A strain of Pseudomonas fluorescens sc3 and its application in the control of bacterial soft rot in crops
A Pseudomonas fluorescens, bacterial technology, applied in the field of Pseudomonas fluorescens SC3 and its application in the control of bacterial soft rot in crops, can solve the problems of field trials, bacterial resistance, and water pollution and other problems, to achieve the effect of reducing the occurrence of bacterial soft rot in crops, good inhibitory effect, and good control effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Isolation and Screening of Pseudomonas SC3
[0037] 1. Strain isolation
[0038] Sample collection: In the rice fields of Sichuan Agricultural University, the plants with rotten bases of rice seedlings were pulled out, together with soil samples from the roots of the rice, stored in bags, and brought back to the laboratory for strain isolation.
[0039] Isolation of bacterial strains: Soil samples were isolated by dilution coating plate method. Specifically, after the collected soil samples were thoroughly mixed by hand, 4.0 g was weighed, soaked in 20 mL of sterile water and shaken, and incubated at 28°C for 20 min on a shaker at 180 rpm for gradient dilution. Take the dilution factor as 10 5 -10 9 100 µL of the diluted solution was spread on the LB plate, air-dried at room temperature, sealed, and incubated upside down in a 28°C incubator for 24 h. Pick a single colony grown on the LB plate, label it, place it in a 2 mL centrifuge tube containing 500 µL ...
Embodiment 2
[0041] Example 2 Identification of Pseudomonas SC3
[0042] 1. Morphological identification
[0043]Strain SC3 is Gram-negative; the colonies on LB nutrient medium are light yellow, round, slightly convex in the middle, smooth in surface and neat in edge; diffuse turbidity in liquid medium, liquefaction of gelatin, production of acid, gas, Produces oxidase, catalase, and no amylase.
[0044] 2. Molecular identification
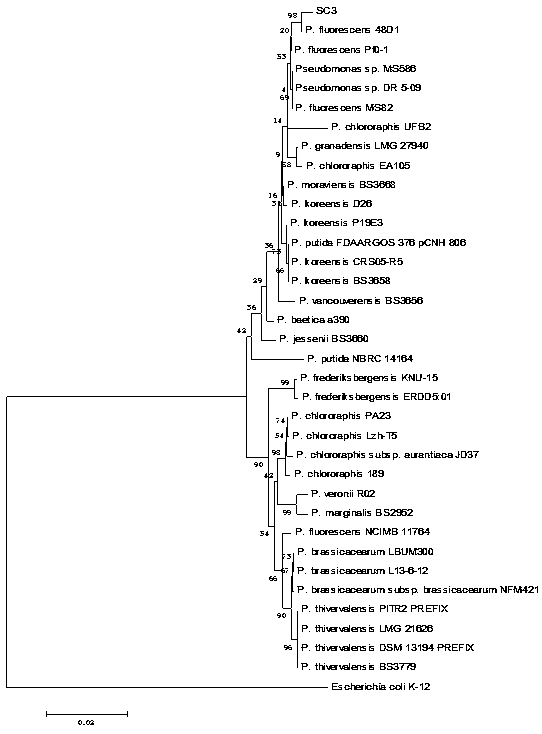
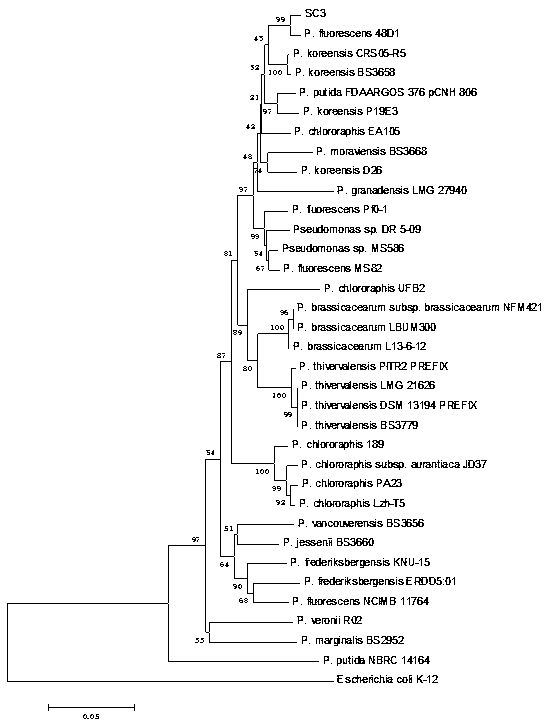
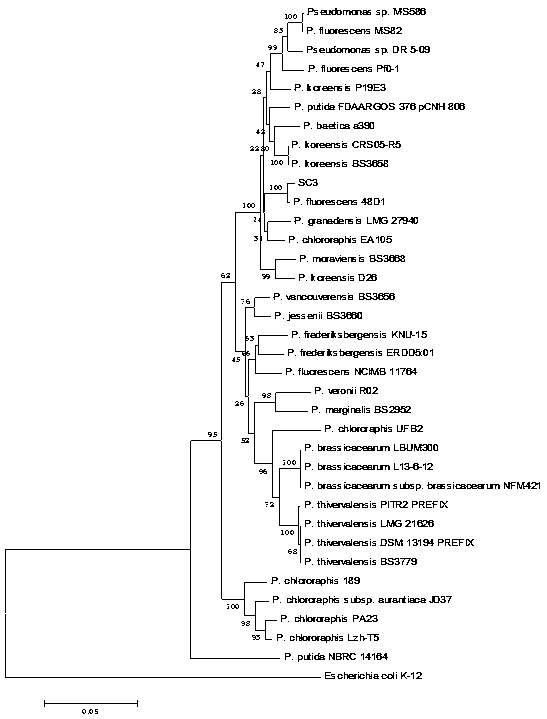
[0045] In order to clarify the taxonomic status of the strain SC3 obtained in Example 1, we used the method of multilocus sequence analysis (Multilocus sequences analysis, MLSA) combined with phylogenetic analysis (Phylogenetic analysis) to identify the SC3 strain.
[0046] 16S rDNA sequence analysis is a molecular biological identification method for bacterial identification. The present invention analyzes the 16S rDNA sequence and combines the conservative housekeeping gene gyrB , wxya and wxya sequence, using MEGA 5.0 software to construct a phyloge...
Embodiment 3
[0048] Example 3 Determination of Pseudomonas SC3 Antibacterial Spectrum
[0049] 1. In order to better study the biocontrol potential of bacterial strain SC3 of the present invention, we have studied the bacteriostatic spectrum of this bacterium. The specific operation is as follows:
[0050] (1) To explore whether SC3 has the ability to inhibit the growth of other pathogenic bacteria. Use the method identical with embodiment 1 bacterial strain screening, change pathogenic bacterium MS2 into the equal amount of rice base rot pathogen EC1, banana bacterial soft rot pathogen MS2 and MS3, bacterial wilt pathogen EP1 (first Replace 15 mL of LB with 15 mL of TTC), Brassicaceae black rot fungus Xcc, rice bacterial blight Xoo, peach rot fungus Pantoea ananatis PP1, yellow skin rot pathogen Pantoea anthophila CL1, the rest of the steps remain unchanged.
[0051] (2) To explore whether SC3 has the ability to inhibit the growth of pathogenic fungi. The fungi tested included ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com