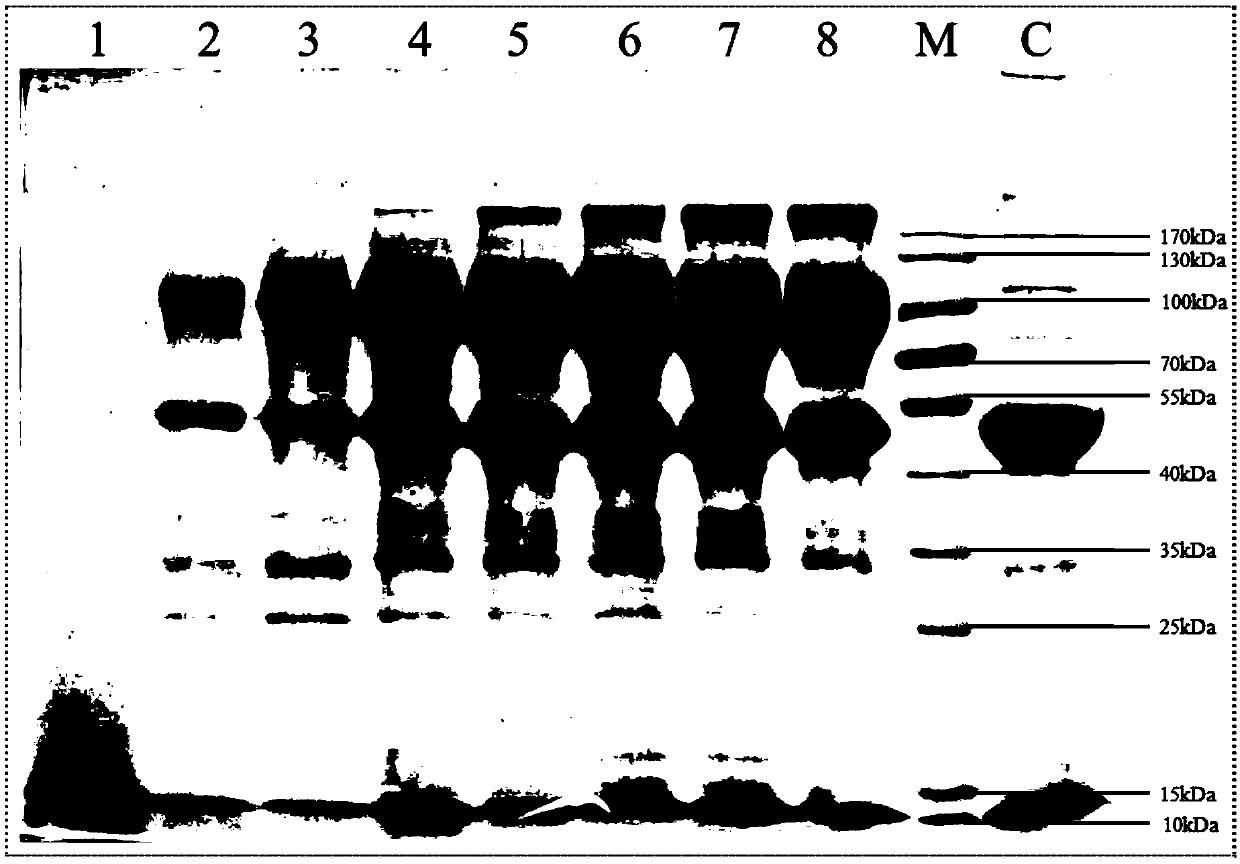
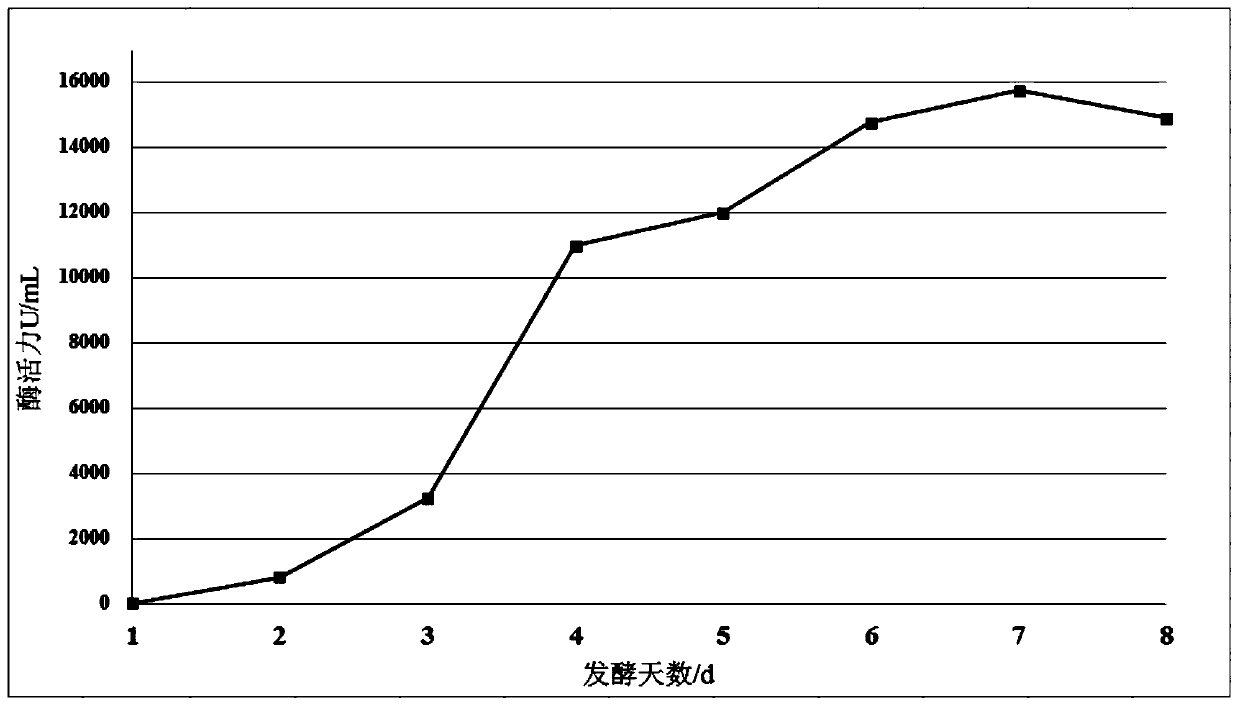
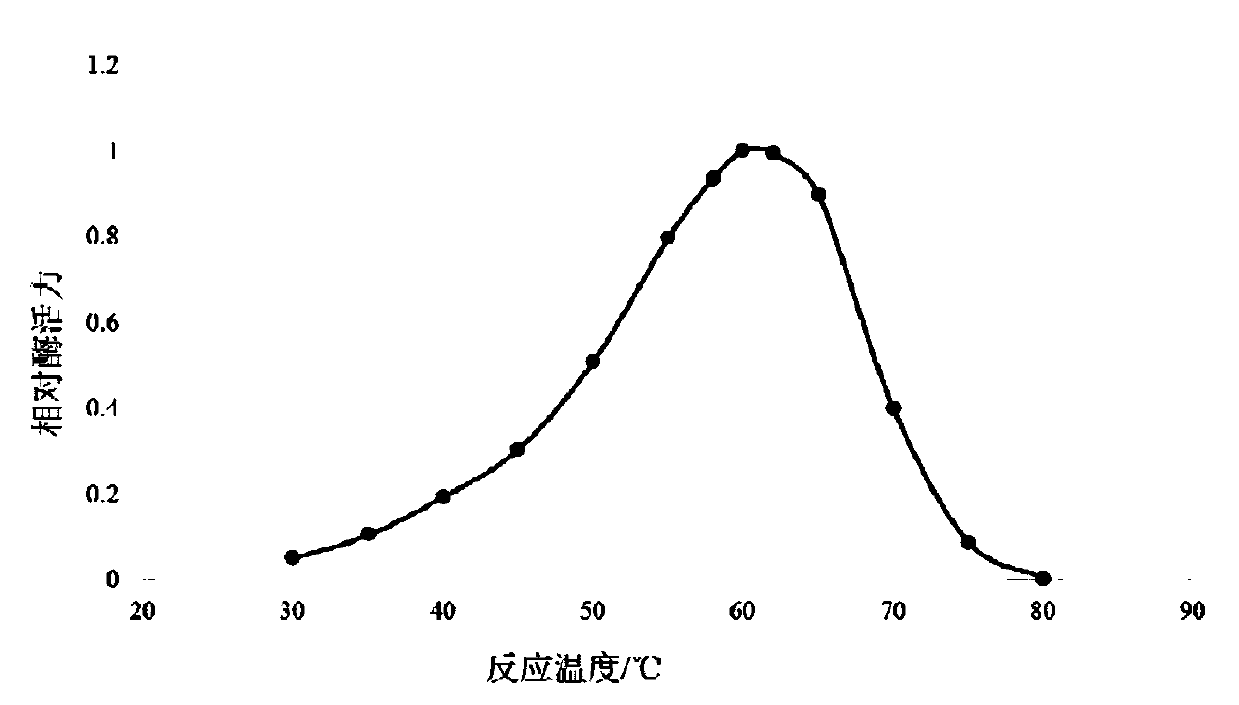
Optimized high-temperature acid trehalinase TreMT1 and coding gene and application thereof
A technology of trehalase and coding gene, which is applied in the field of genetic engineering, can solve the problems of low expression level of recombinant trehalase, achieve good stability, improve utilization rate, and increase yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1, the construction of universal expression vector
[0048] Using the Aspergillus niger genome as a template, perform PCR amplification (Prime STAR premix HS (purchased from takara company)), using primer Ttef-fw (as shown in SEQ ID NO.8) and primer Ttef-rev (as shown in SEQ ID NO. 9) to amplify the tef terminator (as shown in SEQ ID NO.3), use primer amyA-fw (as shown in SEQ ID NO.12) and primer amyA-rev (as shown in SEQ ID NO.13) to amplify Increase the last 1000bp of the gene encoding neutral amylase amyA (shown in SEQ ID NO.5), use the primer pyrG-fw (shown in SEQ ID NO.10) and primer pyrG-rev (shown in Aspergillus nidulans genome) as a template As shown in SEQ ID NO.11) to amplify the pyrG marker (as shown in SEQ ID NO.4), the obtained PCR products are respectively named as Ttef, amyA, pyrG (respectively as SEQ ID NO.3, such as SEQ ID NO.5 , shown in SEQ ID NO.4), using the NEBuilder HiFiDNA Assembly Cloning Kit kit, the four fragments of Ttef, pyrG, am...
Embodiment 2
[0049] Embodiment 2, containing target gene expression vector construction
[0050] Using the optimized nucleotide sequence of the synthetic TreMT1 gene and the Aspergillus niger genome as templates, amplified with primers Tre-fw (shown in SEQ ID NO.16) and primers Tre-rev (shown in SEQ ID NO.17) Obtain the TreMT1 gene sequence (as shown in SEQ ID NO.2), amplify with primer PamyA-fw (as shown in SEQ ID NO.14) and primer PamyA-rev (as shown in SEQ ID NO.15) to obtain Aspergillus niger Amylase promoter PamyA sequence (as shown in SEQ ID NO.7). The two PCR-amplified PCR fragments were connected with the linearized universal expression vector by fusion PCR. The ligation product was transformed into Escherichia coli Match1T1 (purchased from takara company) competent, after 12 hours of culture at 37°C, the transformants were picked and placed in liquid LB+Amp (final concentration 100 μg / ml) medium, and cultured at 37°C with a shaker speed of 200rpm After 12 hours, electrophoresis ...
Embodiment 3
[0051] Embodiment 3, transformation of expression vector pMD20-PamyA-TreMT1-Ttef-pyrG-amyA plasmid in Aspergillus niger
[0052] Prepared according to the procedure provided in (Gomi K, Iimura Y, Hara S. Integrative transformation of Aspergillus oryzae with a plasma containing the Aspergillus nidulans argB gene [J]. Agricultural and biological chemistry, 1987, 51(9): 2549-2555.) The protoplasts of the host fungus Aspergillus niger (ΔpyrG), the expression vector pMD20-PamyA-TreMT1-Ttef-pyrG-amyA plasmid obtained above was transformed into the protoplasts, and the hyperosmotic CD medium (comprising 1M sucrose, 0.3% (w / v)NaNO 3, 0.2% (w / v) KCl, 0.05% (w / v) MgSO 4 .7H 2 O, 0.1% (w / v) K 2 HPO 4 .3H 2 O, 0.001% (w / v) FeSO 4 .7H 2 O, 2% (w / v) agar powder, pH 5.5, the unit of w / v is g / mL), put it into a 30°C incubator, and observe the growth of transformants after 5 days.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com