Maize dna fingerprint library construction and variety molecular identification SNP core site combination-maizesnp384
A DNA fingerprinting, corn technology, applied in the biological field, can solve the problems of inability to combine application of sites, difficult to improve site flux, and increase the number of sites.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1. Preparation of maizeSNP384 and its chip for maize DNA fingerprint database construction and variety molecular identification SNP core site combination
[0049] Based on the 3072 SNP locus data of 300 corn hybrids, 300 domestic and foreign inbred lines, and 240 DH line samples (see the Chinese invention patent with the patent number ZL 2012 8 0000286.2), maizeSNP3072, a general-purpose corn SNP chip product Among them, the core 384 SNP sites suitable for maize authenticity identification and fingerprint database construction were screened and determined according to the test effect, discrimination efficiency, and uniform distribution principle. The specific screening process is divided into three steps: (1) according to the following indicators "3072SNPs are fully compatible with both GoldenGate and Infinium platforms, high data quality, MAF value ≥ 0.2" to obtain 1212 candidate SNPs; (2) according to Illumina design rank score>= 0.7, the three genotypes of AA...
Embodiment 2
[0436] Example 2, using the SNP chip prepared in Example 1 to construct a corn standard DNA fingerprint
[0437] 1. DNA extraction and quality identification
[0438] The conventional CTAB method was used to extract the genomic DNA of all maize varieties used to construct the maize DNA fingerprint database, and remove the RNA. The quality of the extracted DNA was detected by agarose electrophoresis and ultraviolet spectrophotometer, and it was found that the extracted genomic DNA had reached the relevant quality requirements, that is, the agarose electrophoresis showed a single DNA band without obvious dispersion; the ultraviolet spectrophotometer detected A260 / 280 is between 1.8-2.0 (DNA sample has no protein contamination); A260 / 230 is between 1.8-2.0 (DNA sample has low salt ion concentration); 270nm has no obvious light absorption (DNA sample has no phenol contamination); DNA concentration > 50ng / μl; chip detection DNA dosage 500ng / sample. The diluted DNA concentration ...
Embodiment 3
[0443] Embodiment 3, utilizing the SNP chip prepared in embodiment 1 to identify the authenticity of corn varieties
[0444] One, utilize the SNP chip maizeSNP384 prepared in embodiment 1 to distinguish the determination of the authenticity test result judgment standard of maize variety
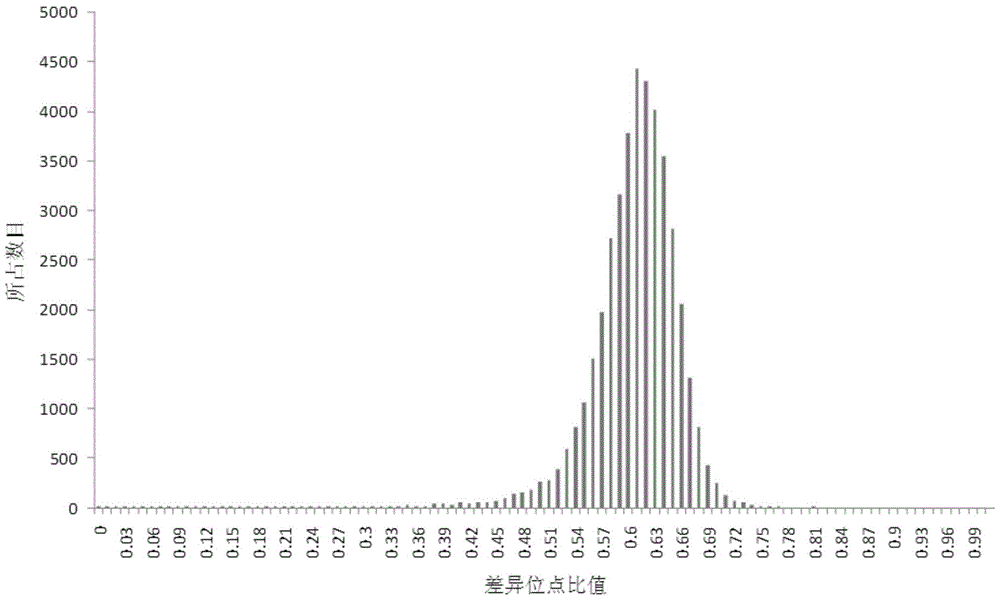
[0445] Different from the principle of site screening in other research fields, in addition to inbred line samples, the present invention also uses a large number of hybrids as evaluation materials to evaluate site specificity, stability and variety discrimination ability, and finally obtains 384 core SNP sites point (for the SNP site screening method, refer to Example 1 for details). The percentage analysis of 300 corn hybrids (1984-2011 nationally approved and popularized varieties) showed that the percentage of difference loci was between 40% and 75%. %; Among them, the percentage of difference loci is about 60% and occupies the most; the number of pairwise comparison samples with the per...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com