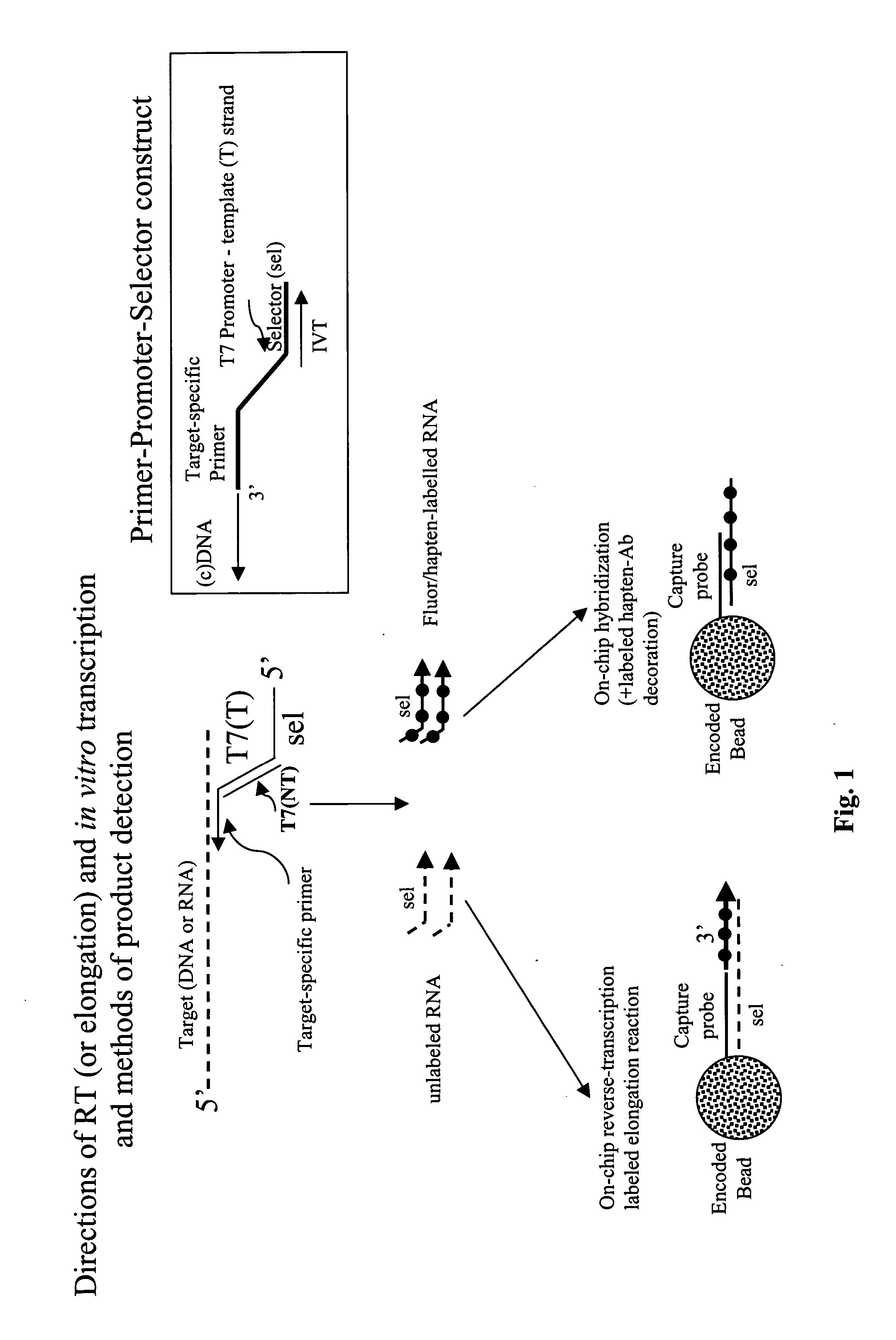
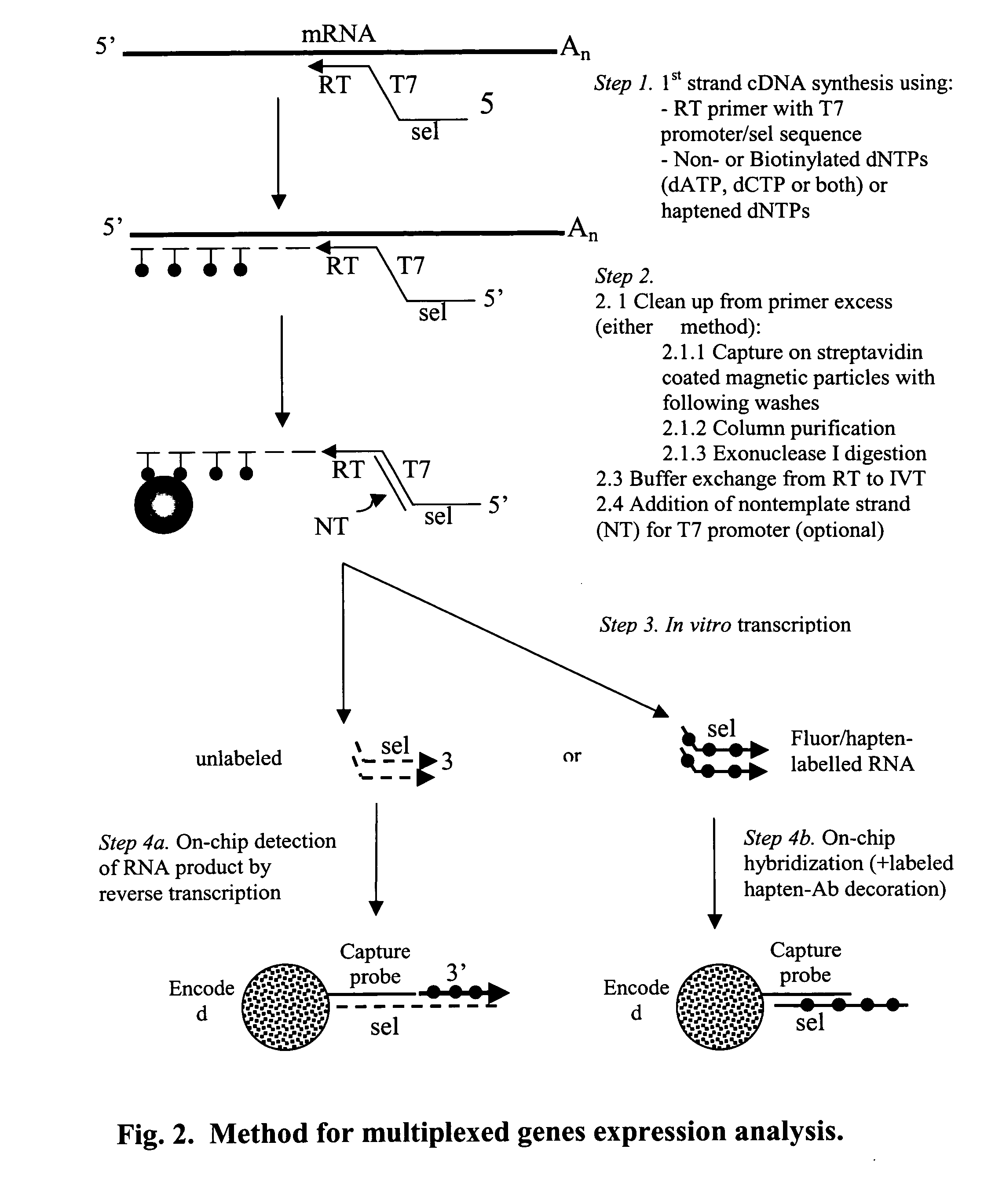
Message abundance and allele copy number determination using IVT with single-stranded primer-promoter-selector constructs
ActiveUS20070065864A1Improve efficiencyImprove performanceMicrobiological testing/measurementFermentationSingle strandHeterozygosity Loss
Disclosed is a single stranded primer-promoter-selector construct comprising (in 3′ to 5′ orientation) a primer subsequence annealing to the target, a T7 or other promoter subsequence (the template strand), and a selector subsequence. The primer can be extended by template mediated elongation, including reverse transcription, or ligation to another oligonucleotide. The promoter sequence is oriented to direct the in-vitro transcription (IVT) opposite to that of primer extension, where the selector subsequence serves as a template for IVT. The selector is associated with the target subsequence of interest and it, and the amplified product are unique subsequences, dissimilar to other sequence present in the sample. The construct's is useful for determination of the presence and relative abundance of designated subsequences in the sample, multiplex gene expression analysis, multiplex allele counting, determination of polymorphic / mutation site, and loss of heterozygosity.
Owner:BIOARRAY SOLUTIONS
Diagnostic methods involving loss of heterozygosity
InactiveUS20130079423A1Short overall survivalExact numberBiocideLibrary screeningDiseaseMolecular analysis
The invention relates generally to methods of molecular analysis and particularly to methods of using genetic copy number variations and loss of heterozygosity in the characterization and treatment of disease.
Owner:MYRIAD GENETICS
Kit and method for detecting chromosome heterozygosity loss on basis of amplicon sequencing
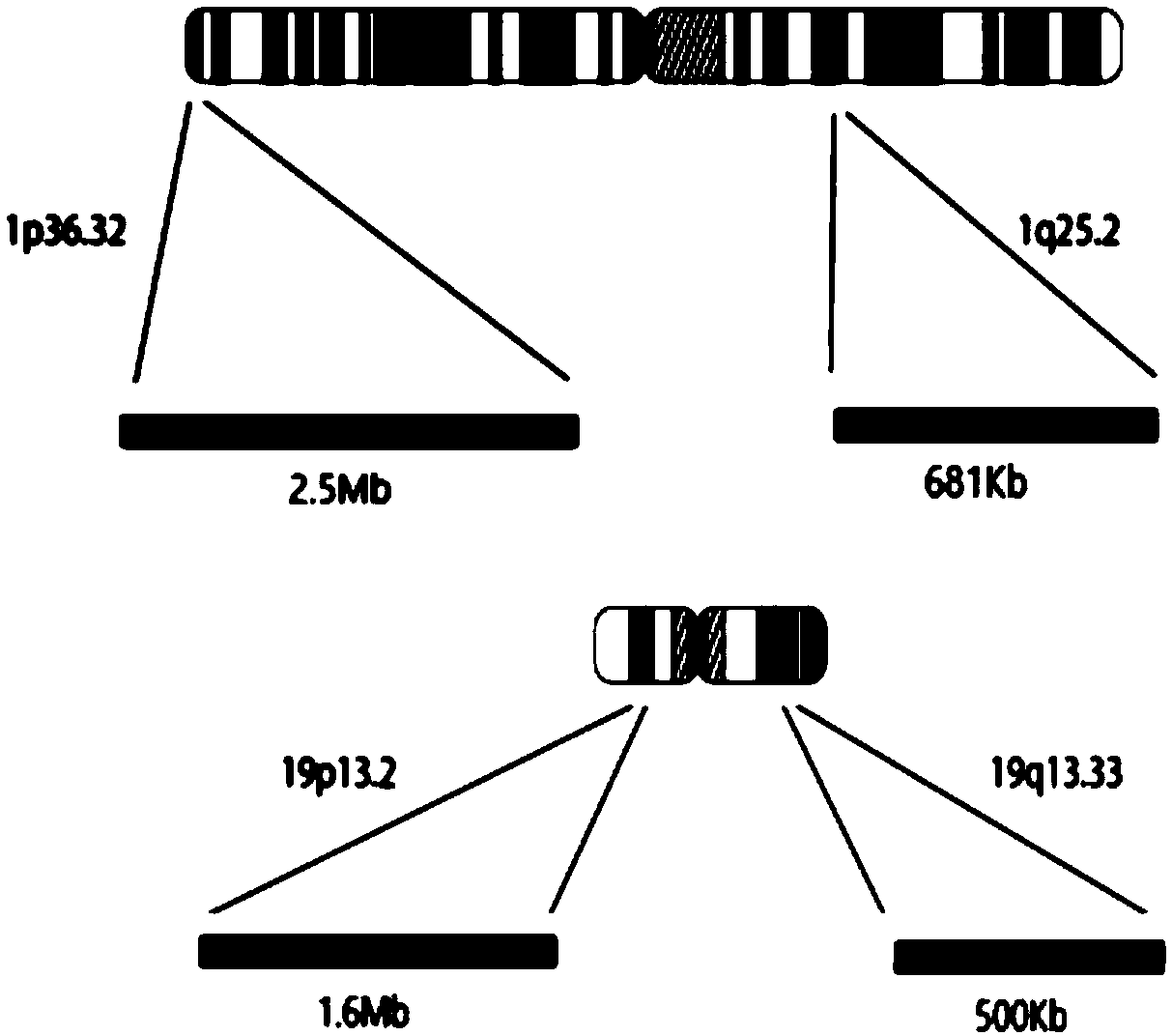
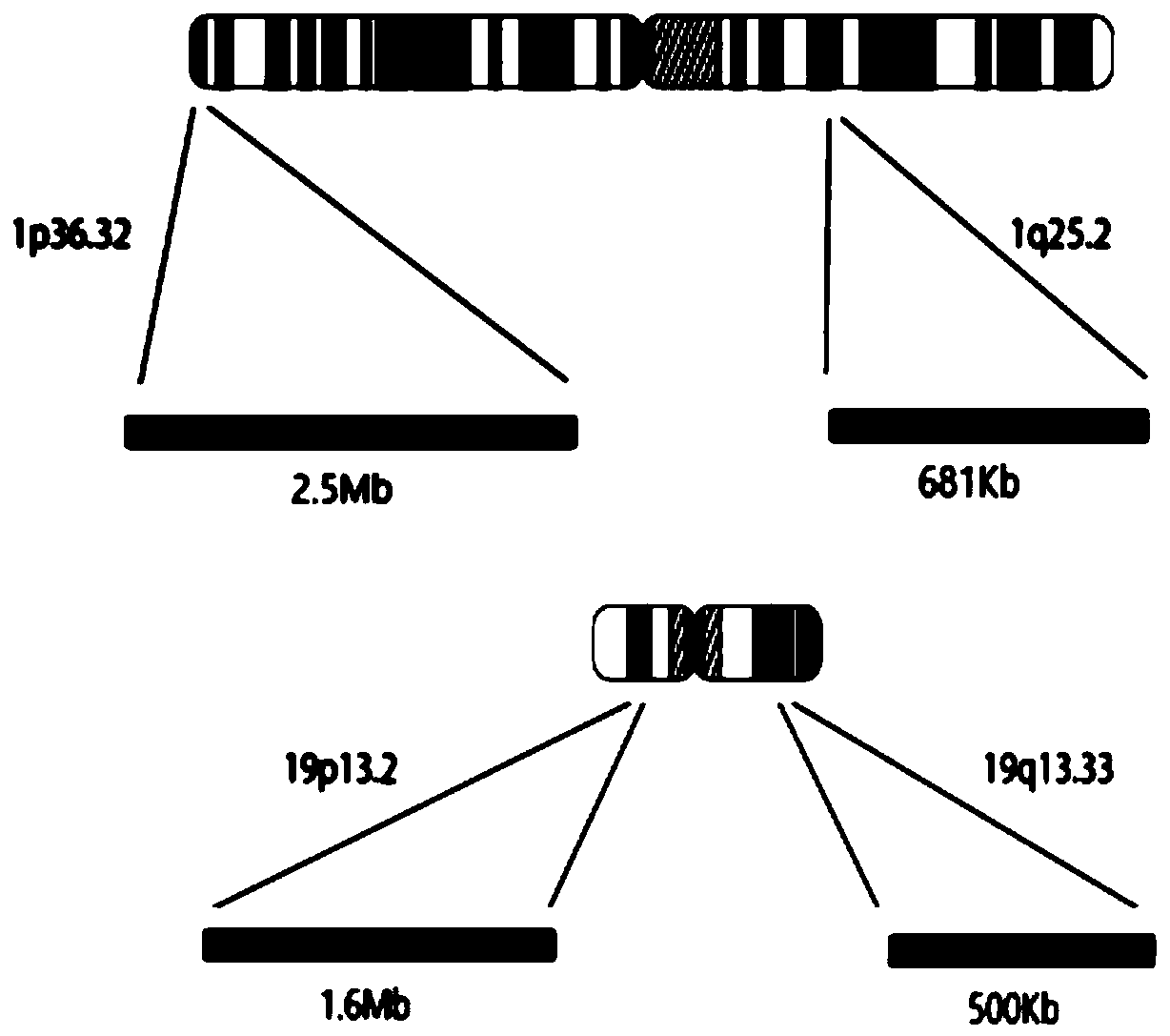
ActiveCN109504770AComprehensive, true and accurate responseReduce the amount of solutionMicrobiological testing/measurementDNA/RNA fragmentationNucleic acid detectionChromosome regions
The invention discloses a kit and method for detecting chromosome heterozygosity loss on the basis of amplicon sequencing, belongs to the field of molecular biology nucleic acid detection, and relatesto the kit and method for detecting chromosome heterozygosity loss by means of amplicon sequencing. The kit contains a mixture of 14 groups of primer pairs used for detecting a 1p36.32 region of a human chromosome 1, 6 groups of primer pairs used for detecting a 1q25.2 region of the human chromosome 1, 14 groups of primer pairs used for detecting a 19p13.2 region of a human chromosome 19 and 7 groups of primer pairs used for detecting a 19q13.33 region of the human chromosome 19, 20 kinds of tag primers used for distinguishing samples, a GC stabilizer, a PCR enzyme mixture, purifying magneticbeads, a quality control standard substance and ddH2O. Through two-time PCR amplification and two-time product purification and sequencing, through a ratio of READS(1p) / READS(1q) to READS(19q) / READS(19p), the heterozygosity loss state of 1p19q in the samples is analyzed. By adopting the method, the variation situation of the 1p19q can be reflected comprehensively, truly and accurately, and gene variation of the 96 samples can be subjected to high-throughput detection and analysis simultaneously, so that the detection cost is greatly reduced.
Owner:艾普拜生物科技(苏州)有限公司
Knockout mouse for the tumor suppressor gene ANX7
InactiveUS7504223B2Inhibit cell proliferationVirusesMicrobiological testing/measurementKnockout animalOncology
This invention provides methods, including a method of assessing the prognosis of a breast cancer patient, comprising assaying for loss of heterozygosity at the 10q21 region of the genome of the patient, a method of identifying a probability that a patient with breast cancer has metastasized breast cancer, a method of determining a survival probability of a patient with breast cancer, and a method of identifying a probability that a patient with prostate cancer has a severe form of prostate cancer. This invention also provides assay complexes, including assay complexes which comprise at least one prostate tissue sample or tissue sample extract, an antibody that specifically binds ANX7, and a label, or which comprise at least one breast tissue sample or tissue sample extract, an antibody that specifically binds ANX7, and a label.
Owner:THE HENRY M JACKSON FOUND FOR THE ADVANCEMENT OF MILITARY MEDICINE INC
Universal platform for car therapy targeting a novel antigenic signature of cancer
PendingUS20210230251A1Peptide/protein ingredientsAntibody mimetics/scaffoldsAntigenEffector Immune Cell
A nucleic acid molecule comprising a nucleotide sequence encoding an inhibitory chimeric antigen receptor (i CAR) capable of preventing or attenuating undesired activation of an effector immune cell, wherein the i CAR comprises an extracellular domain that specifically binds to a single allelic variant of a polymorphic cell surface epitope absent from mammalian tumor cells due to loss of heterozygosity (LOH) but present at least on all cells of related mammalian normal tissue; and an intracellular domain comprising at least one signal transduction element that inhibits an effector immune cell is provided. Vectors and transduced effector immune cells comprising the nucleic acid molecule and methods for treatment of cancer comprising administering the transduced effector immune cells are further provided.
Owner:GAVISH GALILEE BIO APPL +1
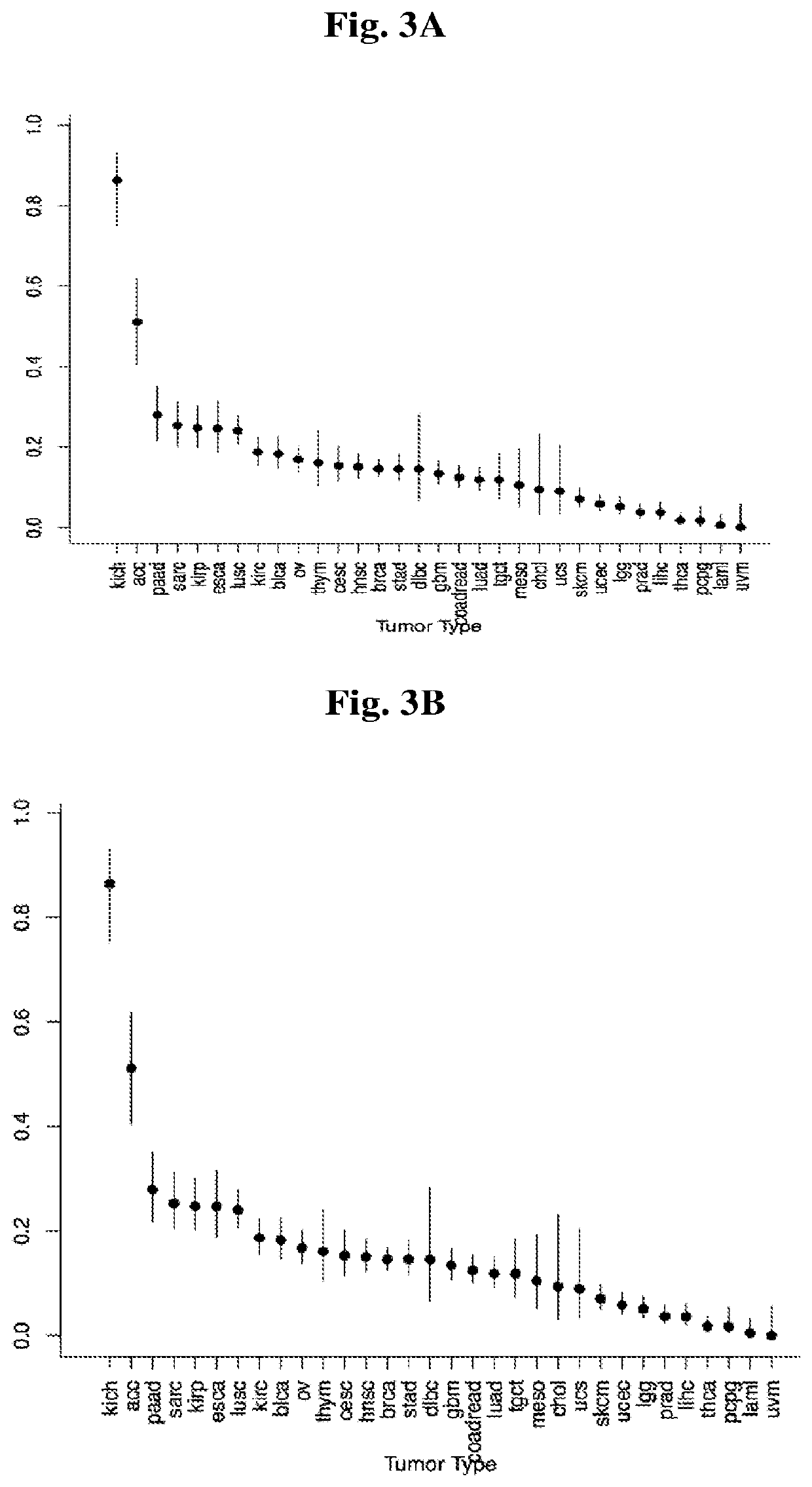
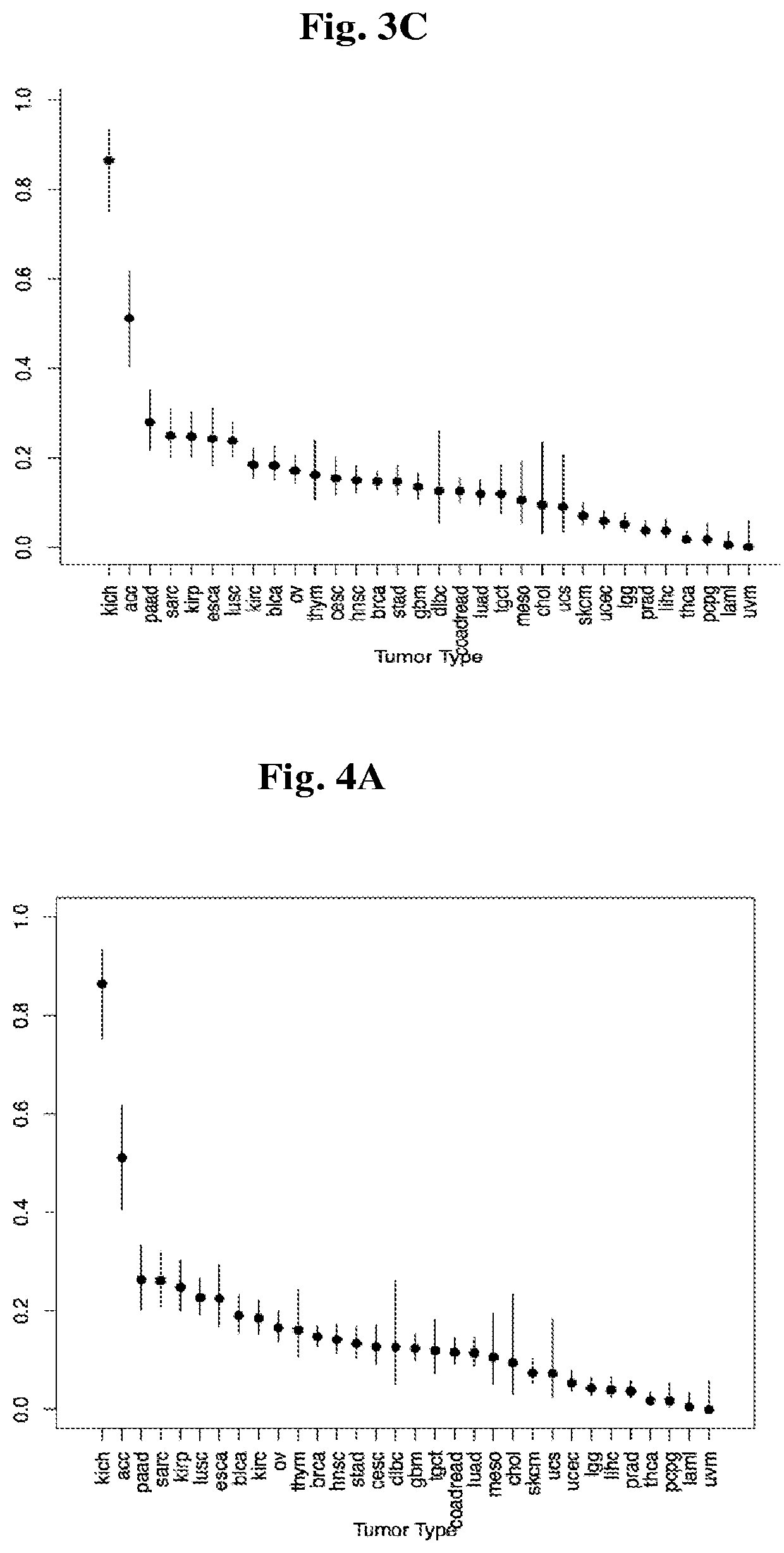
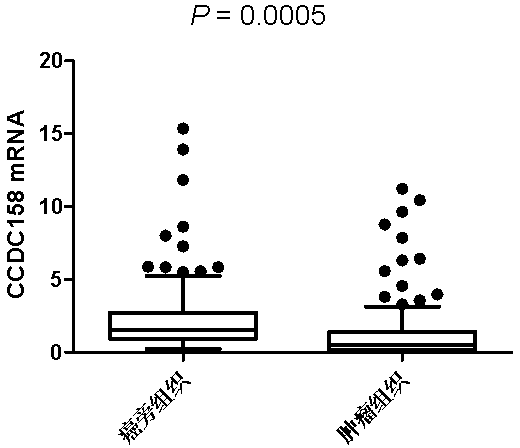
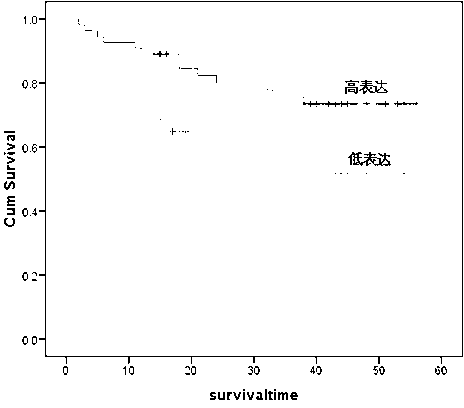
Application of CCDC158 gene in preparation of liver cancer treatment or diagnosis medicament
InactiveCN103290112ASlow growth rateMicrobiological testing/measurementGenetic material ingredientsNeoplasm diagnosisPharmaceutical drug
The invention discloses the application of CCDC158 gene in preparation of liver cancer treatment or diagnosis medicament. The CCDC158 causes the loss of heterozygosity and reduced expression in a liver cancer tissue, and moreover has significant correlation with the prognosis of the liver cancer; and in the liver cancer cells, the cancer cell growth and the colony-forming ability can be reduced and the cancer cell apoptosis is induced by upregulating the expression of the CCDC158. The above shows that the gene not only has a significant effect on the occurrence and the development of the liver cancer, but also has a function of inhibiting the biology of cancer cells. Therefore, the CCDC158 can be used for preparing a new anti-tumor medicament and a kit for tumor diagnosis and prognosis evaluation.
Owner:SUN YAT SEN UNIV
Method of Detecting Precancerous Lesions
The present invention relates to methods of detection of precancerous lesions and / or cancer. The present invention also relates to the presence of DNA replication stress in precancerous lesions. The present invention further relates to the detection of loss of heterozygosity at common fragile sites and phosphorylated substrates of DNA damage activated kinases.
Owner:HALAZONETIS THANOS D
Gene family with transformation modulating activity
InactiveUS6930175B1Low percentagePeptide/protein ingredientsMicrobiological testing/measurementAbnormal tissue growthSuppressor mutation
pp32 is a member of a highly conserved family of differentiation-regulated nuclear proteins that is highly expressed in nearly all human prostatic adenocarcinomas of Gleason Grade≧5. This contrasts with the low percentage of prostate tumors that express molecular alterations in proto-oncogens or demonstrate tumor suppressor mutation or loss of heterozygosity. By analysis of specimens of human prostatic adenocarcinoma and paired adjacent normal prostate from three individual patients, the inventors have shown that normal prostate continues to express normal pp32, whereas three of three sets of RT-PCR-amplified transcripts from prostatic adenocarcinomas display multiple cancer-associated coding sequence changes. The cancer-associated sequence changes appear to be functionally significant. Normal pp32 exerts antineoplastic effects through suppression of transformation. In contrast, cancer-associated pp32 variants augment, rather than inhibit, transformation.
Owner:THE JOHNS HOPKINS UNIVERSITY SCHOOL OF MEDICINE
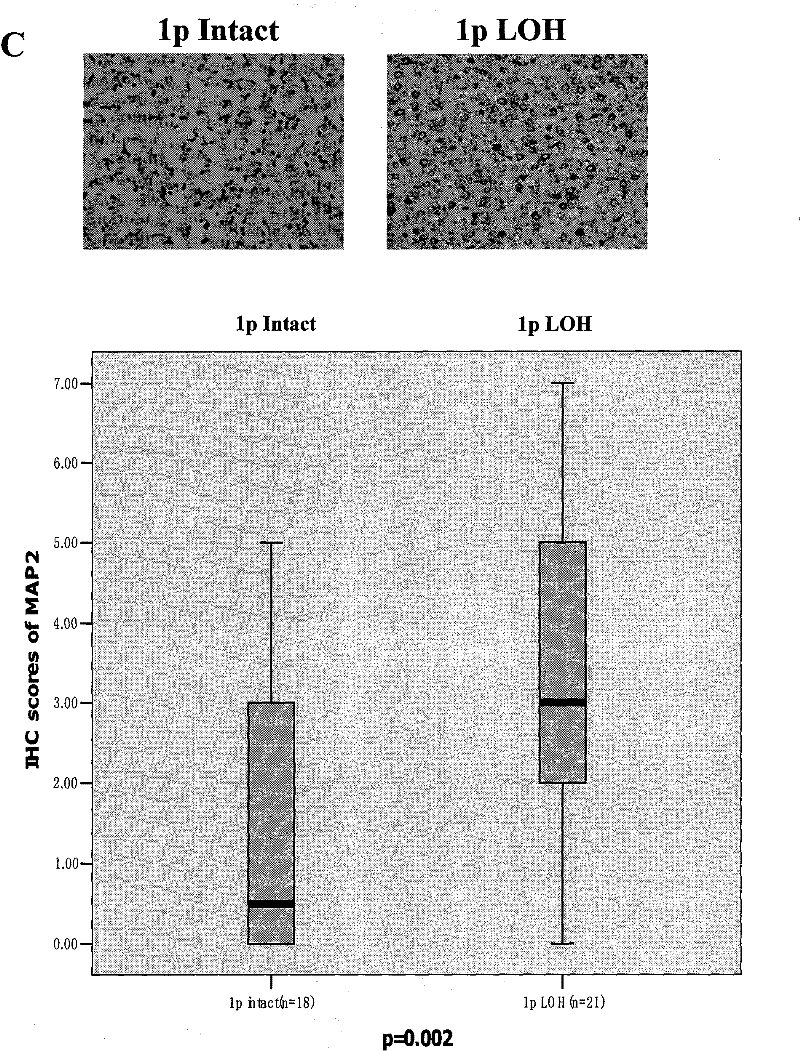
Oligoden droglioma (OG) marker MAP2 (microtubuleassociated protein 2) protein and use thereof
InactiveCN102408478AMaterial analysis by electric/magnetic meansBiological testingOligodendroglial TumorFhit gene
The invention belongs to the field of biotechnology and medicine, relates to oligoden droglioma (OG) markers, and particularly relates to a protein marker MAP2 (microtubuleassociated protein 2) protein which can differentiate OG that clinically has sensitivity to chemotherapy and 1P loss of heterozygosity (1P LOH), and use thereof. According to the invention, the protein marker MAP2 protein which can differentiate the OG that clinically has sensitivity to chemotherapy and 1P loss is determined through the iTRAQ-LC-MS / MS technology. The MAP2 protein has 90% accuracy for the prognosis of the 1P loss. The marker not only can explain the mechanism for the sensitivity to chemotherapy of an OG sample with the 1P LOH, but also can prognose the sensitivity to chemotherapy of an OG patient with the 1P LOH through a decision tree model. The MAP2 protein can be further used for preparing a clinical kit so as to offer help for the diagnosis, the prognosis and the treatment of the OG.
Owner:FUDAN UNIV
Method of improving prediction of response for cancer patients treated with immunotherapy
PendingUS20200157642A1Easy to predictReliable detectionMicrobiological testing/measurementRegimenOncology
A method of determining a therapeutic regimen in a patient with cancer comprising determining in a sample from the patient the tumor mutation burden (TMB) and loss of heterozygosity (LOH), wherein high TMB in combination with no LOH is indicative of a positive outcome when treated with a checkpoint inhibitor and high TMB with LOH is indicative of a poor outcome, is provided herein.
Owner:PERSONAL GENOME DIAGNOSTICS INC
A kit and method for detecting chromosome heterozygosity loss based on amplicon sequencing
ActiveCN109504770BComprehensive, true and accurate responseReduce the amount of solutionMicrobiological testing/measurementDNA/RNA fragmentationNucleic acid detectionAmplicon sequencing
The invention discloses a kit and method for detecting chromosome heterozygosity loss on the basis of amplicon sequencing, belongs to the field of molecular biology nucleic acid detection, and relatesto the kit and method for detecting chromosome heterozygosity loss by means of amplicon sequencing. The kit contains a mixture of 14 groups of primer pairs used for detecting a 1p36.32 region of a human chromosome 1, 6 groups of primer pairs used for detecting a 1q25.2 region of the human chromosome 1, 14 groups of primer pairs used for detecting a 19p13.2 region of a human chromosome 19 and 7 groups of primer pairs used for detecting a 19q13.33 region of the human chromosome 19, 20 kinds of tag primers used for distinguishing samples, a GC stabilizer, a PCR enzyme mixture, purifying magneticbeads, a quality control standard substance and ddH2O. Through two-time PCR amplification and two-time product purification and sequencing, through a ratio of READS(1p) / READS(1q) to READS(19q) / READS(19p), the heterozygosity loss state of 1p19q in the samples is analyzed. By adopting the method, the variation situation of the 1p19q can be reflected comprehensively, truly and accurately, and gene variation of the 96 samples can be subjected to high-throughput detection and analysis simultaneously, so that the detection cost is greatly reduced.
Owner:艾普拜生物科技(苏州)有限公司
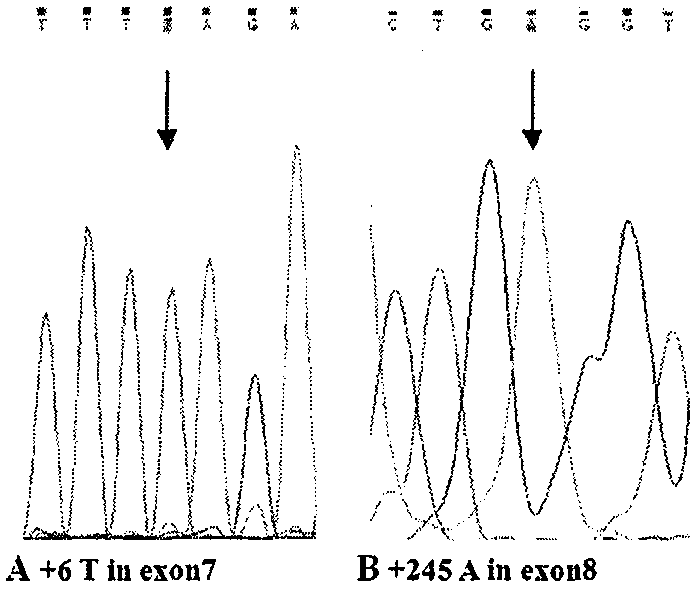
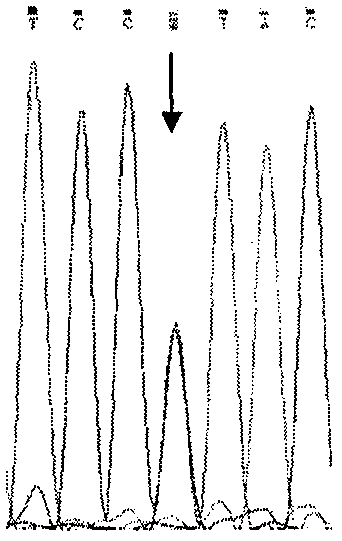
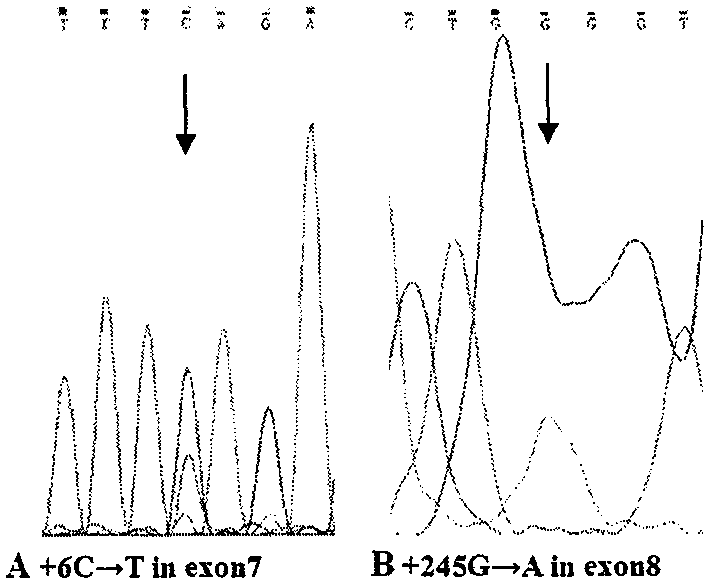
Simple and convenient method and process for detecting SMA by DNA sequencing
PendingCN110066860AGene level clearSimple methodMicrobiological testing/measurementDiseaseMedical unit
The invention relates to a simple and convenient method and specific process for detecting SMA. The key point of the technical scheme is that DNA sequencing is carried out on SMN intron 6-exon 8 in the PCR products; if exons 7 and 8 of SMN1 are homozygous deletions, only homozygous peaks of +6T on exon 7 of SMN2 and +245A on exon 8 of SMN2 are found; the method can be used for screening the molecular background of the copy number of the same SMN2 and the phenotypic deviation of the SMA; the +6 position on the exon 7 and +245 position on exon 8 of SMN appear nesting peak; if the phenotype is normal, the subject is normal people; if the subject is a patient, the point mutation of SMN1 can be preliminarily screened. The main application is as follows: the gene level of 98.6 percent of patients can be determined; the molecular background of the same SMN2 copy number and SMA phenotypic deviation can be screened; the remaining few SMA patients with loss of heterozygosity and point mutation are able to screen for point mutations in SMN1. The method is very suitable for basic medical units and is beneficial to further research on the mechanism of the disease.
Owner:刘维亮 +3
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com