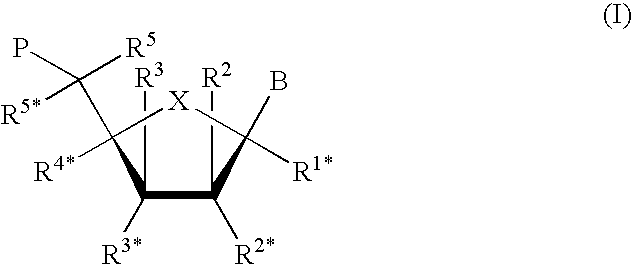
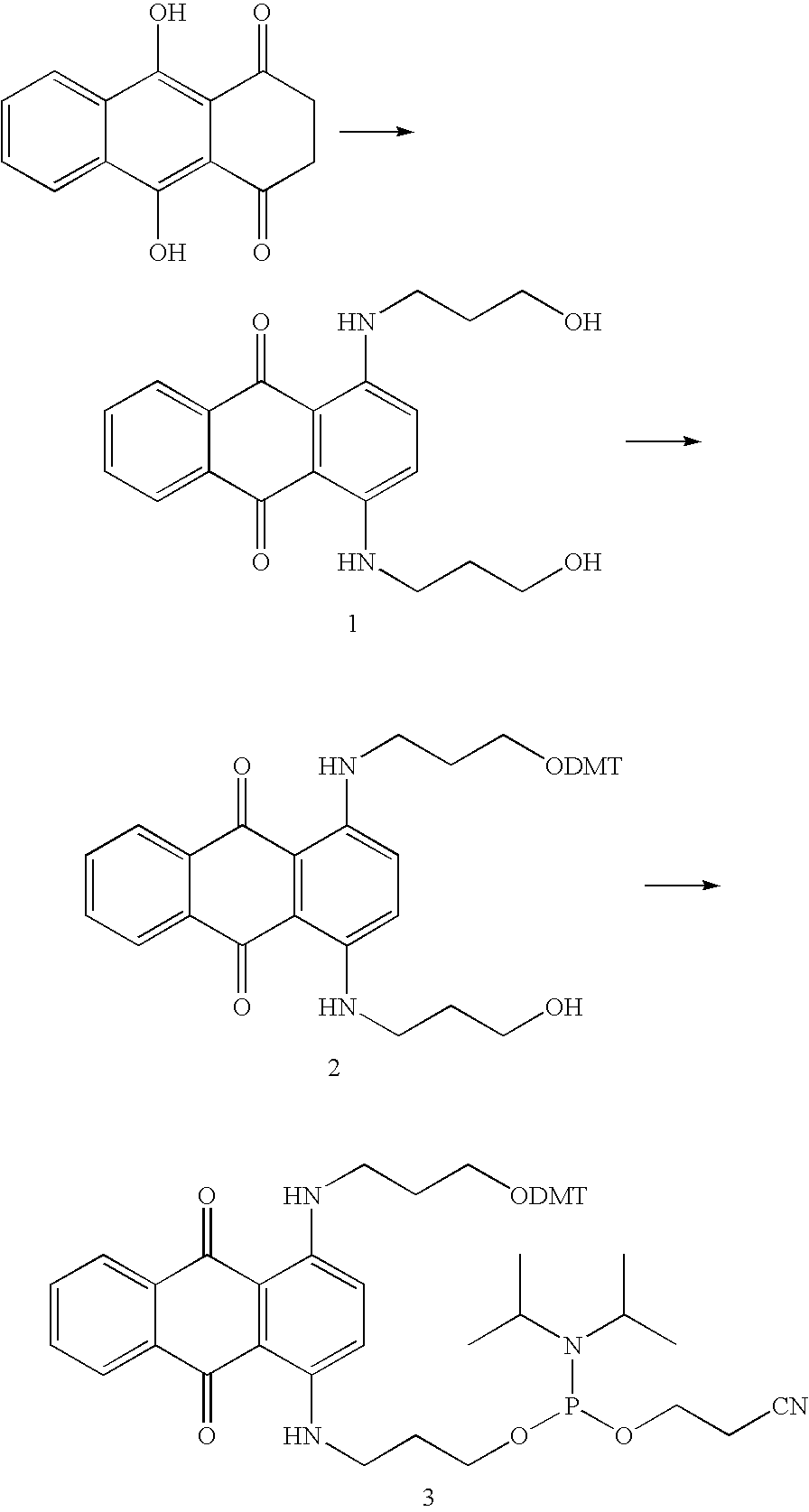
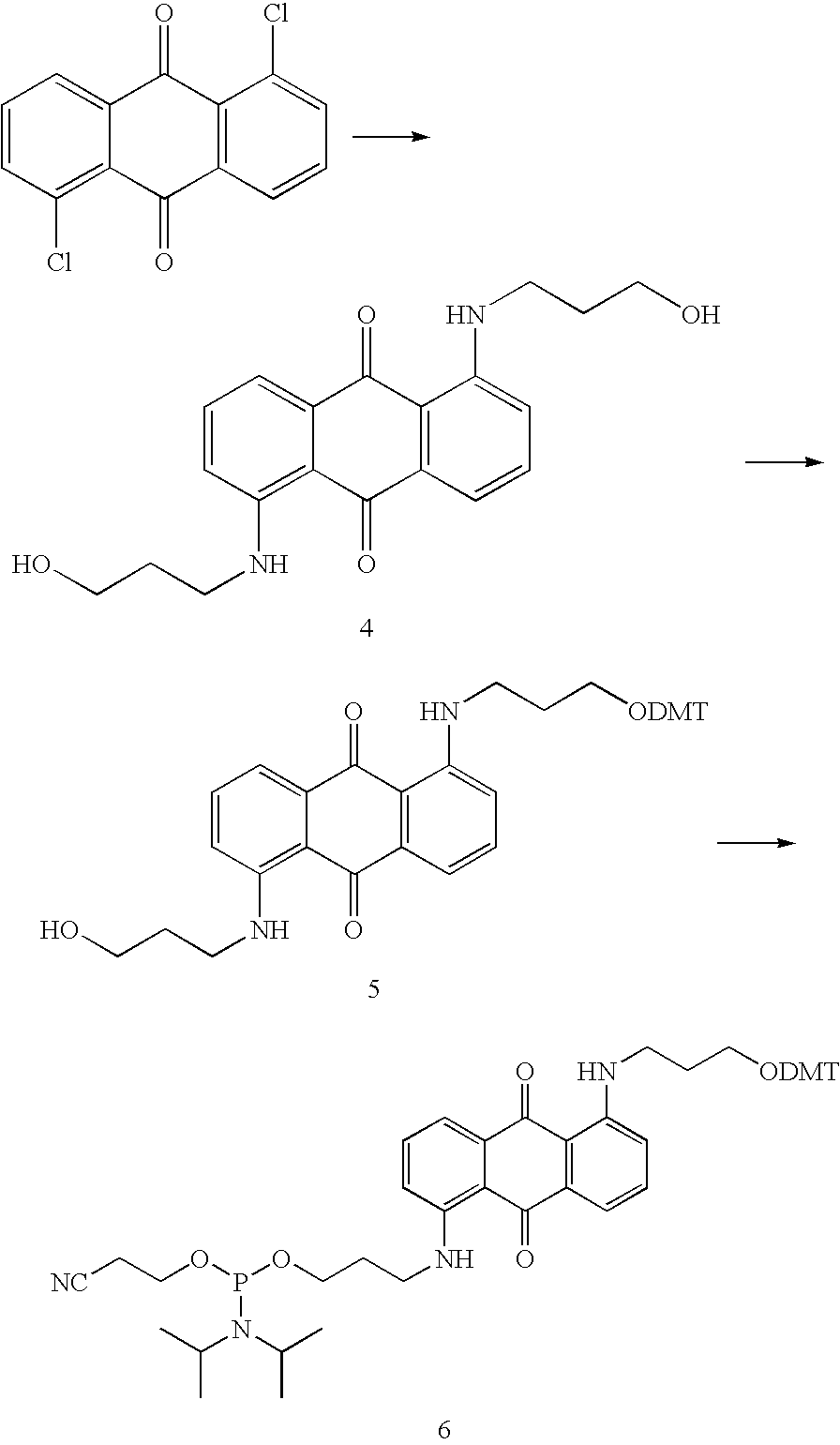
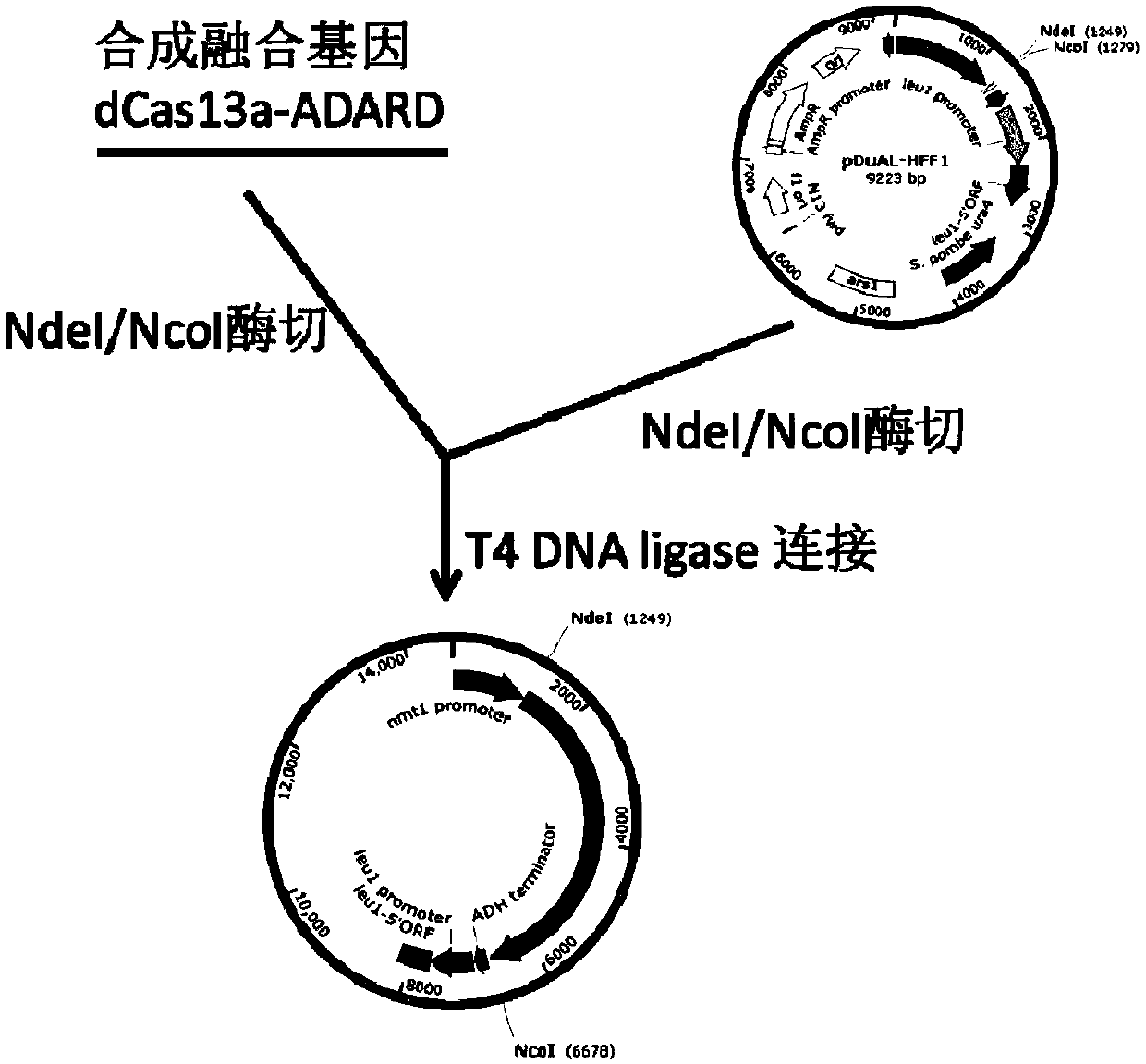
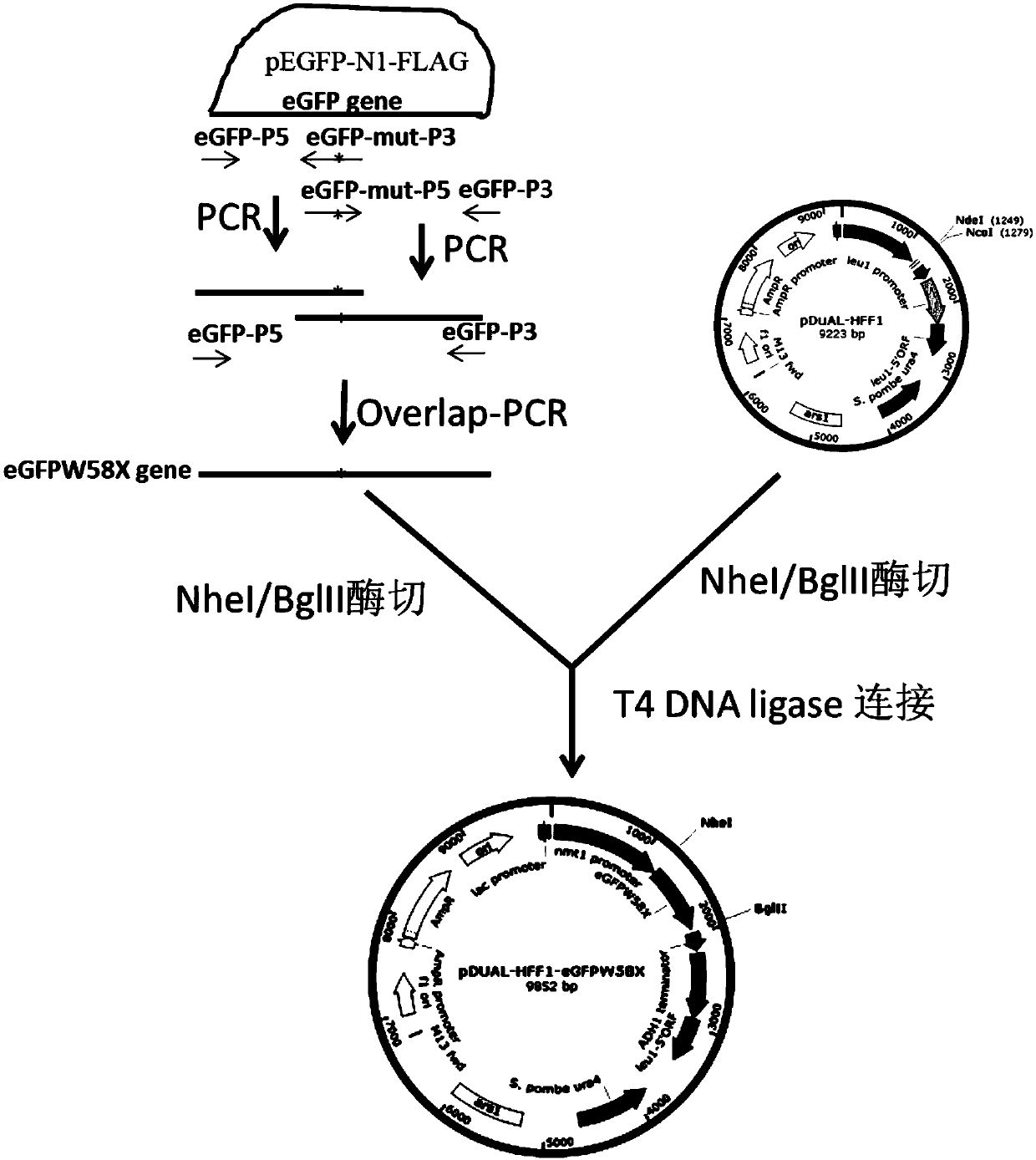
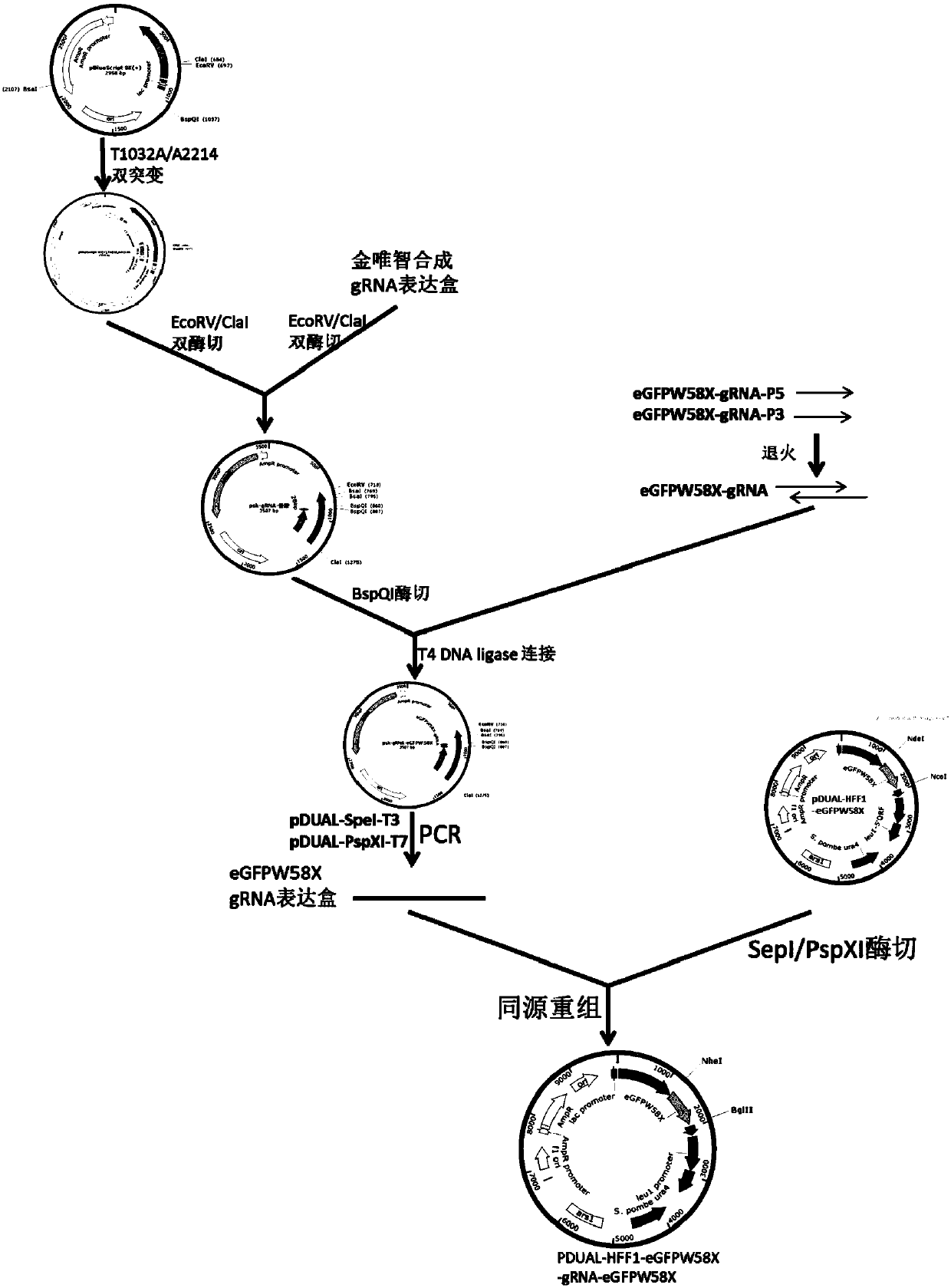
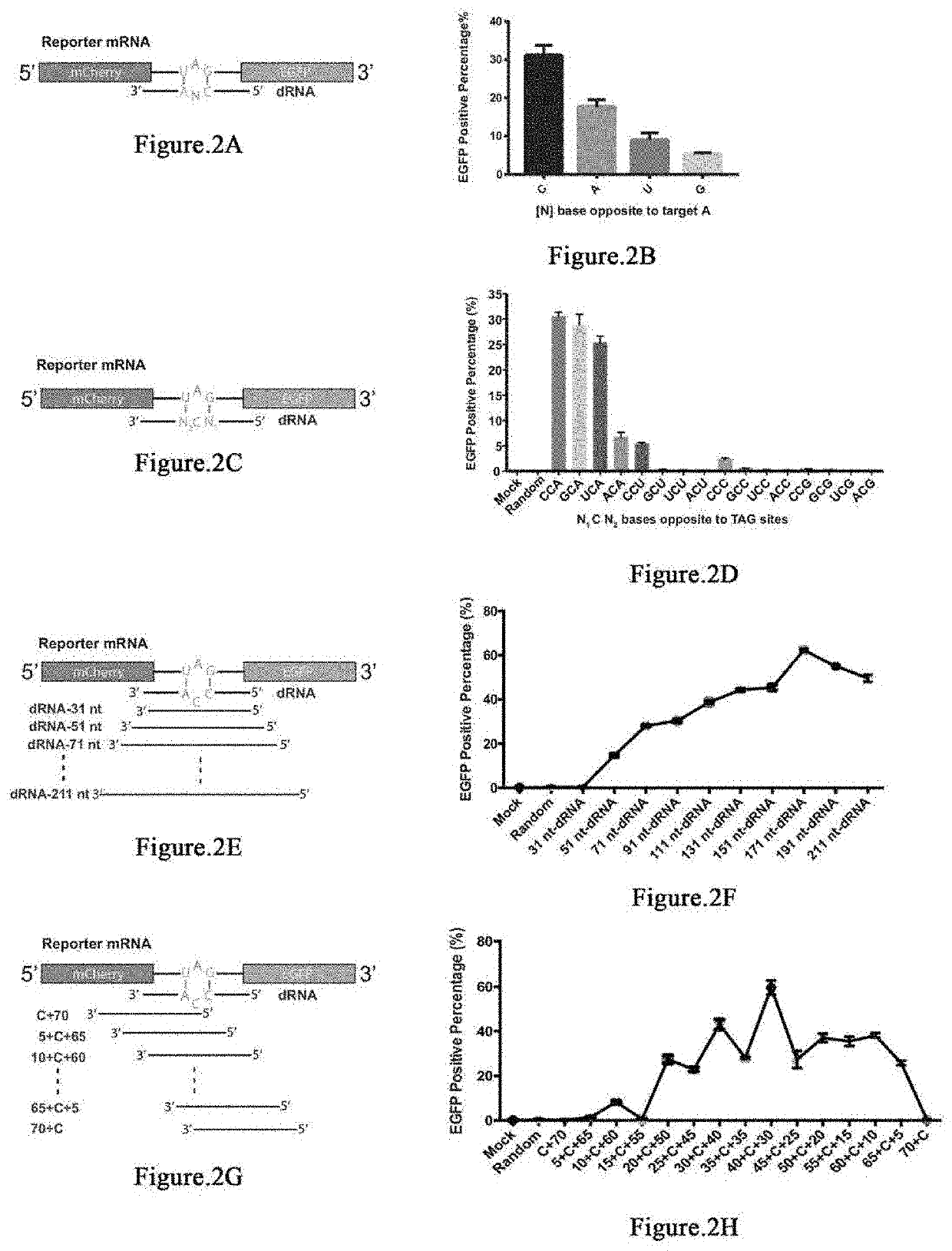
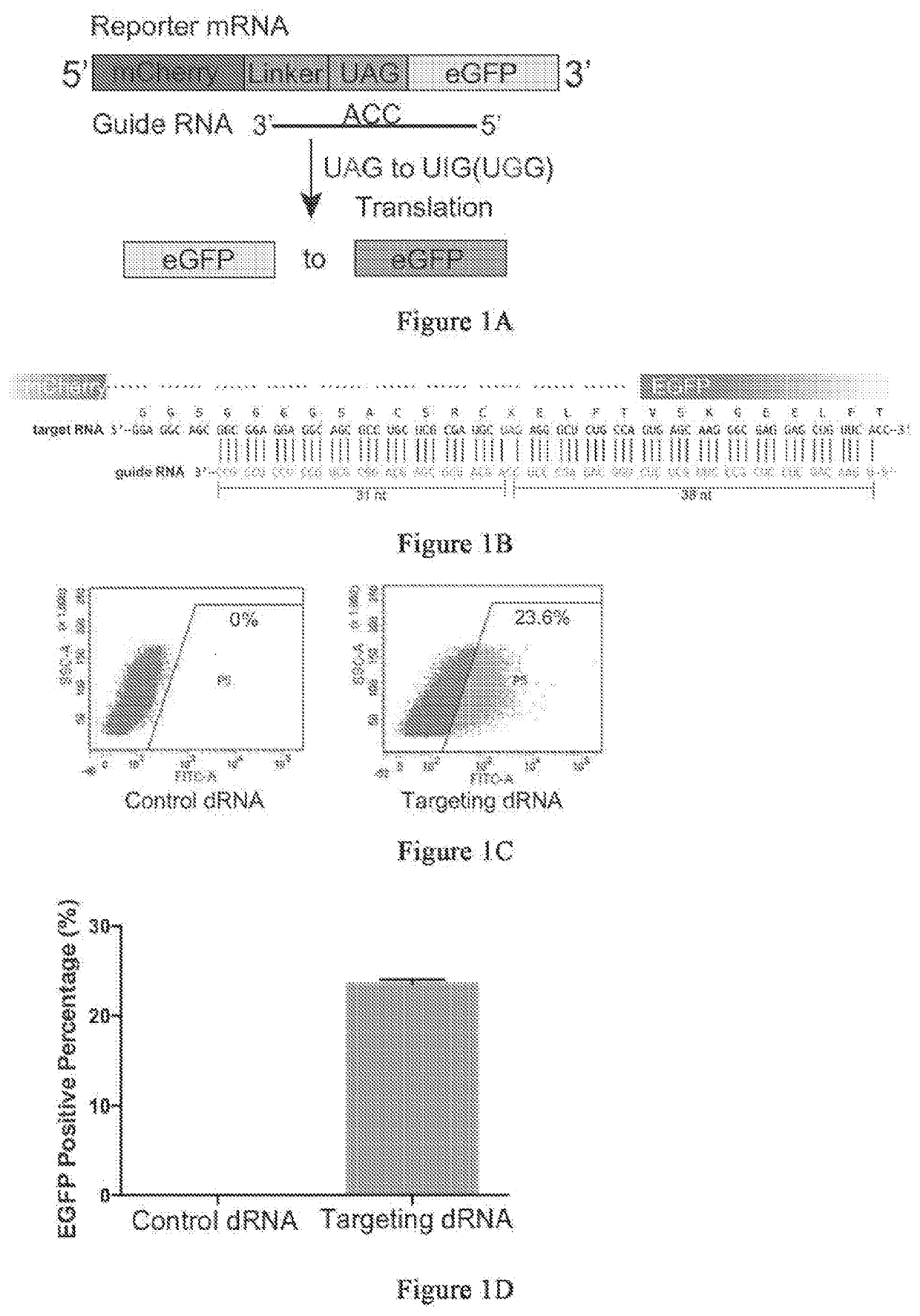
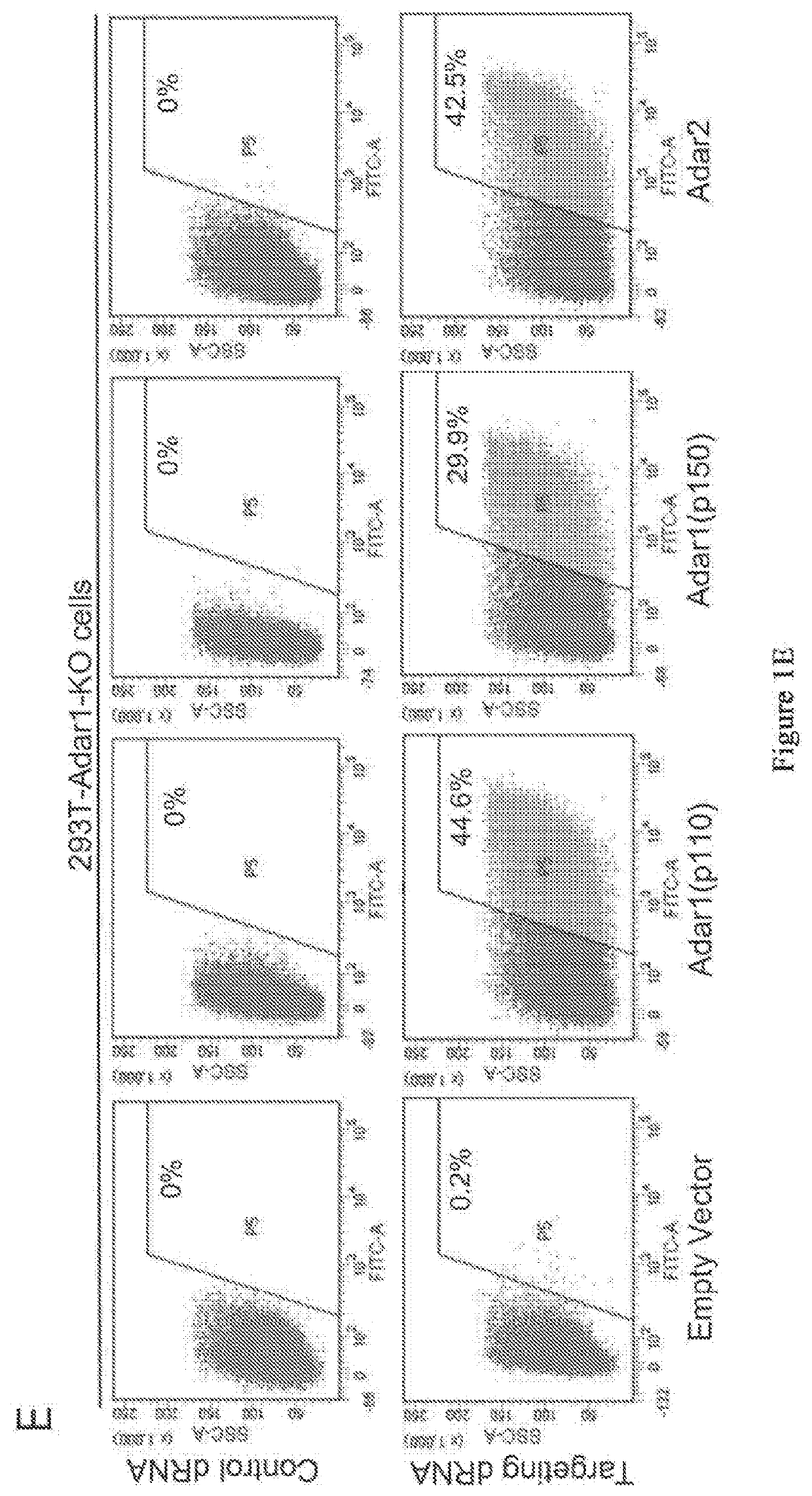
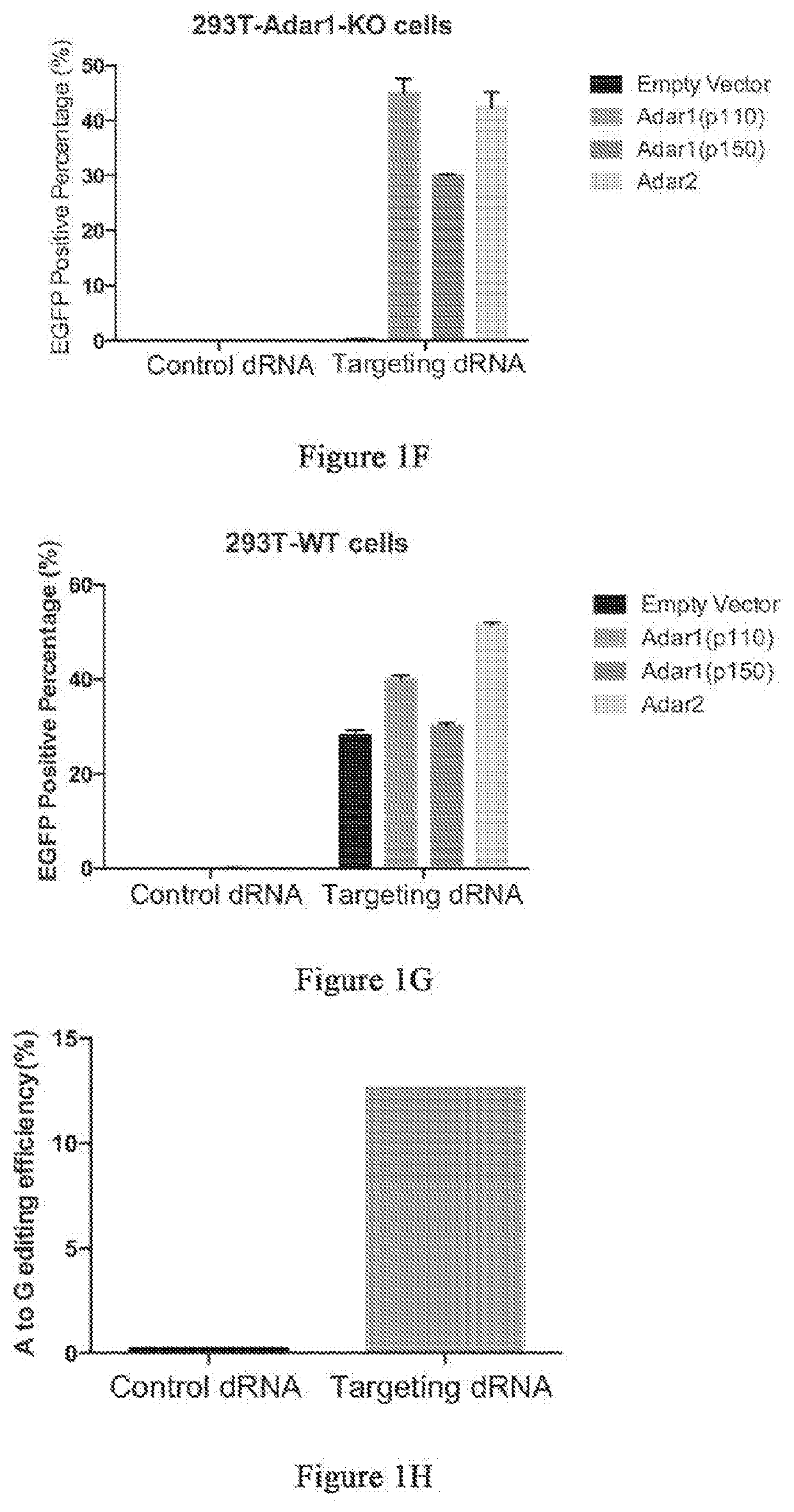
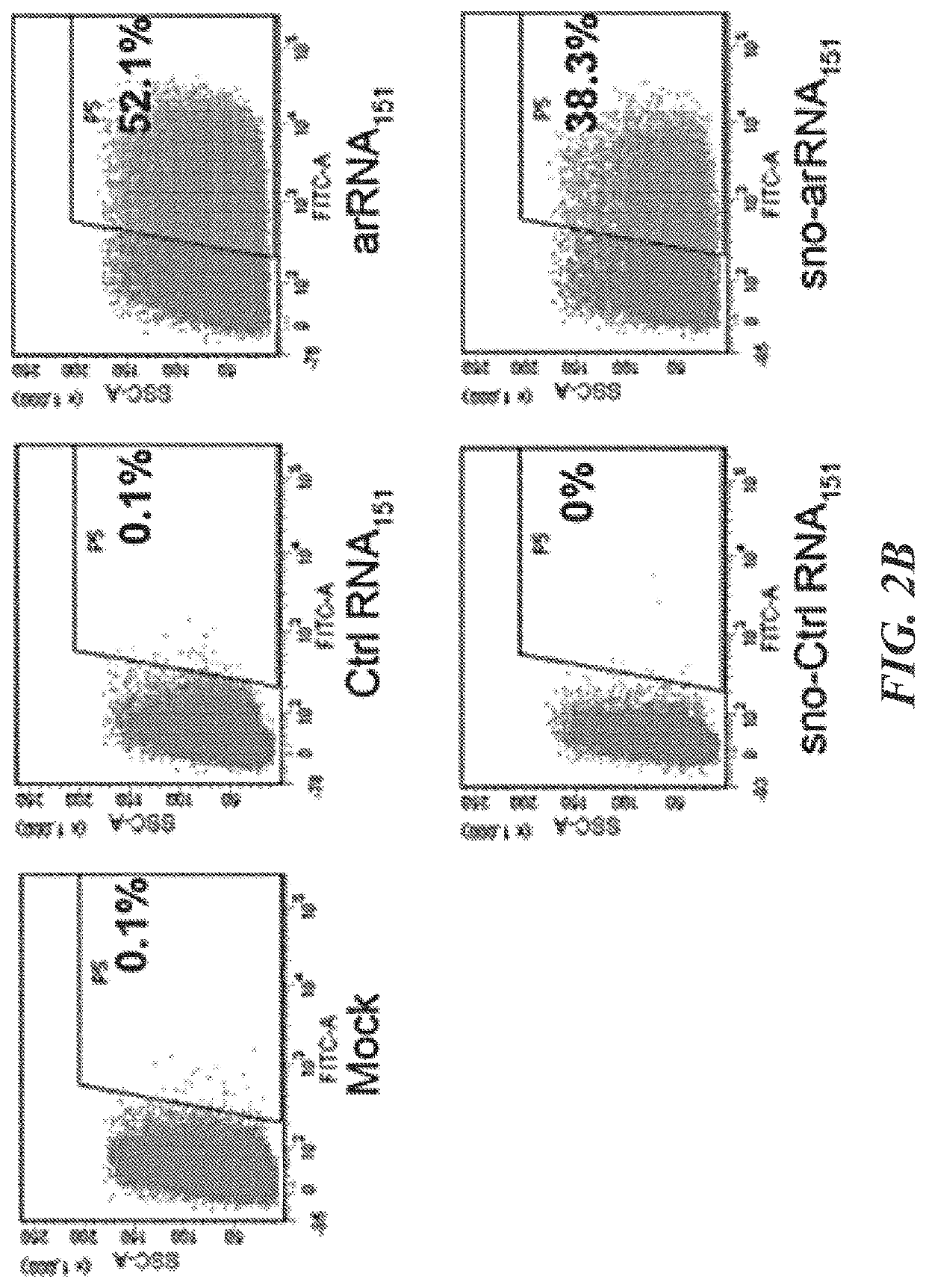
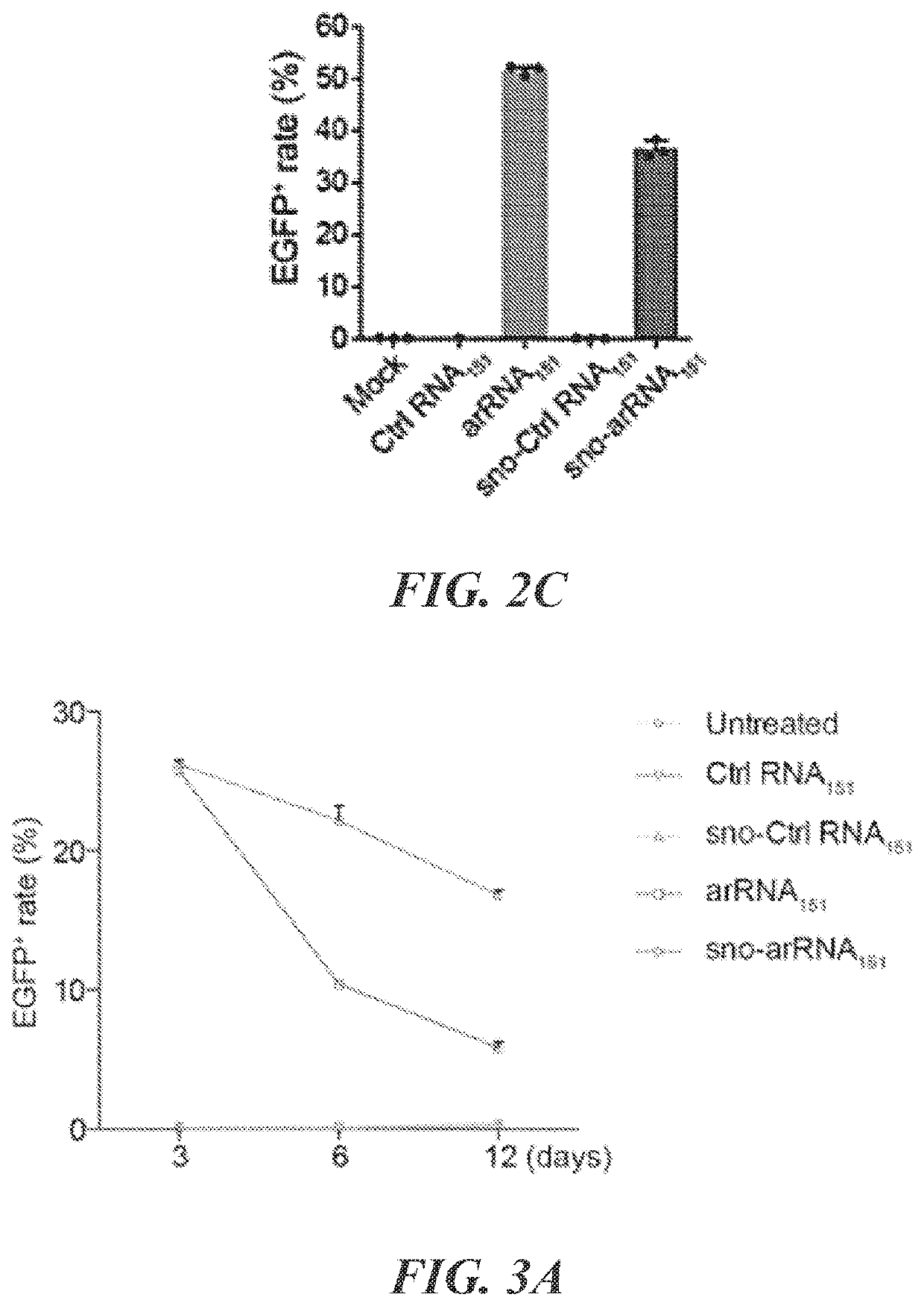
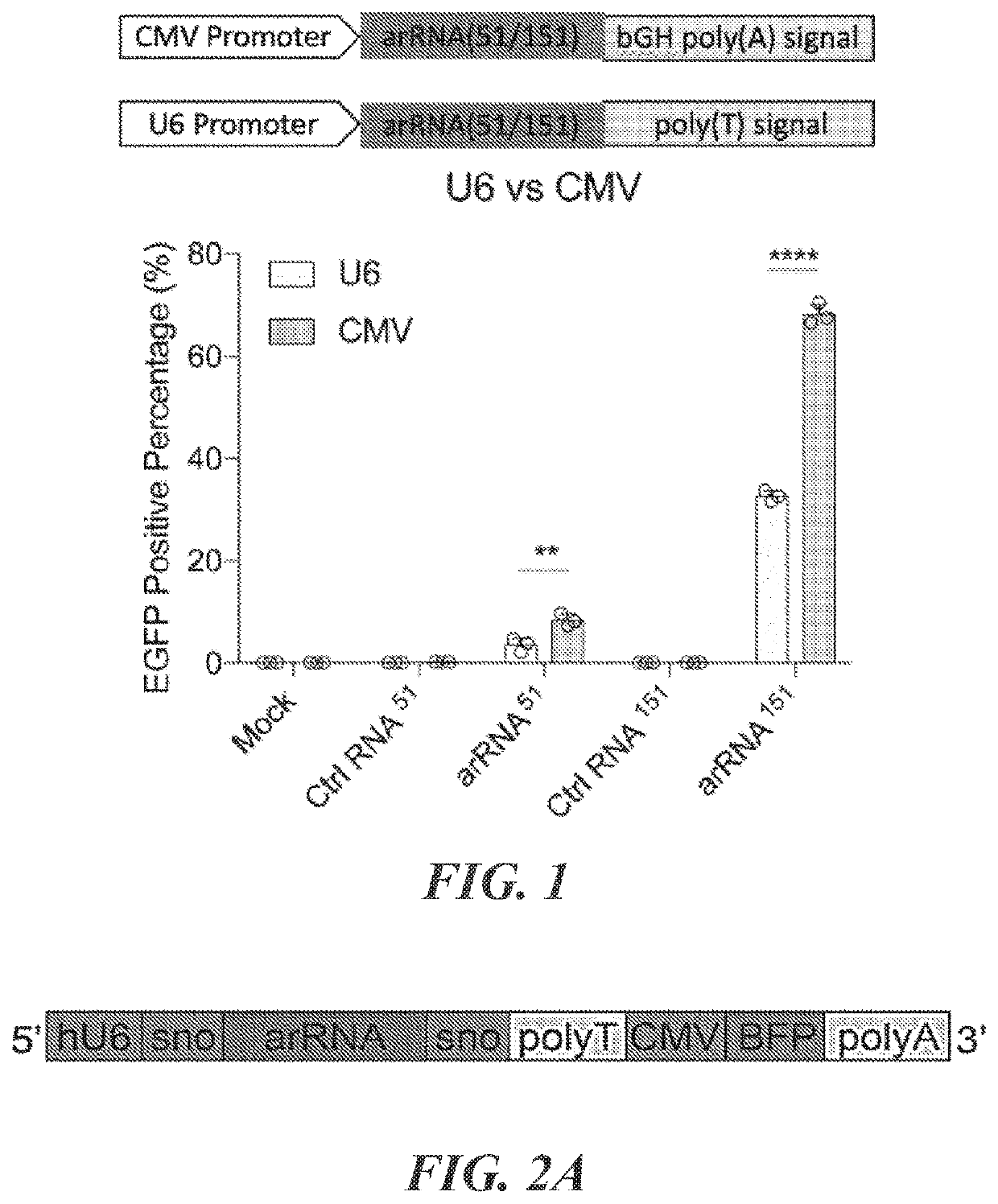
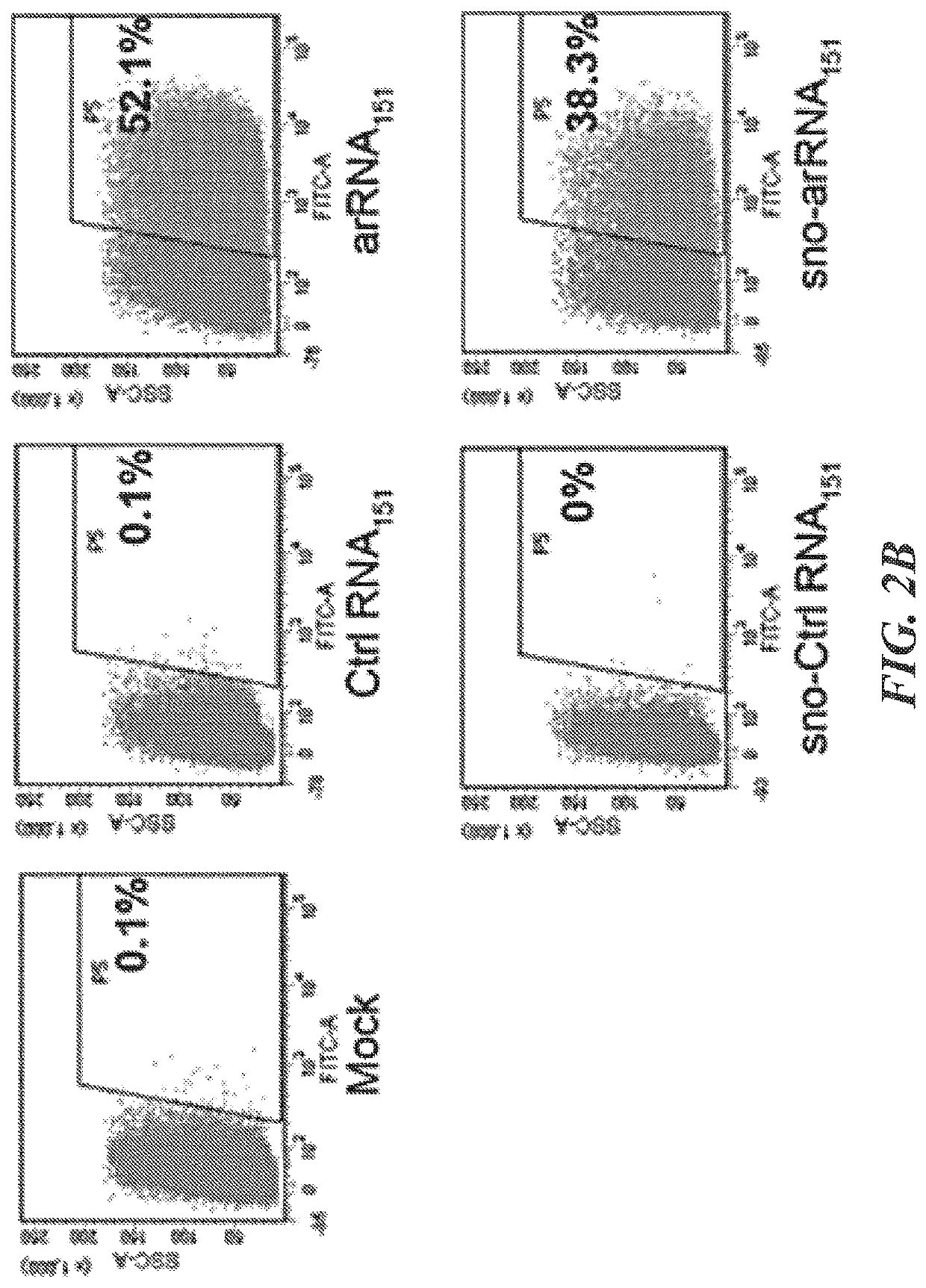
Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
69 results about "Mitochondrial RNA editing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
RNA editing has been observed in some tRNA, rRNA, mRNA, or miRNA molecules of eukaryotes and their viruses, archaea, and prokaryotes. RNA editing occurs in the cell nucleus and cytosol, as well as within mitochondria and plastids. In vertebrates, editing is rare and usually consists of a small number of changes to the sequence of affected molecules.
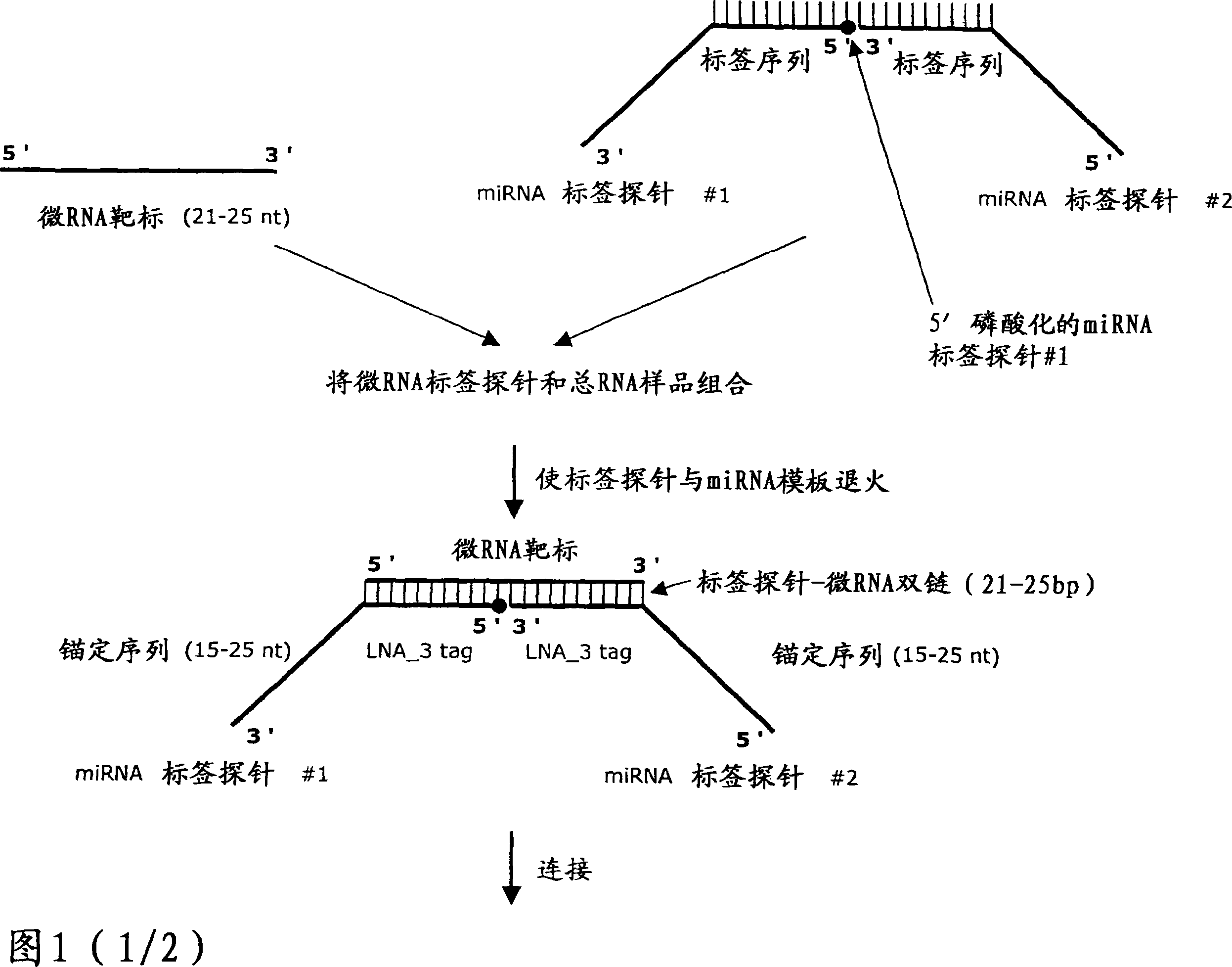
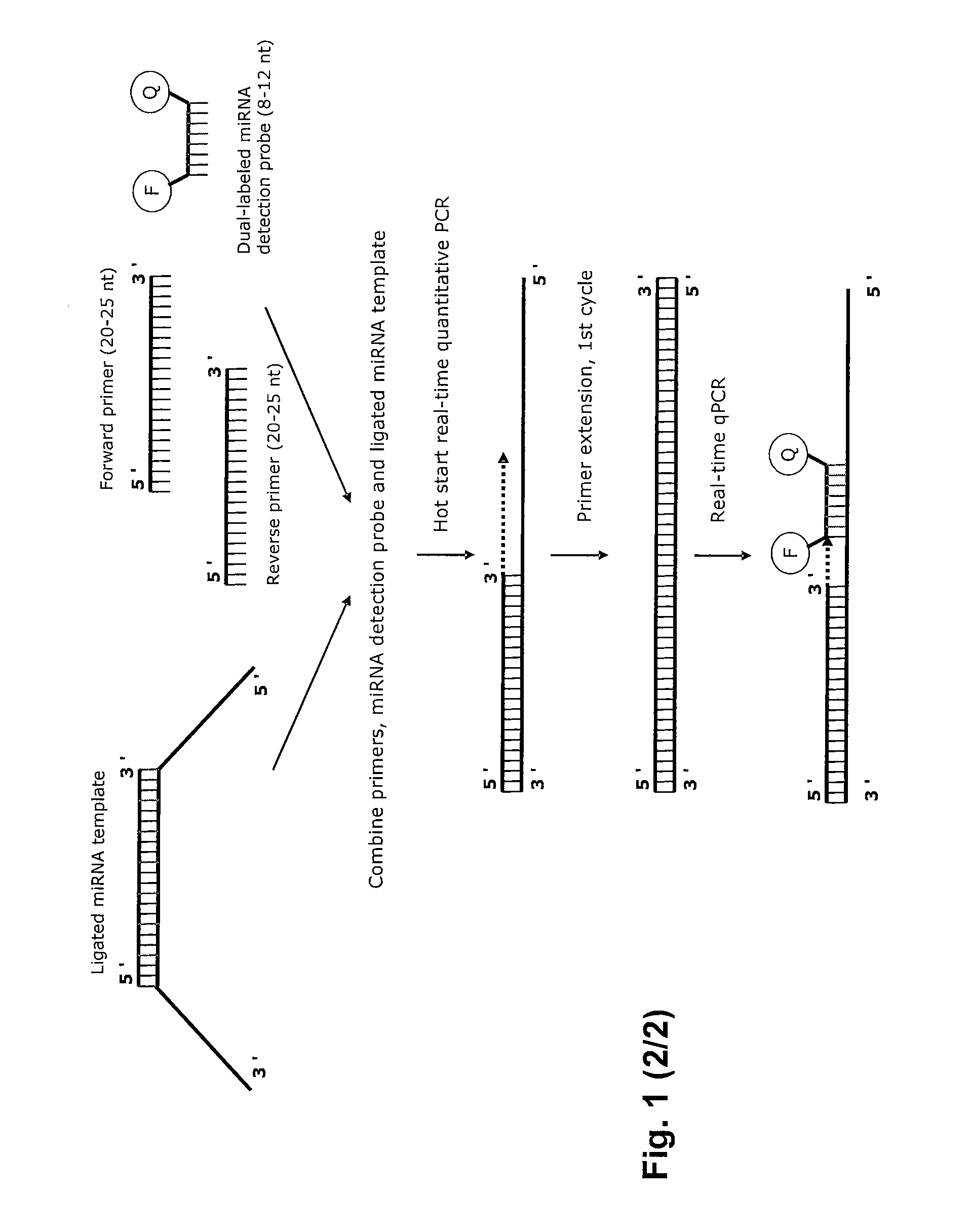
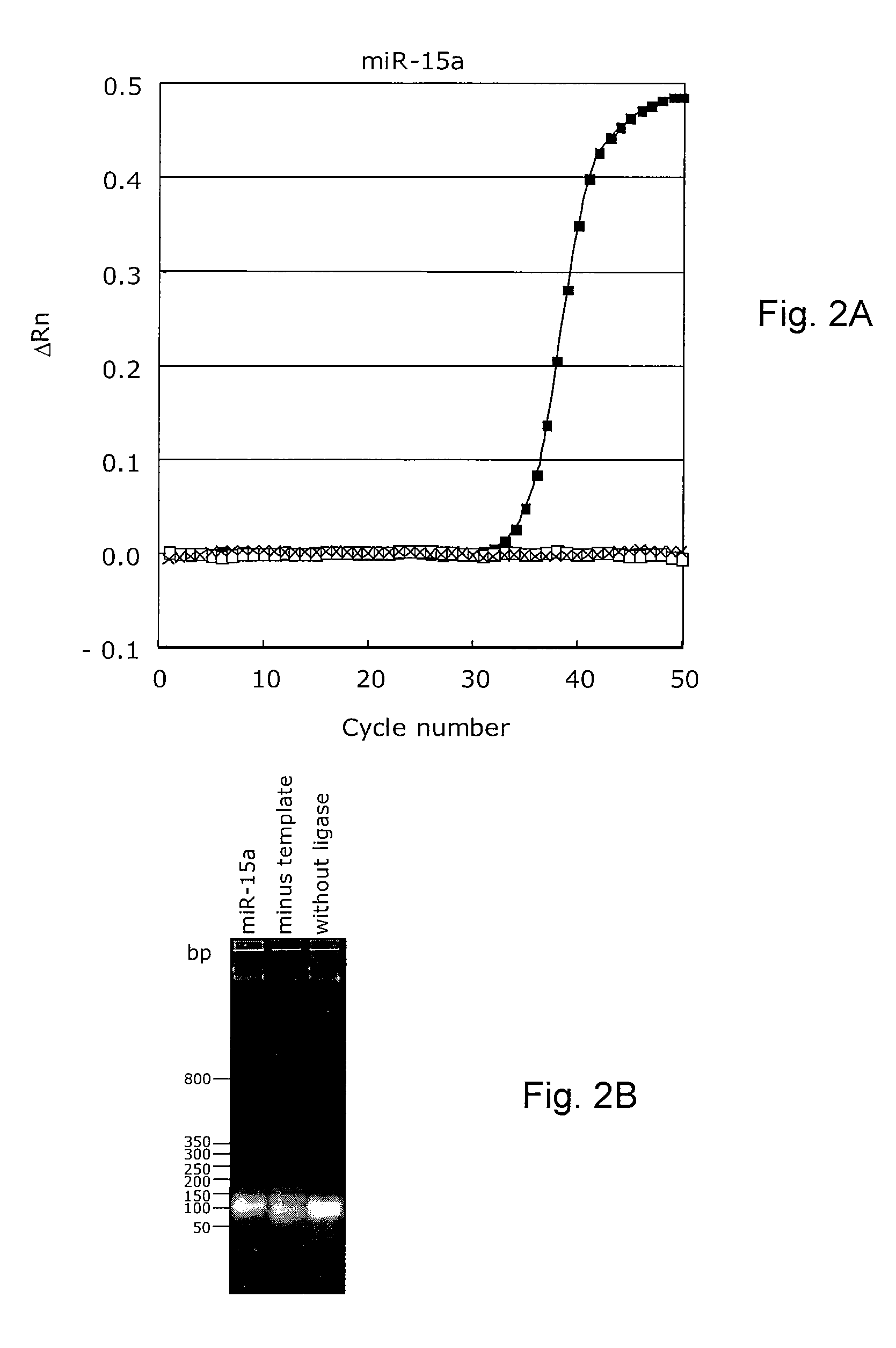
Novel methods for quantification of microRNAs and small interfering RNAs
ActiveUS20050272075A1Highly sensitive and specific hybridizationHighly sensitive and specific and ligationSugar derivativesMicrobiological testing/measurementMicroRNAAllele
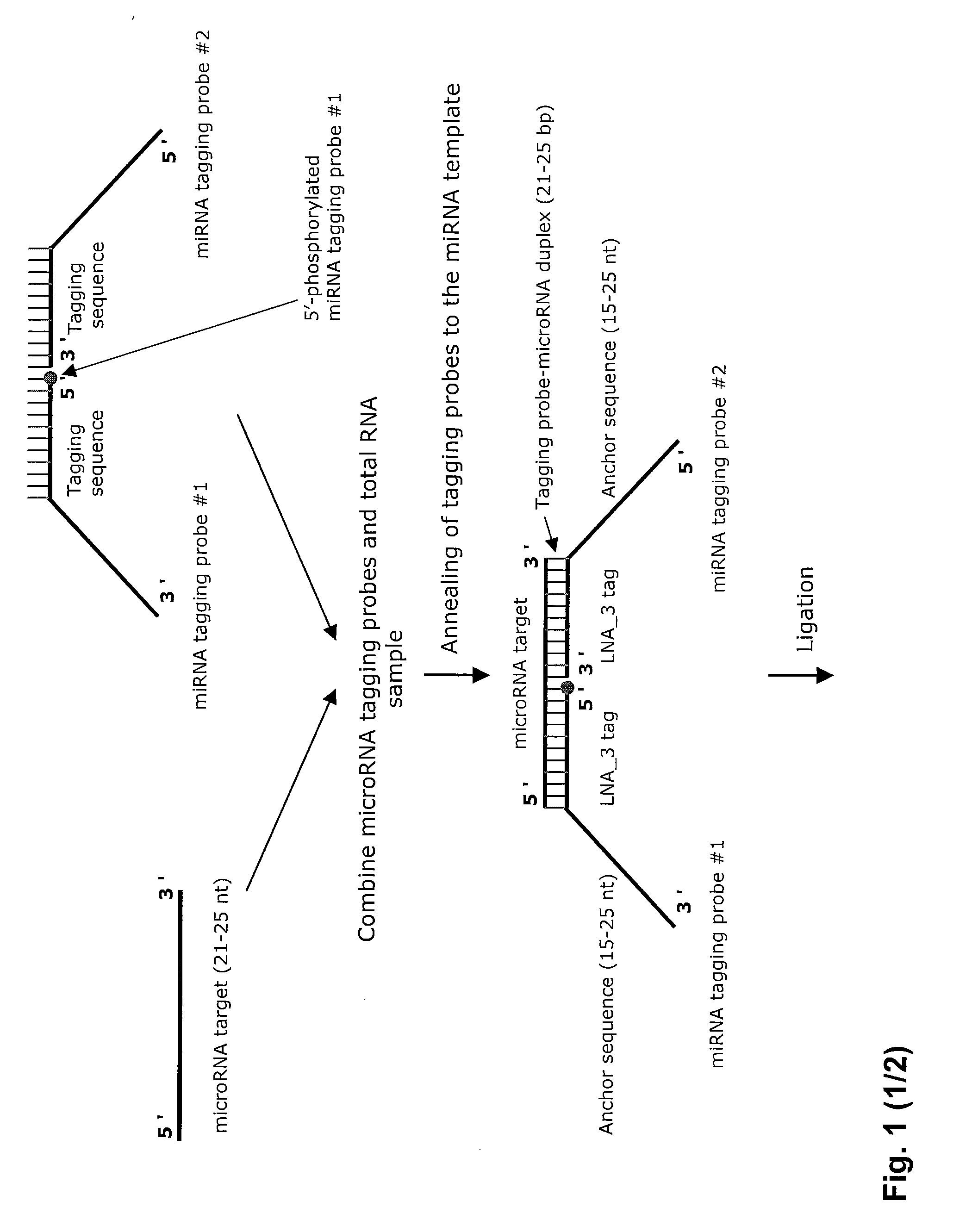
The invention relates to ribonucleic acids, probes and methods for detection, quantification as well as monitoring the expression of mature microRNAs and small interfering RNAs (siRNAs). The invention furthermore relates to methods for monitoring the expression of other non-coding RNAs, mRNA splice variants, as well as detecting and quantifying RNA editing, allelic variants of single transcripts, mutations, deletions, or duplications of particular exons in transcripts, e.g., alterations associated with human disease such as cancer. The invention furthermore relates to methods for detection, quantification as well as monitoring the expression of deoxy nucleic acids.
Owner:QIAGEN GMBH
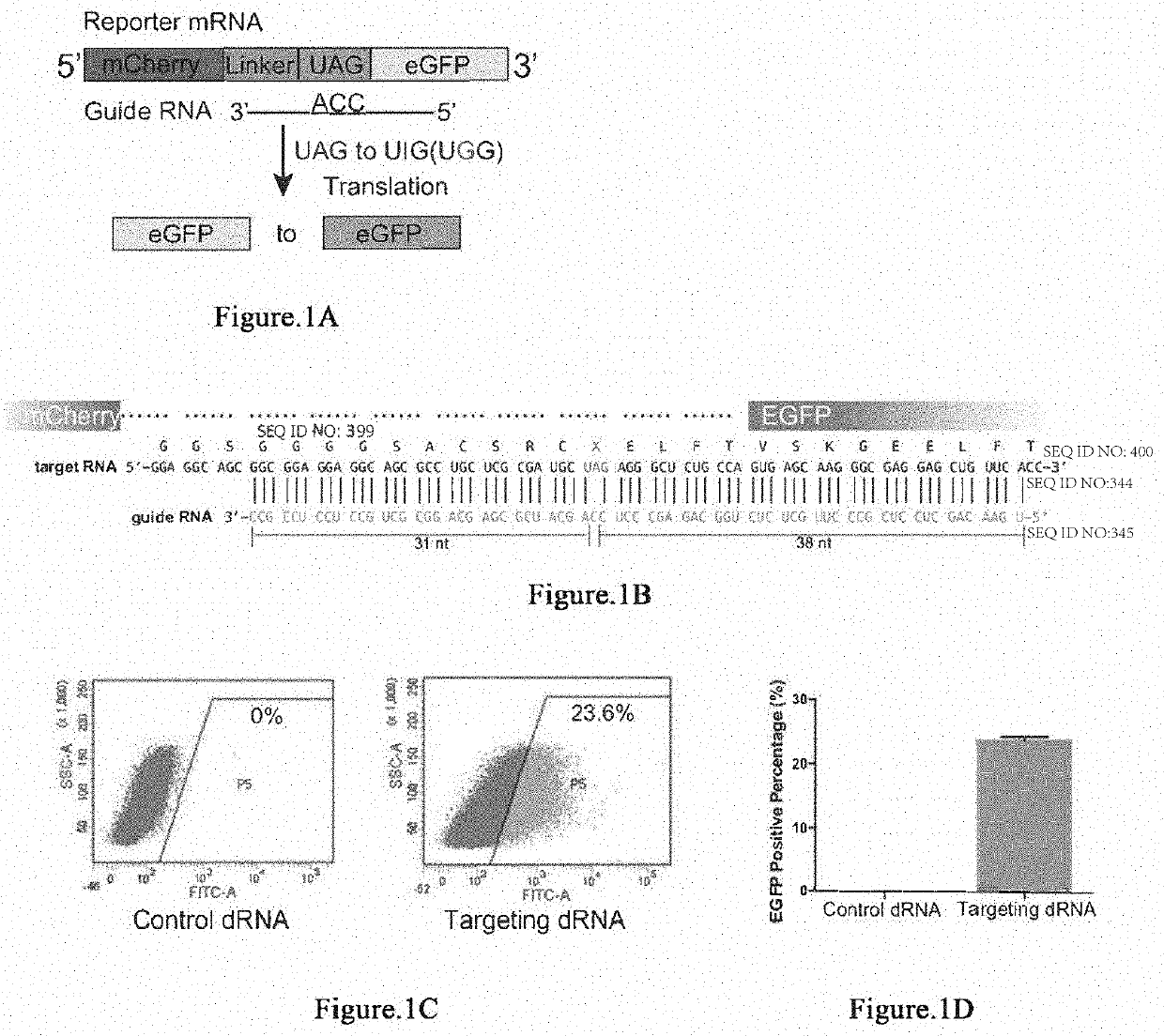
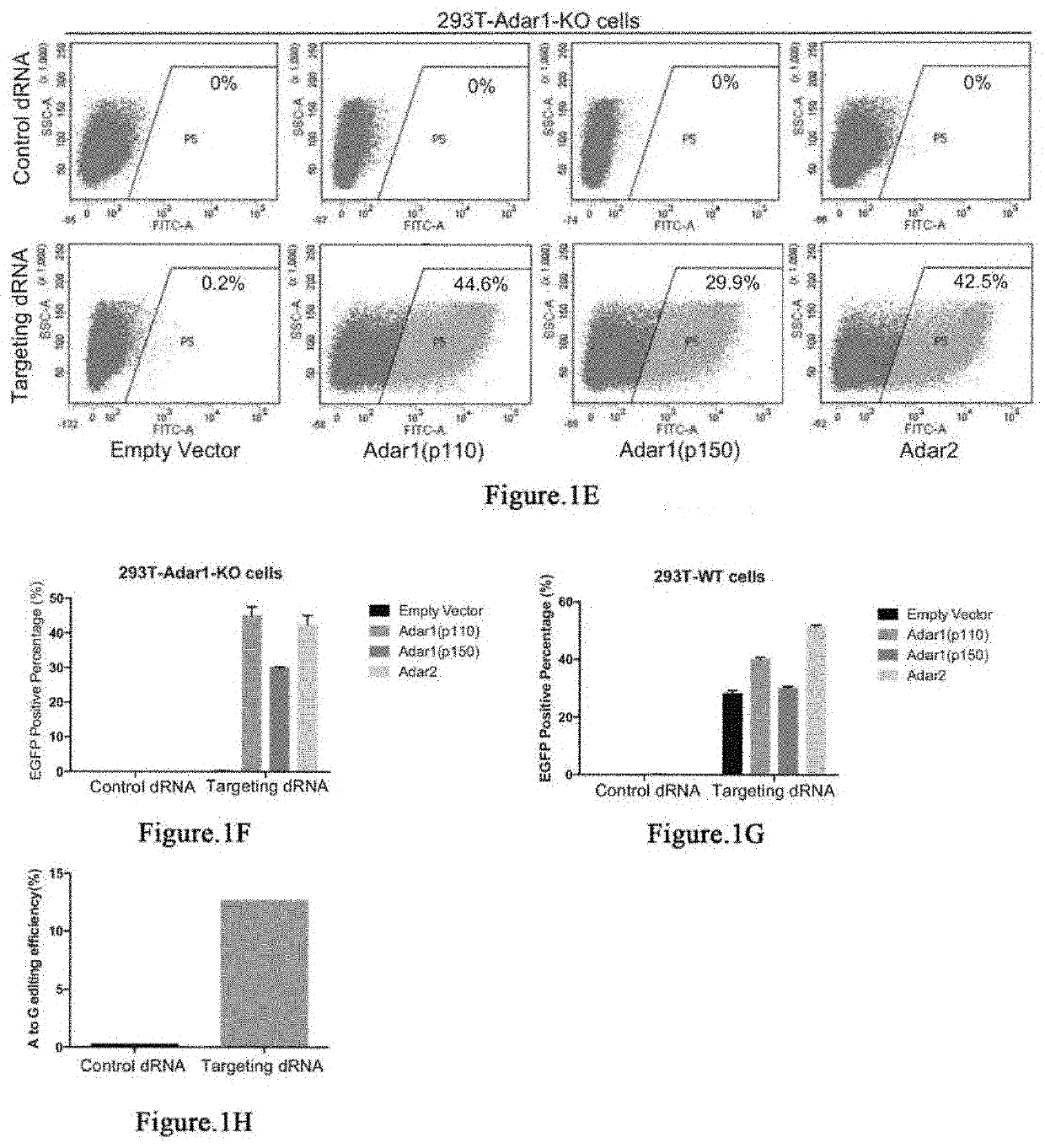
RNA site-directed editing technology based on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas13a
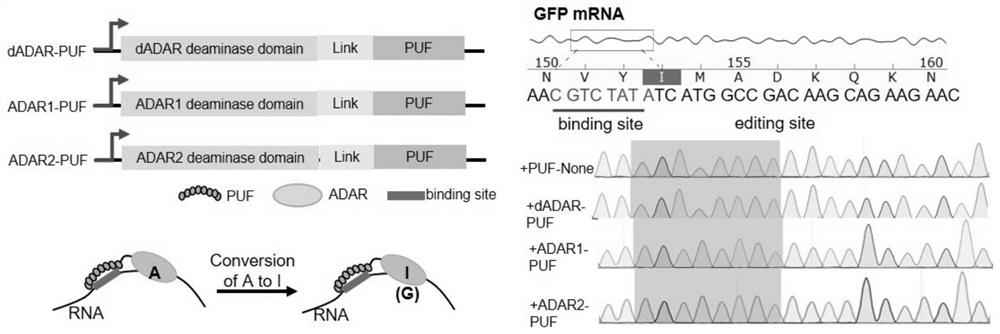
The invention relates to a site-directed RNA (Ribonucleic Acid) editing technology based on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas13a, and discloses an RNA site-directed editing method based on the CRISPR-Cas13a. By use of the method disclosed by the invention, during RNA site-directed editing, under the guidance of a crRNA, a Cas13a mutant takes a catalytic structural domain of ADAR2 (Adenosine Deaminase Acting on RNA 2) to an appointed RNA position to finish the site-directed editing from A to I. The Cas13a mutant does not have ssRNA cleavage activity, has ssRNA binding activity and is blended with the catalytic structural domain of the ADAR2. On the basis, the design of the crRNA is also optimized.
Owner:CAS CENT FOR EXCELLENCE IN MOLECULAR PLANT SCI
Site-directed RNA editing
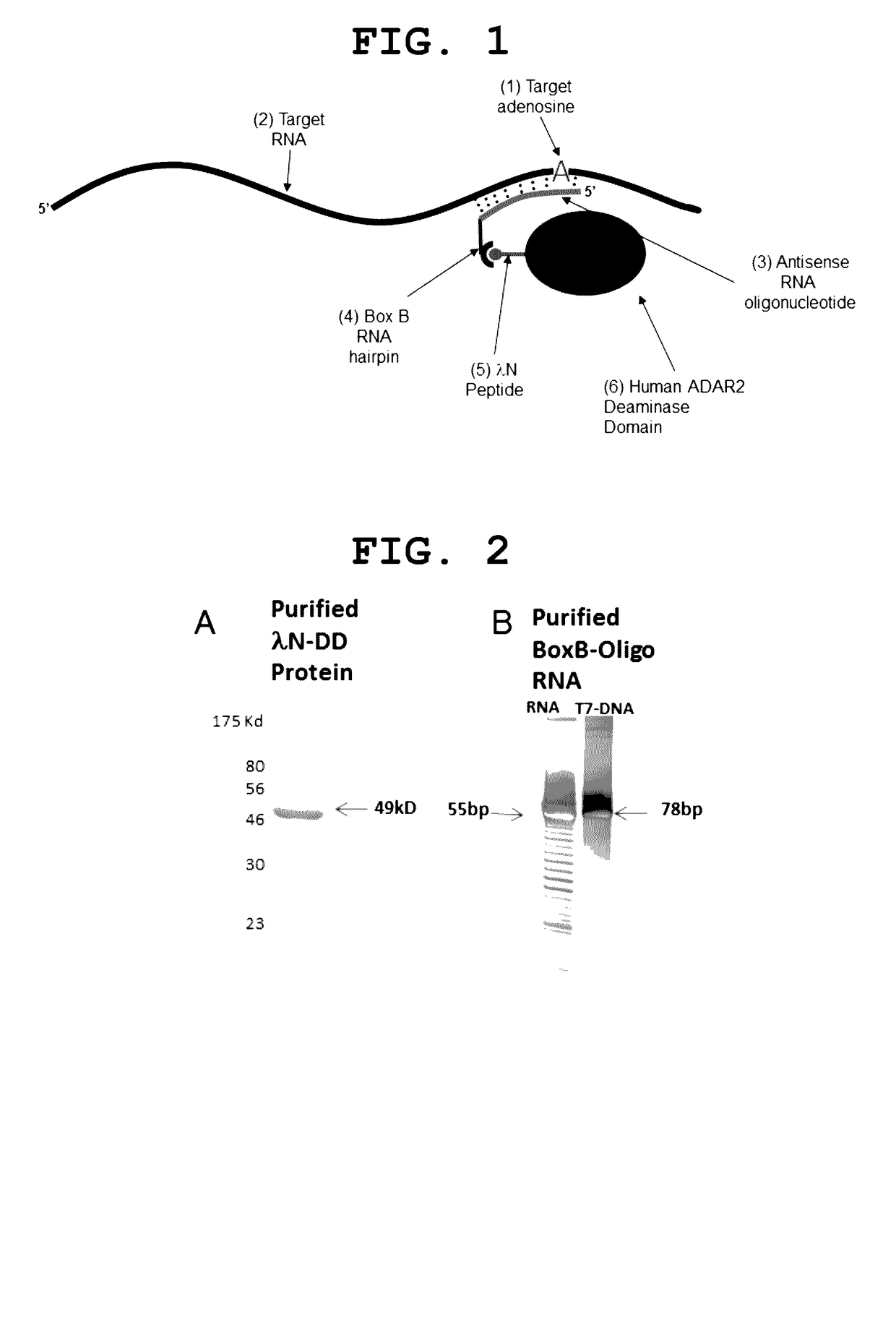
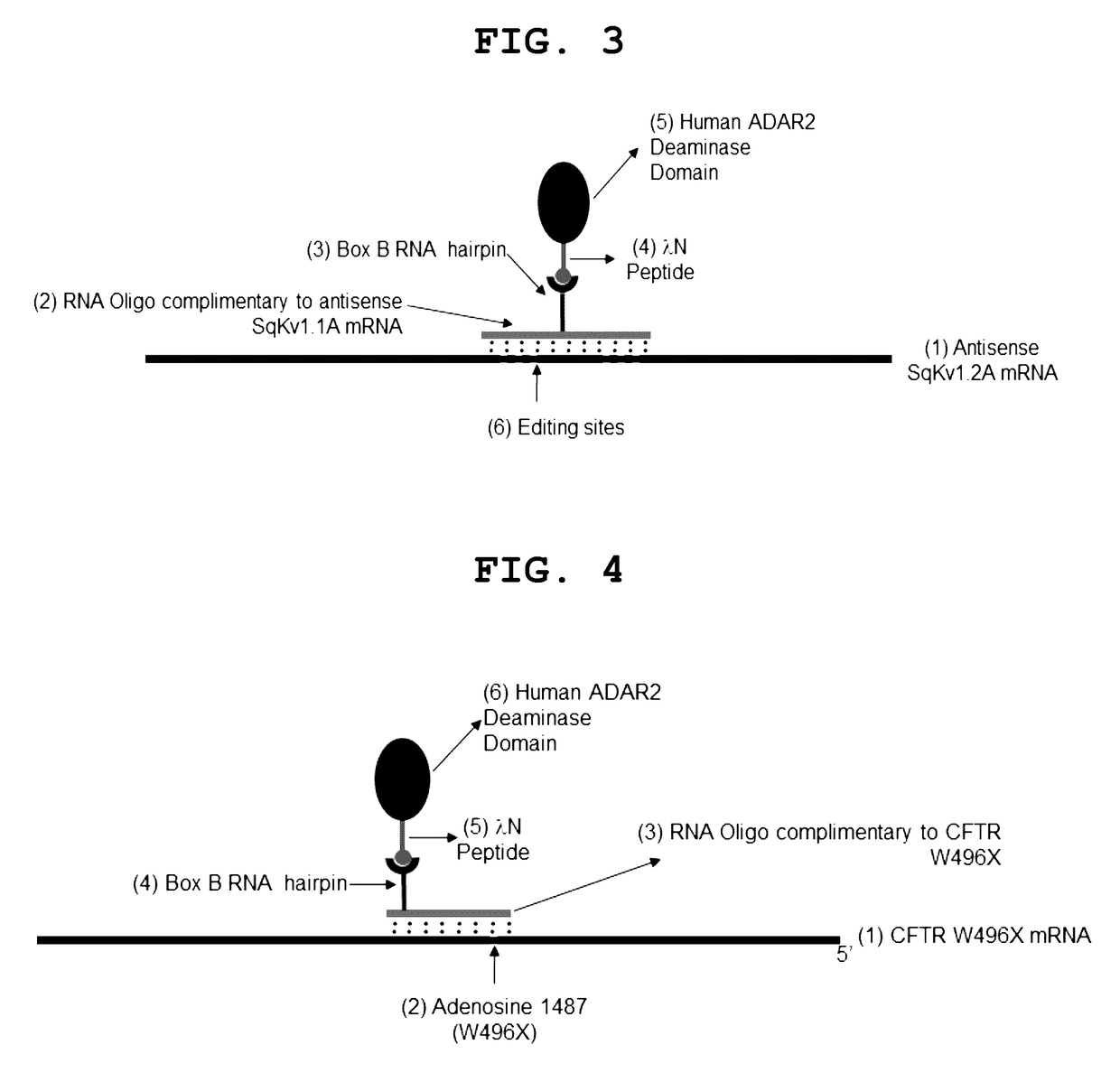
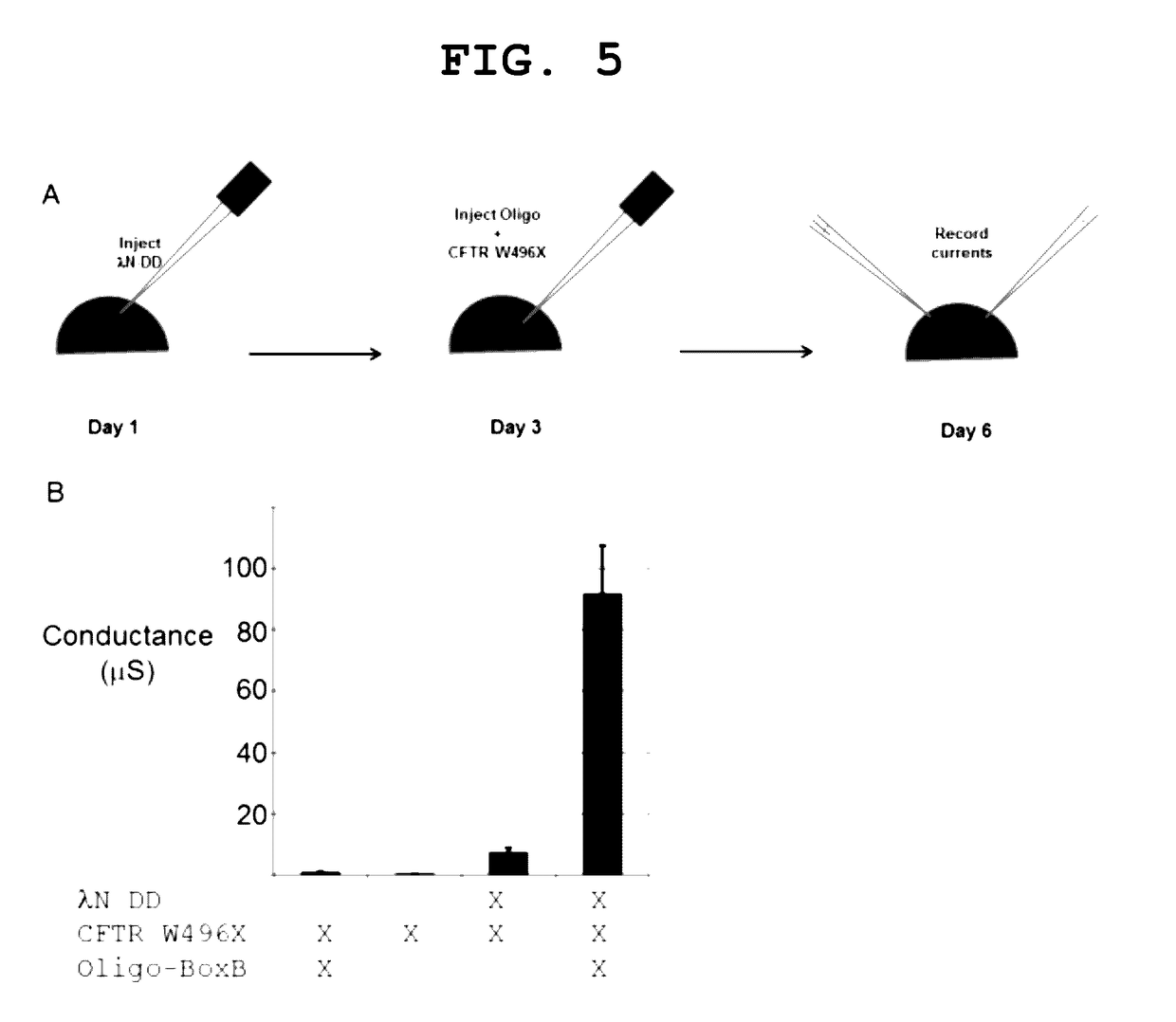
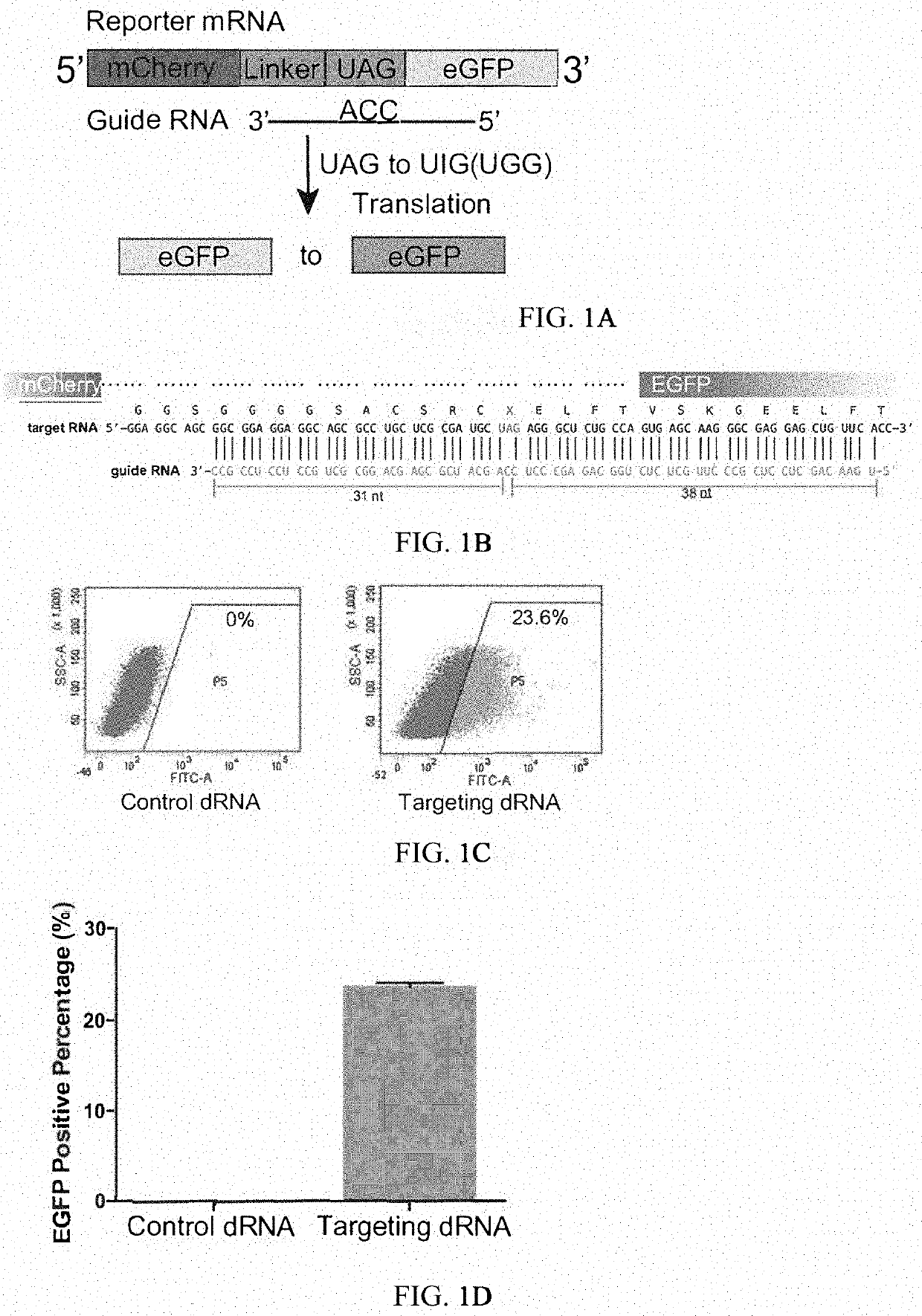
The invention provides a way to target RNA editing by adenosine deamination to a chosen adenosine within RNA. An antisense RNA oligonucleotide is used for targeting the entire complex to a specific address on the RNA molecule. A Box B RNA and a λ N-peptide are used as a linkage between the antisense RNA oligonucleotide and a deaminase domain of human ADAR2 used to catalyze the deamination of the specific adenosine residue. These elements make up two molecules: the antisense RNA Oligo Box B RNA hairpin forms a single unit, as does the λ N-peptide-deaminase domain of human ADAR2.
Owner:UNIVERSITY OF PUERTO RICO
Methods for quantification of microRNA and small interfering RNA
The invention relates to ribonucleic acids, probes and methods for detection, quantification as well as monitoring the expression of mature microRNAs and small interfering RNAs (siRNAs). The invention furthermore relates to method s for monitoring the expression of other non-coding RNAs, mRNA splice variants, as well as detecting and quantifying RNA editing, allelic variants of single transcripts, mutations, deletions, or duplications of particular exons in transcripts, e.g., alterations associated with human disease such as cancer. The invention furthermore relates to methods for detection, quantification a s well as monitoring the expression of deoxy nucleic acids.
Owner:QIAGEN GMBH
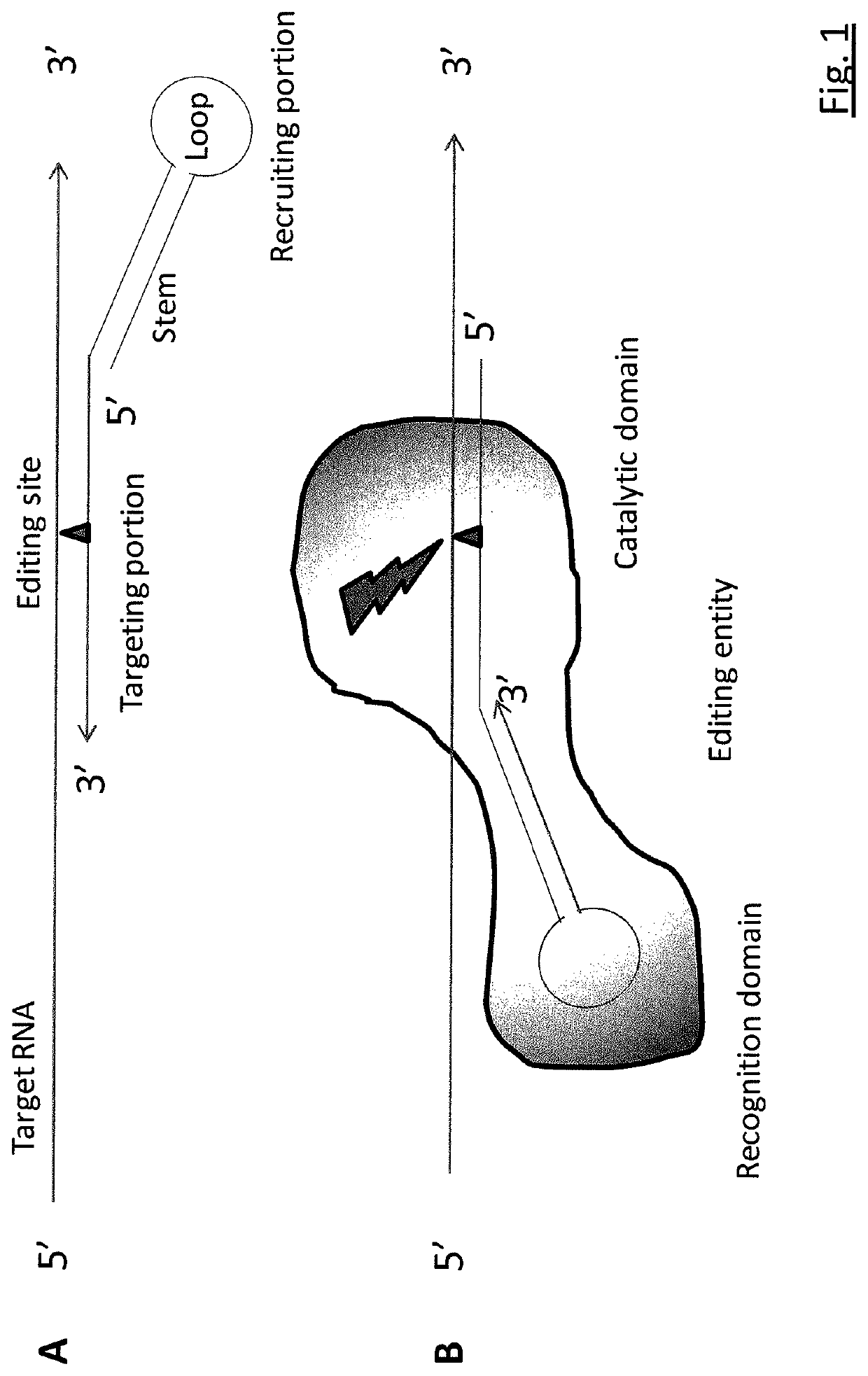
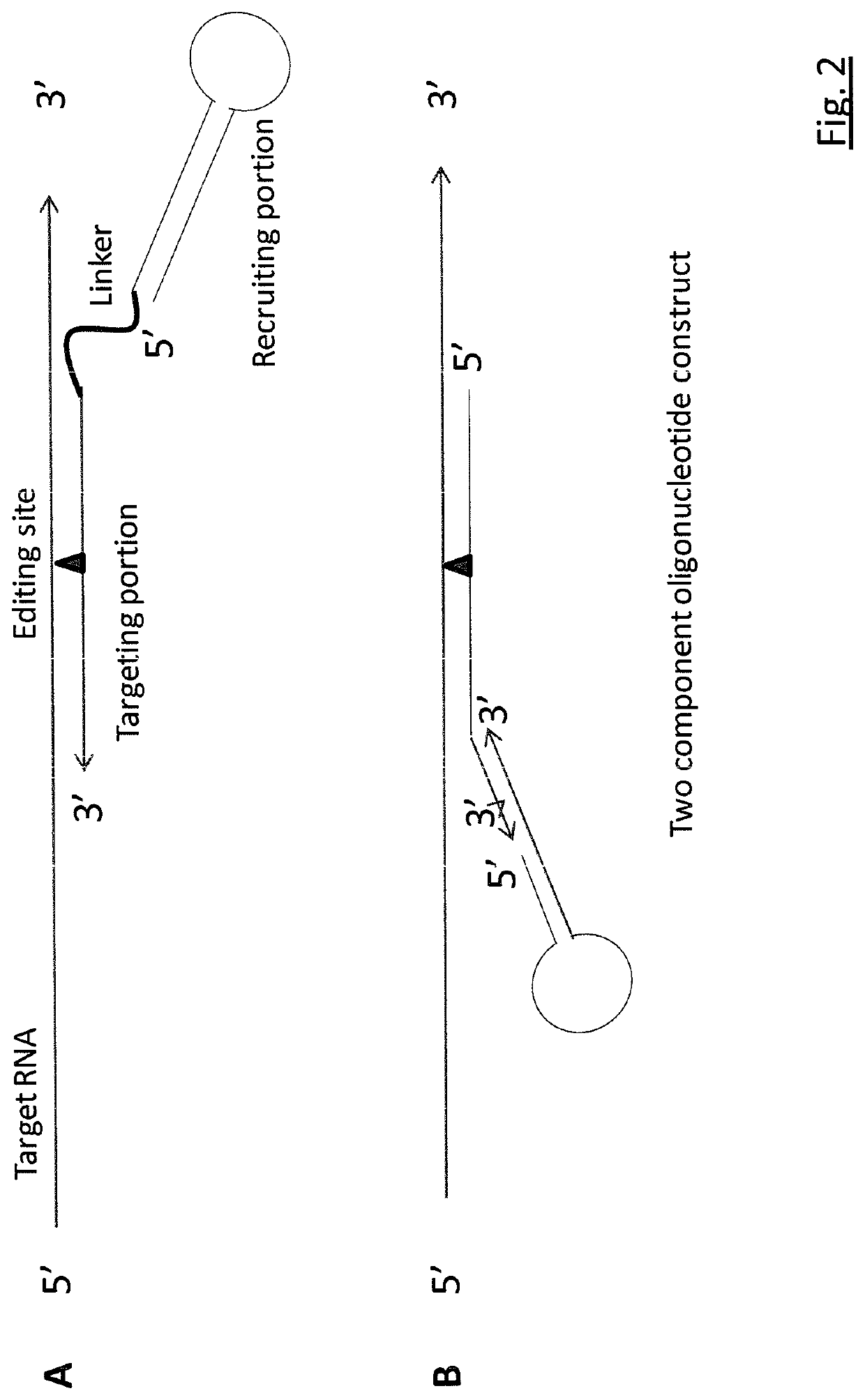
Targeted RNA editing
ActiveUS20190040383A1Improve organ functionMicrobiological testing/measurementStable introduction of DNANucleotideNucleic acid sequencing
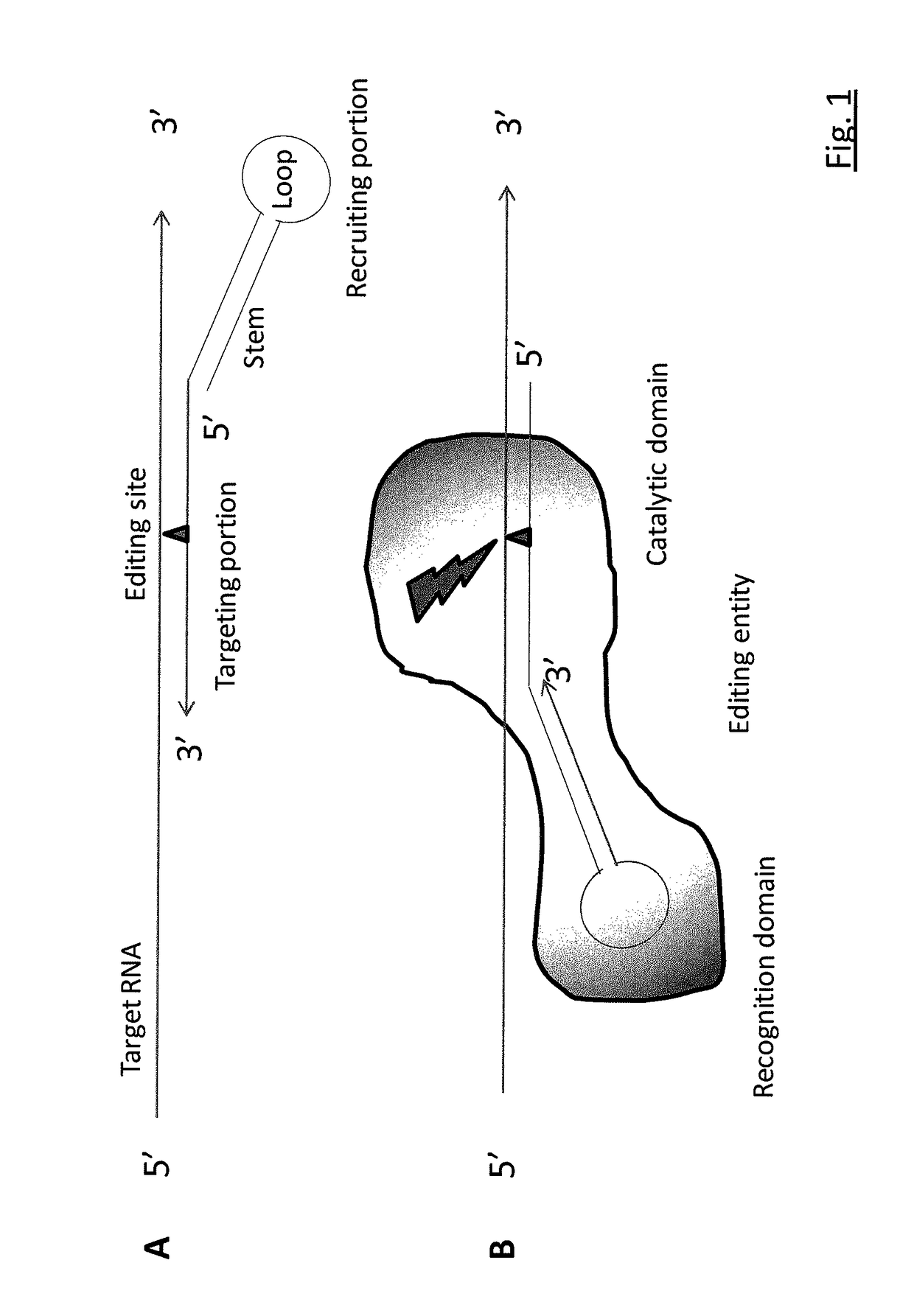
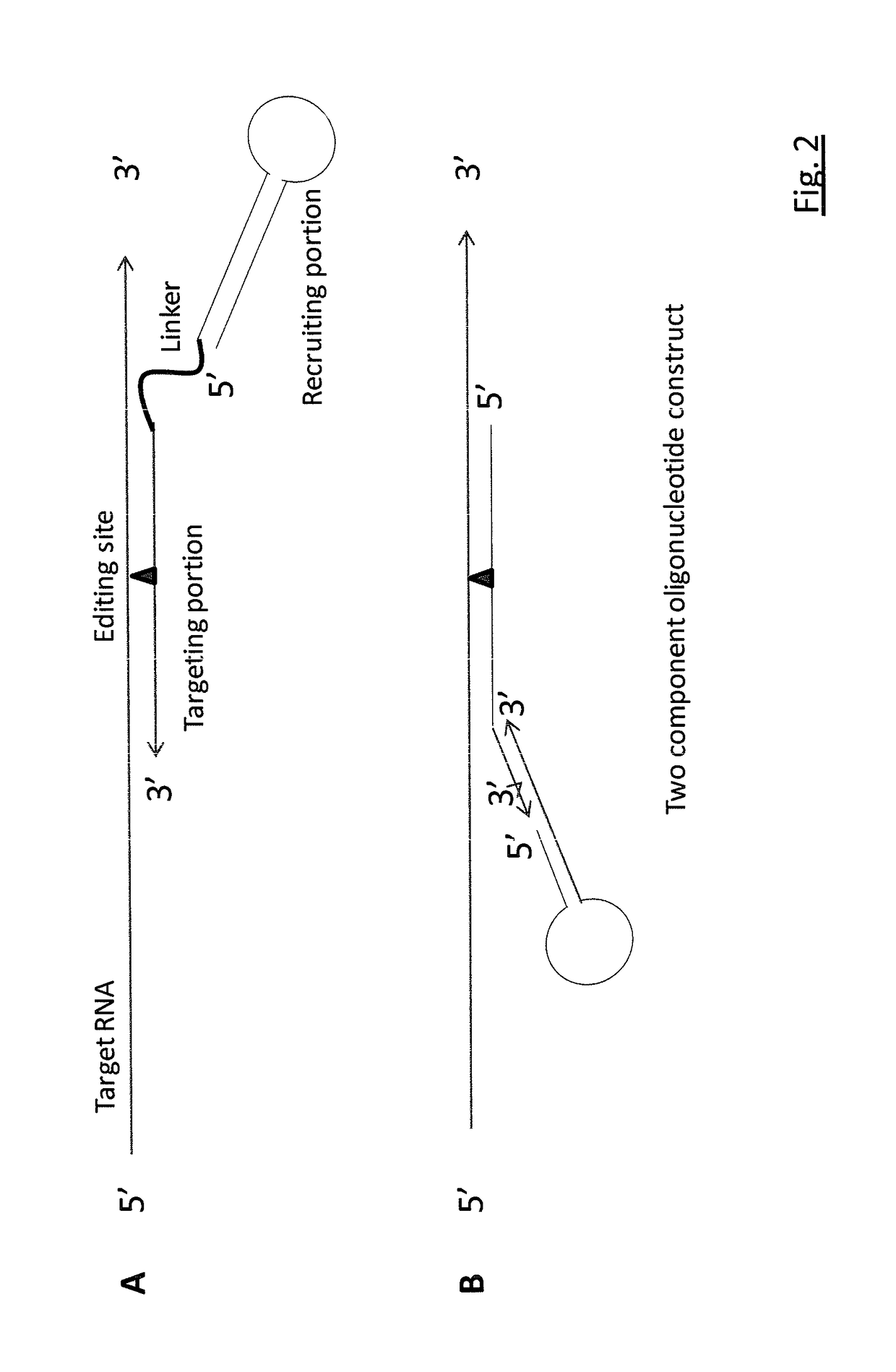
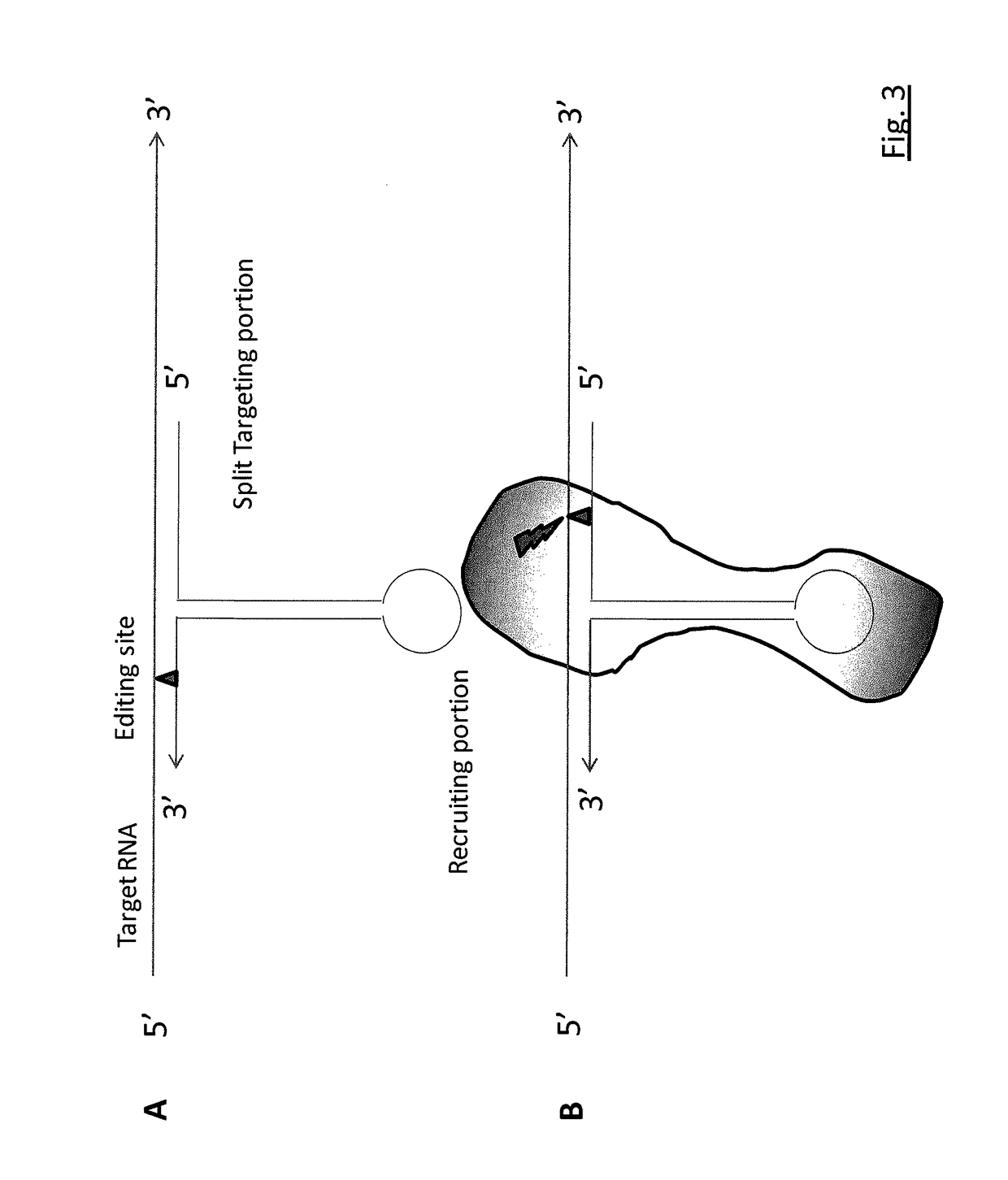
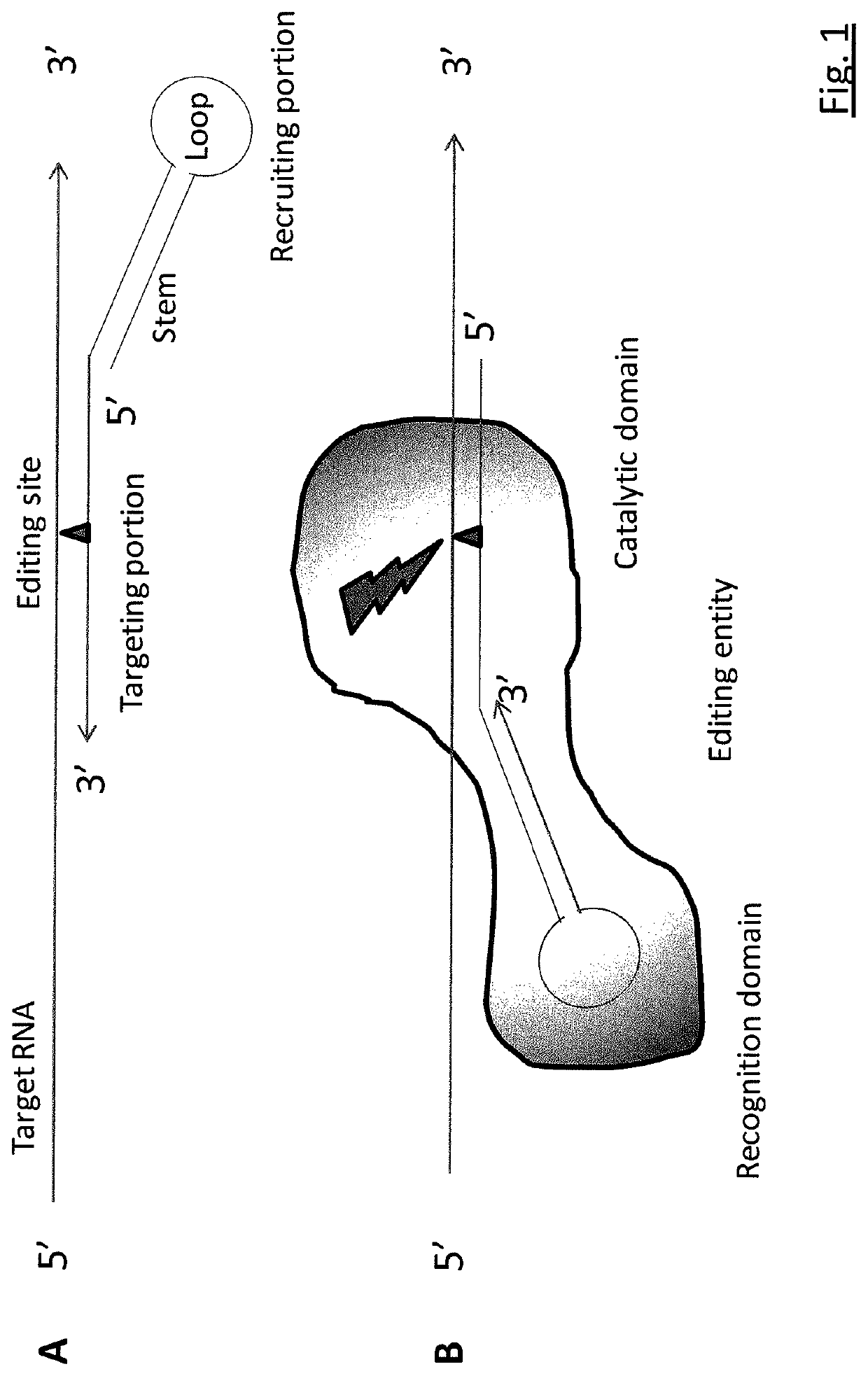
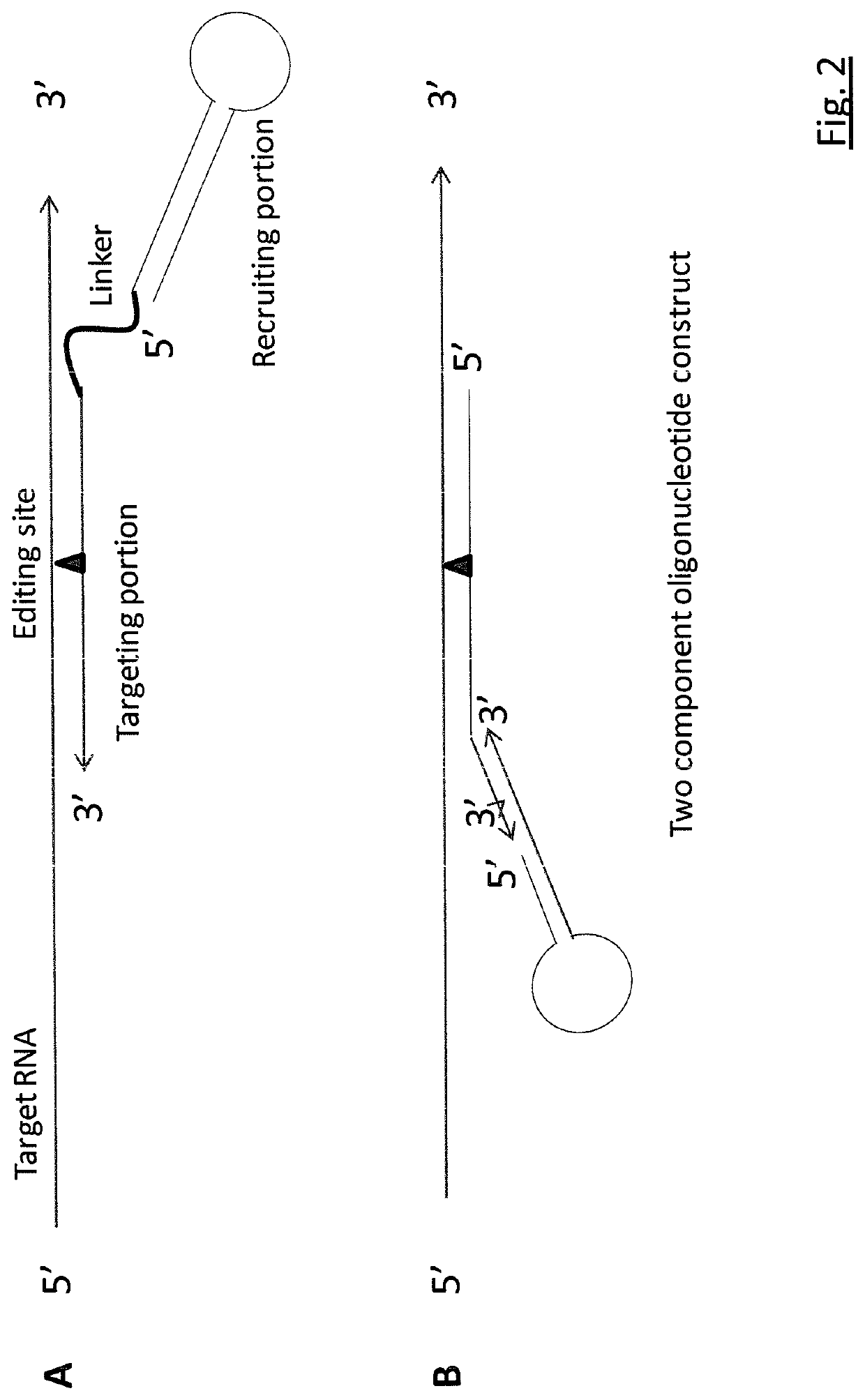
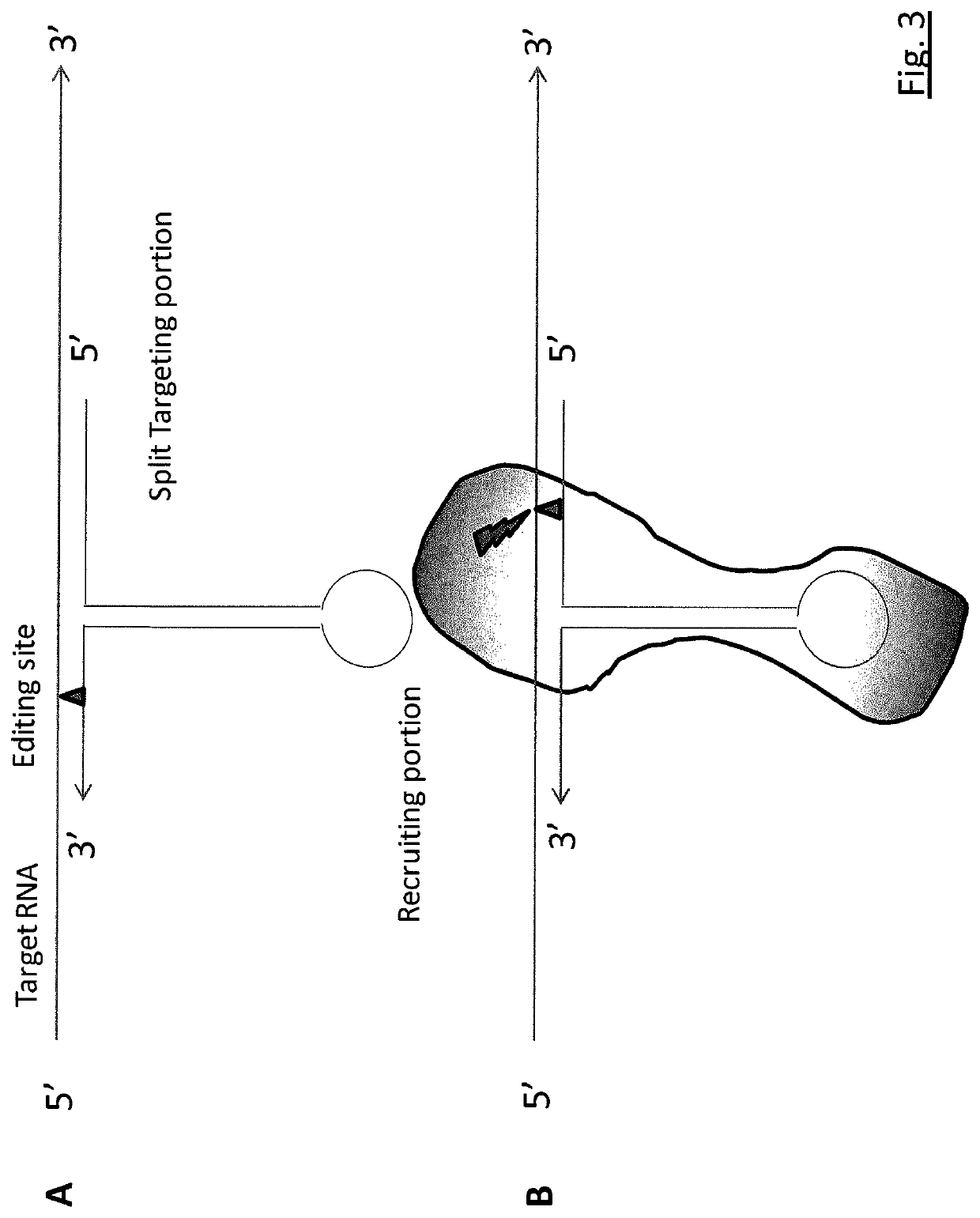
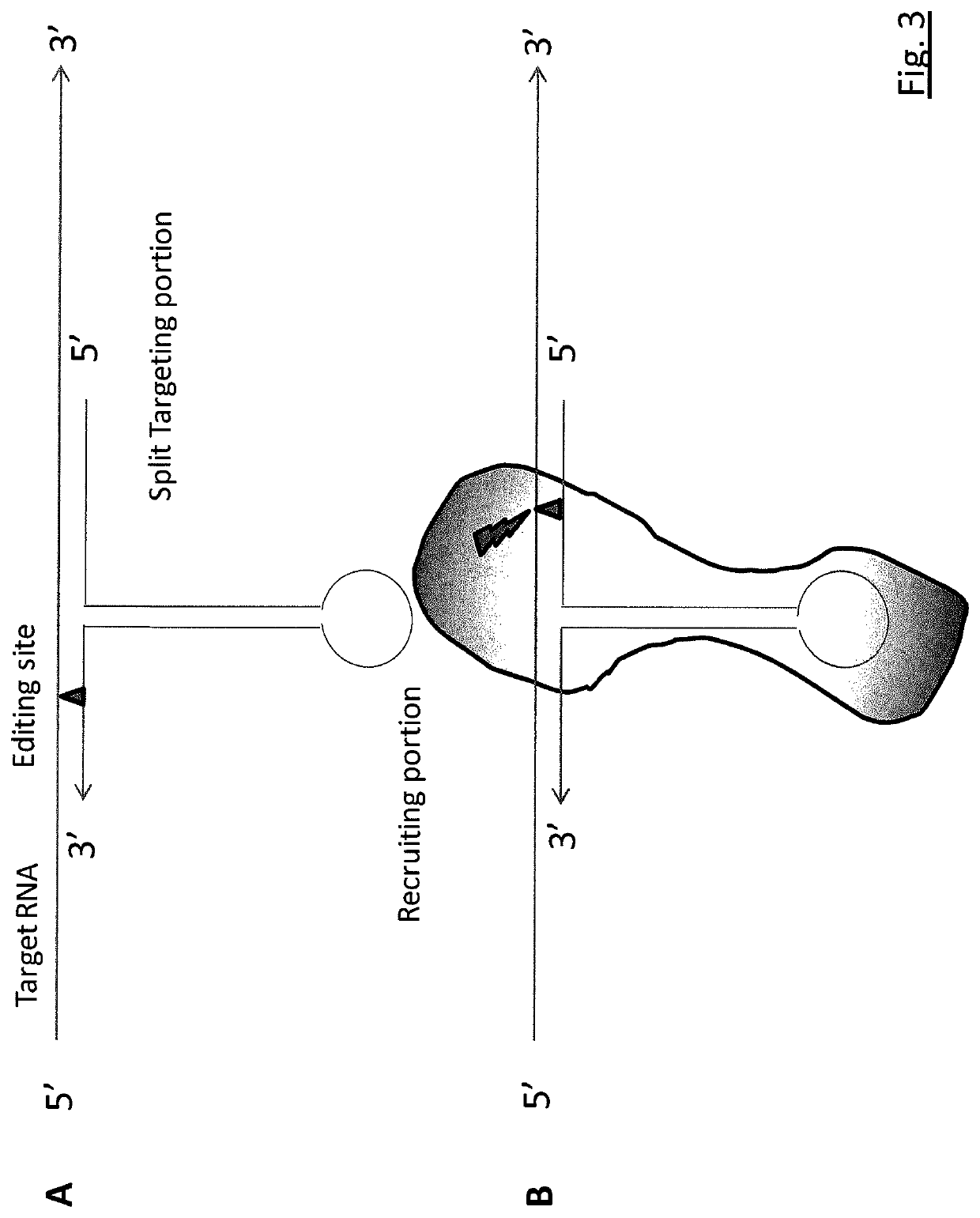
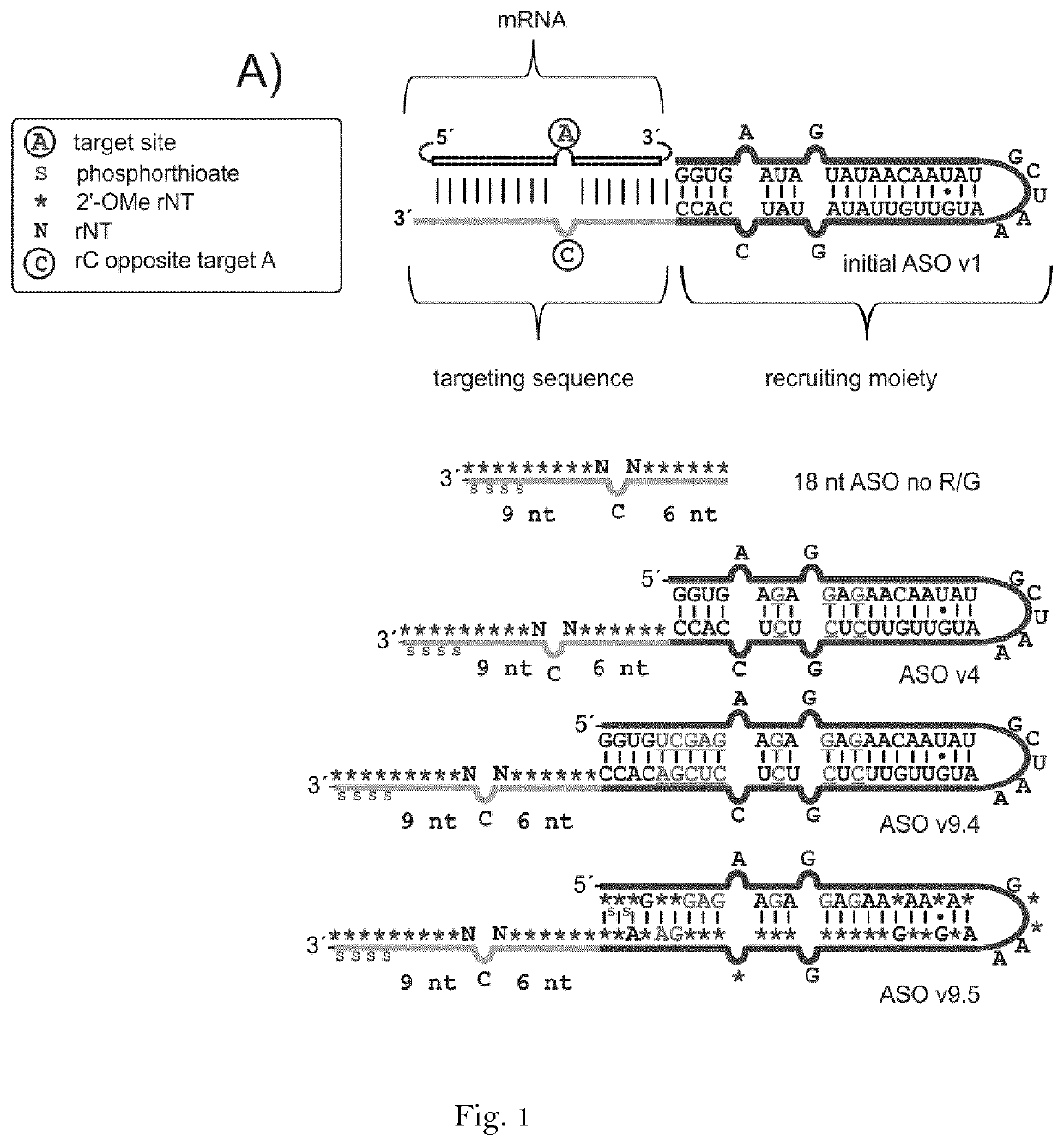
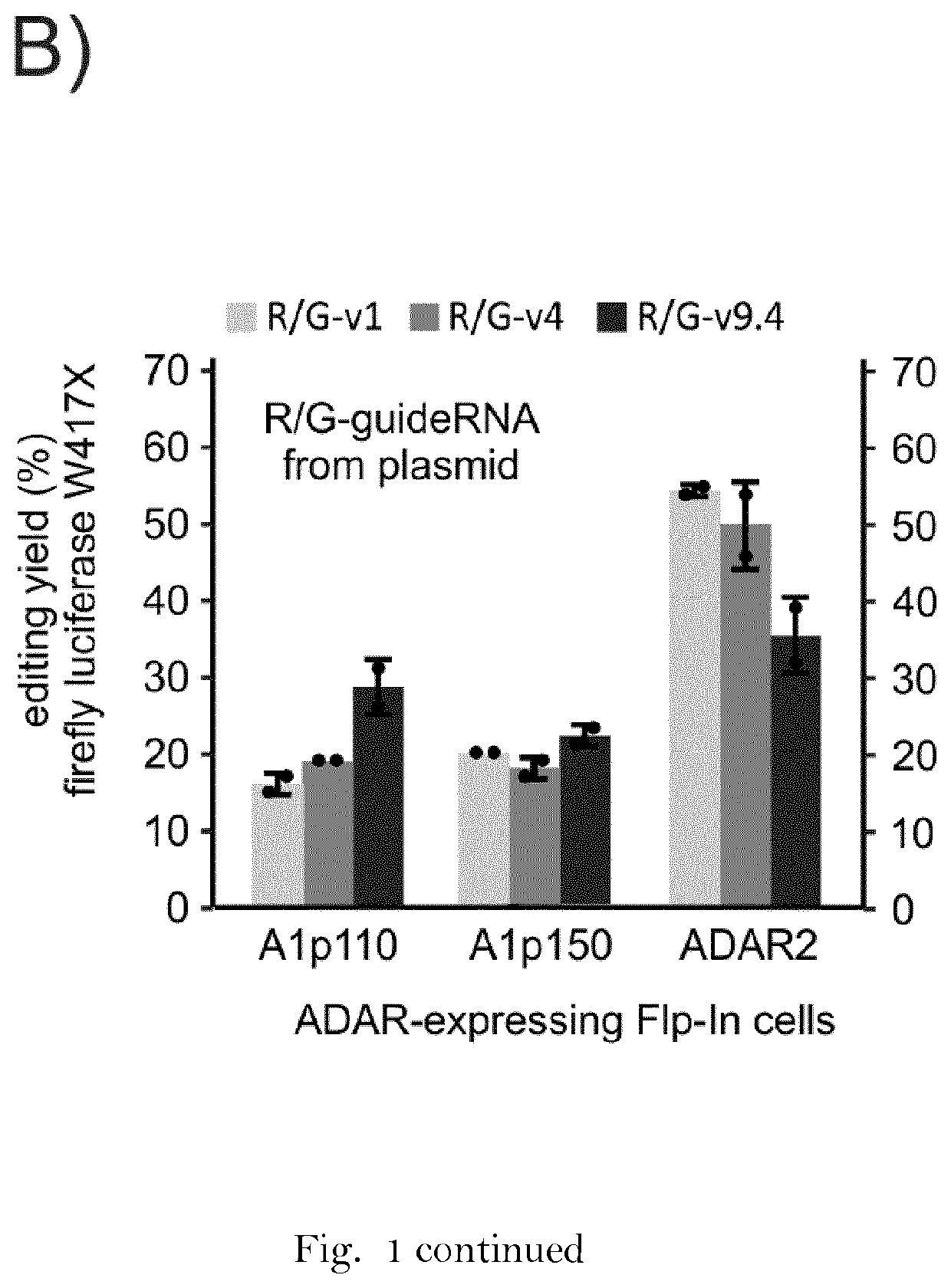
RNA editing is achieved using oligonucleotide constructs comprising (i) a targeting portion specific for a target nucleic acid sequence to be edited and (ii) a recruiting portion capable of binding and recruiting a nucleic acid editing entity naturally present in the cell. The nucleic acid editing entity, such as ADAR, is redirected to a preselected target site by means of the targeting portion, thereby promoting editing of preselected nucleotide residues in a region of the target RNA which corresponds to the targeting portion.
Owner:PROQR THERAPEUTICS II BV
RNA editing system based on CRISPR-Cas13 and application of RNA editing system
ActiveCN111206053APrecise time and space controlRealize precise time and space controlNervous disorderPeptide/protein ingredientsOptogeneticsEnzyme
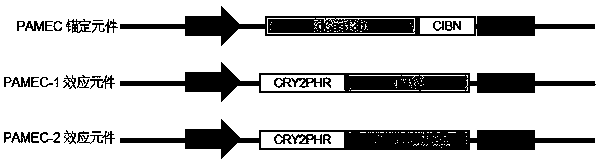
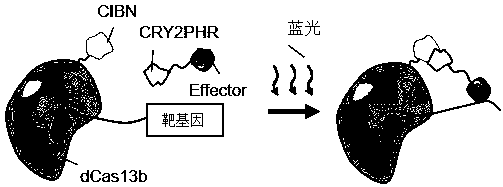
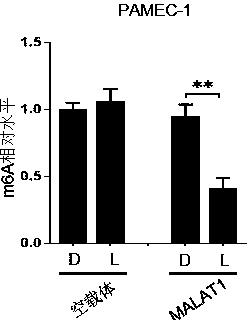
The invention provides an RNA editing system based on CRISPR-Cas13 and application of the RNA editing system. The RNA editing system comprises a targeting element, an editing effect element and a guide element, wherein the targeting element comprises a stimulus-responsive protein A and an inactivated Cas13 enzyme (dCas13), and the editing effect element comprises a stimulus-responsive protein B and an RNA editing effect protein; the stimuli-responsive protein A and the stimuli-responsive protein B are combined with each other under the action of stimuli and are uncombined after the stimuli disappear; the concept of light-operated RNA editing is provided for the first time, optogenetics and RNA editing are combined, the light-operated RNA editing system based on the CRISPR-Cas system is constructed, accurate space-time regulation and control of RNA editing are achieved, and the system has wide application prospects and huge market value.
Owner:TIANJIN HOSPITAL
Targeted RNA editing
ActiveUS20200199586A1Improve organ functionMicrobiological testing/measurementStable introduction of DNANucleotideNucleic acid sequencing
Owner:PROQR THERAPEUTICS II BV
Novel methods for quantification of micrornas and small interfering rnas
ActiveUS20110076675A1Solve low usageHighly hybridizationSugar derivativesMicrobiological testing/measurementNon-coding RNAAllele
The invention relates to ribonucleic acids, probes and methods for detection, quantification as well as monitoring the expression of mature microRNAs and small interfering RNAs (siRNAs). The invention furthermore relates to methods for monitoring the expression of other non-coding RNAs, mRNA splice variants, as well as detecting and quantifying RNA editing, allelic variants of single transcripts, mutations, deletions, or duplications of particular exons in transcripts, e.g., alterations associated with human disease such as cancer. The invention furthermore relates to methods for detection, quantification as well as monitoring the expression of deoxy nucleic acids.
Owner:QIAGEN GMBH
Targeted RNA editing
ActiveUS10676737B2Improve organ functionMicrobiological testing/measurementStable introduction of DNANucleotideNucleic acid sequencing
RNA editing is achieved using oligonucleotide constructs comprising (i) a targeting portion specific for a target nucleic acid sequence to be edited and (ii) a recruiting portion capable of binding and recruiting a nucleic acid editing entity naturally present in the cell. The nucleic acid editing entity, such as ADAR, is redirected to a preselected target site by means of the targeting portion, thereby promoting editing of preselected nucleotide residues in a region of the target RNA which corresponds to the targeting portion.
Owner:PROQR THERAPEUTICS II BV
Oligonucleotide complexes for use in RNA editing
ActiveUS20190352641A1Improve propertiesAvoid widthHydrolasesDNA/RNA fragmentationNucleotideAdenosine
Owner:PROQR THERAPEUTICS II BV
Methods and kits for diagnosing schizophrenia
Methods and kits for diagnosing schizophrenia are disclosed. The diagnostic method comprises analyzing in a biological sample of a subject a level of A-to-I RNA editing of at least one CNS-expressed gene as set forth in Table 1, wherein an amount of the A-to-I RNA editing of the at least one gene below a predetermined level is indicative of schizophrenia in the subject.
Owner:RAMOT AT TEL AVIV UNIV LTD +1
A feature analysis method for RNA editing sites
ActiveCN105528532AEasy to analyzeQuick analysisSpecial data processing applicationsData setFree energies
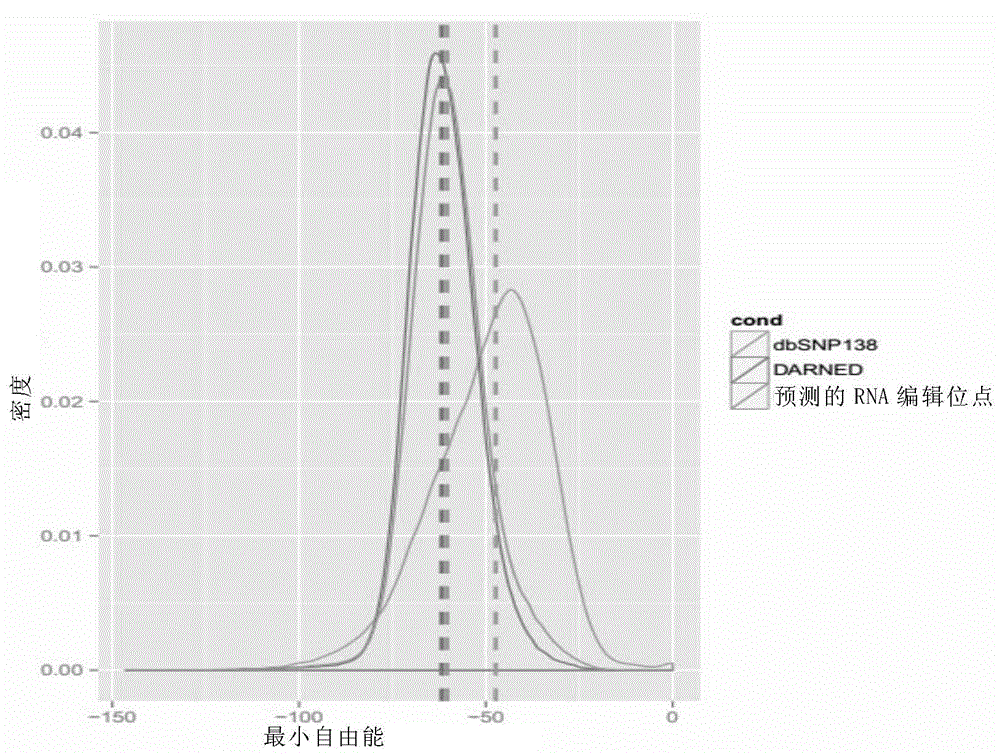
The invention provides a feature analysis method for RNA editing sites. The method comprises the steps of performing sequencing on to-be-analyzed samples and obtaining DNA and RNA data; analyzing the obtained data to obtain an RNA editing site data set; obtaining an RNA secondary structure free energy distribution curve of RNA editing site upstream and downstream sequences in the RNA editing site data set through statistics. The method can conveniently and rapidly analyze the basic features of RNA editing site data.
Owner:BGI SHENZHEN CO LTD
RNA fixed-point editing by artificially constructing RNA editing enzyme and related application
InactiveCN111793627ALow immunogenicityReduce system complexityFusion with RNA-binding domainHydrolasesPathogenicityFunctional protein
Owner:SHANGHAI INST OF BIOLOGICAL SCI CHINESE ACAD OF SCI
Antisense oligonucleotides for RNA editing
InactiveUS20210079393A1Depleting amountConvenient verificationOther foreign material introduction processesGene therapyRiboseADAR
The invention relates to editing oligonucleotides (EONs) that carry 2′-0-methoxyethyl (2′-MOE) ribose modifications at specified positions and that do not carry such modifications on positions that would lower RNA editing efficiency. The selection of positions that should or should not carry a 2′-MOE modification is based on computational modelling that revealed steric clashes between the 2′-MOE modification and mammalian ADAR enzymes.
Owner:PROQR THERAPEUTICS II BV
Apobec3a cytidine deaminase induced RNA editing
Provided are methods for identifying agents which can induce or inhibit C>U deamination in RNA driven by apolipoprotein B editing catalytic proteins. The method comprises contacting APOBEC3A or APOBEC3G with a suitable RNA substrate and determining the extent of C>U deamination under conditions which induce APOBEC driven C>U deamination.
Owner:HEALTH RES INC
VI-B type CRISPR/Cas13 gene editing system and application thereof
The invention relates to the technical field of gene editing, in particular to a VI-B type CRISPR / Cas13 gene editing system and application thereof. The VI-B type CRISPR / Cas13 gene editing system disclosed by the invention comprises an Lt1Cas13b protein and CRISPR RNA; and the Lt1Cas13b protein is RNA endonuclease, and the Lt1Cas13b protein has an amino acid sequence as shown in SEQ ID NO: 1 or anamino acid sequence which is at least 80% the same as the amino acid sequence as shown in the SEQ ID NO: 1. The invention digs out the novel VI-B type CRISPR / Cas13 gene editing system. The system canbe widely applied to scenes needing to identify, combine and edit prokaryote or eukaryote RNA, and provides a new application choice for an RNA editing tool.
Owner:ZHUHAI SHU TONG MEDICAL TECH CO LTD
Oligonucleotide complexes for use in RNA editing
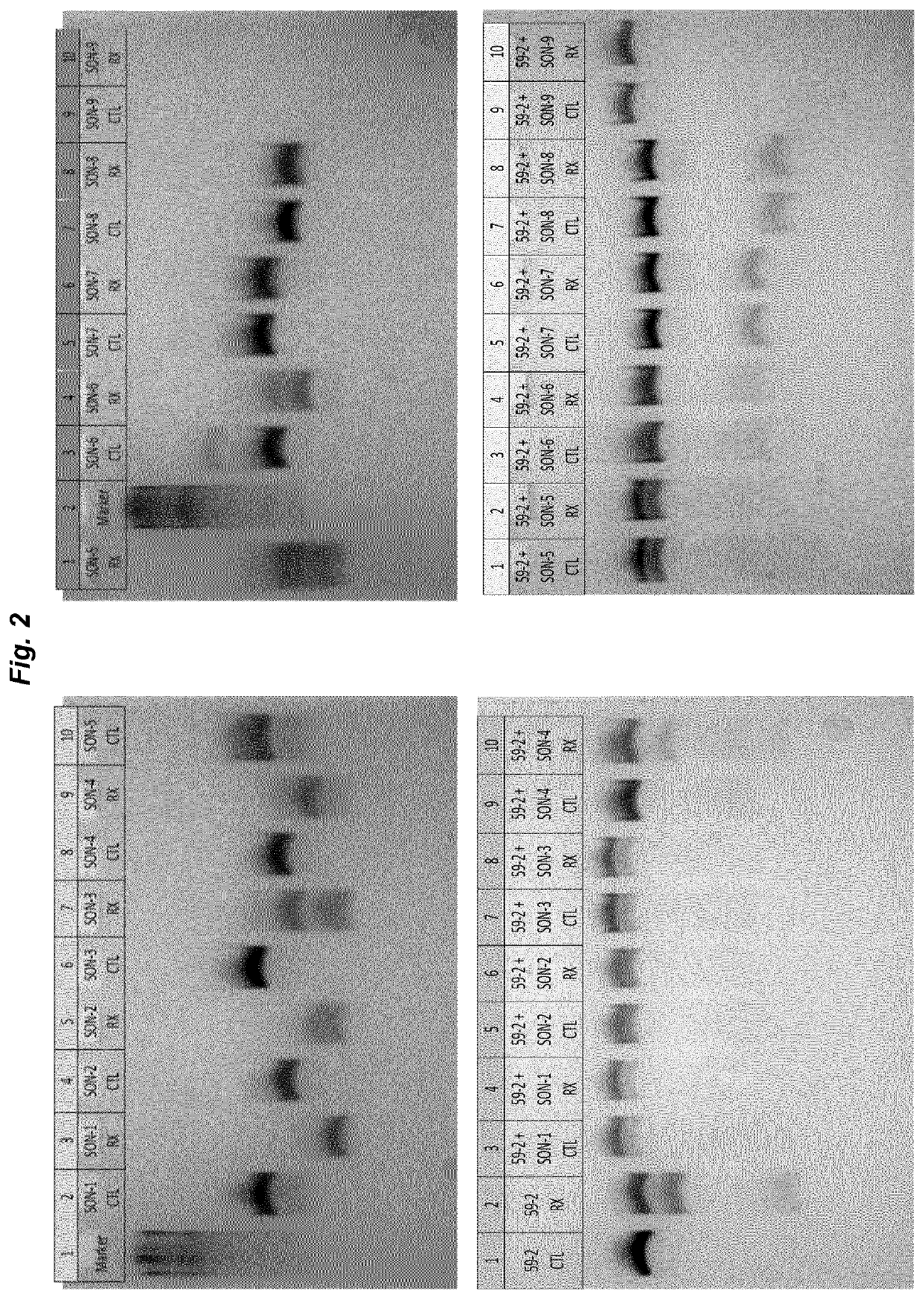
The invention relates to double stranded oligonucleotide complexes comprising an antisense oligonucleotide (AON) and a complementary sense oligonucleotide (SON), for use in the deamination of a target adenosine in a sense target RNA sequence in a cell by an ADAR enzyme, wherein at least the nucleotide in the AON that is directly opposite the target adenosine in the target RNA sequence does not have a 2′-O-alkyl modification and the SON comprises nucleotides that are at least complementary to all nucleotides in the AON that do not have a 2′-O-alkyl modification. The invention further relates to methods of RNA editing using the AON / SON complexes of the invention.
Owner:PROQR THERAPEUTICS II BV
Artificial nucleic acids for RNA editing
PendingUS20220073915A1Improve efficiencyUndesired off-target editing can nevertheless be avoidedHydrolasesDNA/RNA fragmentationDiagnoses diseasesIn vitro diagnostic
The present invention concerns artificial nucleic acids for site-directed editing of a target RNA. In particular, the present invention provides artificial nucleic acids capable of site-directed editing of endogenous transcripts by harnessing an endogenous deaminase. Further, the present invention provides artificial nucleic acids for sited-directed editing of a target RNA, which are chemically modified, in particular according to a modification pattern as described herein. The invention also comprises a vector encoding said artificial nucleic acid and a composition comprising said artificial nucleic acid. Moreover, the invention provides the use of the artificial nucleic acid, the composition or the vector for site-directed editing of a target RNA or for in vitro diagnosis. In addition, the artificial nucleic acid, the composition or the vector as described herein are provided for use as a medicament or for use in diagnosis of a disease or disorder.
Owner:UNIV TUBINGEN
RNA editing biomarkers for diagnosis, pharmacological screening and prognostication in cancer
InactiveUS20170191057A1Not consistentOrganic active ingredientsGenetically modified cellsDiseaseBiology
Owner:RGT UNIV OF CALIFORNIA
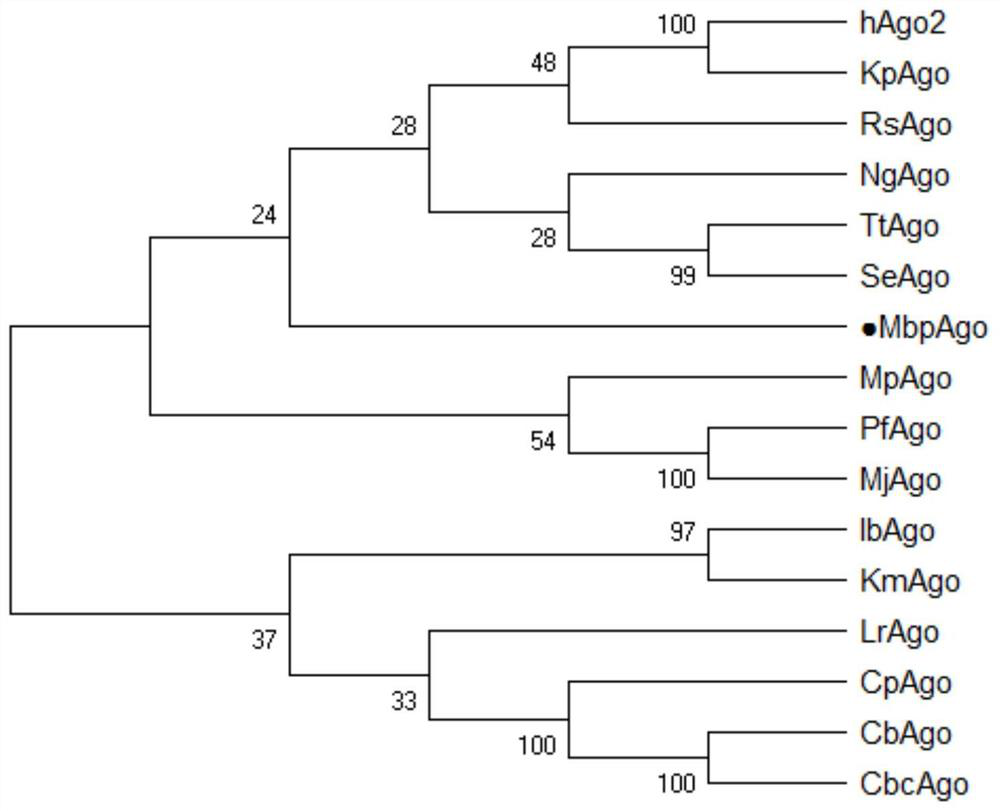
Prokaryote-derived Mbp_Argonaute protein and application thereof
ActiveCN113373129AHave binding activityHas nuclease activityHydrolasesFermentationProkaryote organismsPlant cell
The invention discloses a prokaryote-derived Mbp_Argonaute protein and application thereof. The Mbp_Argonaute protein has an amino acid sequence shown in SEQ ID NO: 1 or as shown in a sequence which has at least 50% or at least 80% of homology with SEQ ID NO: 1. According to the invention, an Argonaute protein gene derived from a cold-resistant prokaryotic organism Mucilaginibacter paaluis is synthesized, the protein is named as MbpAgo, and researches find that the MbpAgo has binding activity to single-stranded guide DNA and has nuclease activity to target RNA and / or target DNA complementarily paired with the single-stranded guide DNA, therefore, the MbpAgo can be used for targeted RNA editing in vivo and in vitro, and then specific site modification of genetic material. The MbpAgo not only can modify RNA with a high-grade structure, but also does not influence an endogenous RNAi pathway of animal and plant cells, provides a brand new powerful tool for RNA editing, and is high in cutting activity and good in specificity.
Owner:HUBEI UNIV
Method for high-flux sequencing for establishing of human miRNA (micro-ribonucleic acid) sequencing library
ActiveCN108130366AGuaranteed uniformityHigh detection sensitivityMicrobiological testing/measurementDNA/RNA fragmentationTotal rnaHigh flux
The invention discloses a method for high-flux sequencing for establishing of a human miRNA (micro-ribonucleic acid) sequencing library. The method comprises the following steps of S1, extracting human total RNA, and reversing into cDNA (complementary deoxyribonucleic acid); S2, using the cDNA as a template, using a probe to perform mmPCR (multi-multiplex polymerase chain reaction) pre-amplification and mmPCR amplification on miRNA in a sample, and respectively connecting a 3' terminal and a 5' terminal to both ends of an amplified product; S4, connecting a tag sequence with a mmPCR amplifiedproduct; S5, performing gel electrophoresis on the sample added with the tag sequence, recycling a pure electrophoresis product, and building the miRNA library; S6, mixing the obtained multiple miRNAlibraries, and utilizing a next generation sequencing technique to perform high-flux sequencing. The method has the advantages that the detection sensitivity of RNA editing and modifying on the precursor miRNA is greatly improved, the uniformity of the RNA-targeted sequencing libraries is improved, the library building and sequencing cycle is shortened, the cost is reduced, and the application prospect is larger.
Owner:SUN YAT SEN UNIV +1
Technique for realizing precisely fixed-point RNA shearing in fish embryos
PendingCN110669795AEffective positioningImprove efficiencyMicrobiological testing/measurementStable introduction of DNABiotechnologyFish embryo
The invention provides a technique for realizing precisely fixed-point RNA shearing in fish embryos. A CasRx-mediated RNA editing technique is used, only mRNA of CasRx and specific targeted guidance RNA need to be provided, precisely fixed-point RNA knockdown can be realized in the fish embryos, and the technique has the advantages such as high efficiency, high specificity, convenience and low cost. The problems of faced low efficiency of a current existing siRNA technique and high cost and prone off-target of a Morpholino-technique-mediated gene knockout technique are solved.
Owner:FUZHOU UNIV
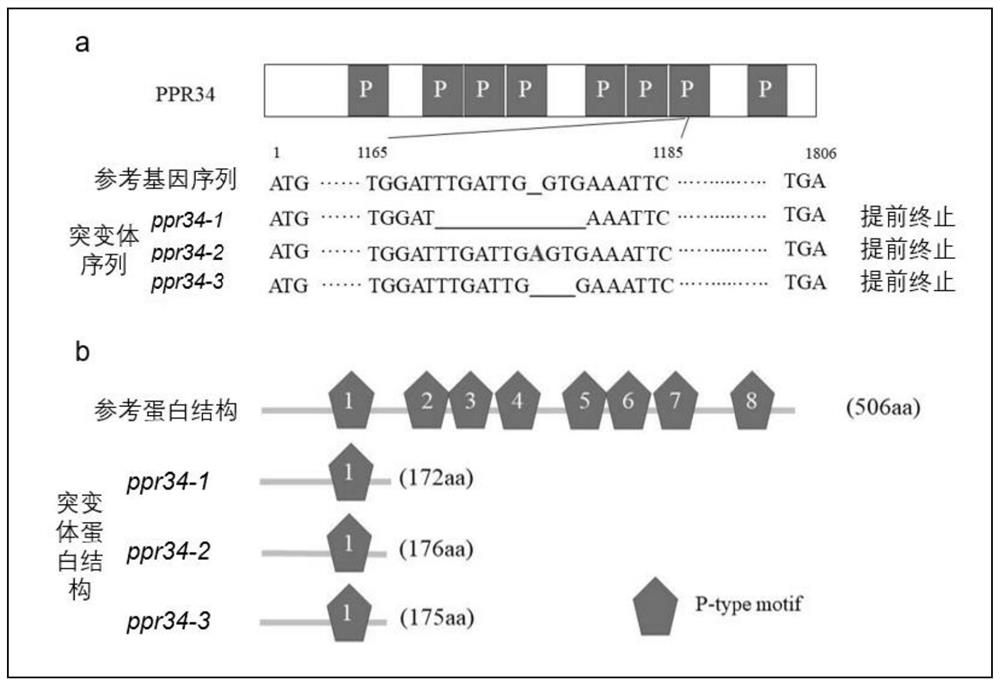
Rice OsPPR34 gene as well as encoding protein and application thereof
The invention provides an OsPPR34 gene which is separated and cloned from rice DNA and is related to a rice plant type and drought resistance. The protein coded by the gene contains a conservative P structural domain of a PPR (pentatricopeptide repeat) family, and participates in RNA editing of a mitochondrial gene. The OsPPR34 gene regulates and controls the plant height of the rice through a gibberellin way, meanwhile, the root tropism is changed, the deep root ratio of a knockout mutant of the OsPPR34 gene is remarkably increased, and the OsPPR34 gene shows high drought resistance in a root canal experiment. The OsPPR34 gene regulates and controls the plant height and the drought resistance of the rice, the plant height of a knockout mutant of the OsPPR34 gene is reduced, and the OsPPR34 gene has a high depth-to-root ratio and can be applied to improvement of the plant type of the cultivated rice and improvement of the drought resistance of the cultivated rice.
Owner:SHANGHAI AGROBIOLOGICAL GENE CENT
Chemically modified oligonucleotides for RNA editing
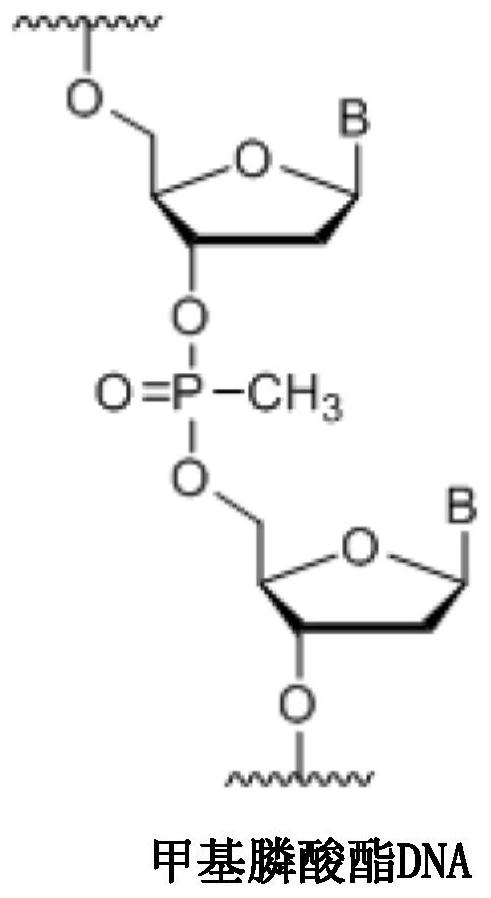
The invention relates to single-stranded RNA editing antisense oligonucleotides (AONs) for binding to a target RNA molecule for deaminating a target nucleotide, preferably an adenosine, present in the target RNA molecule and recruiting, in a cell, preferably a human cell, an enzyme with nucleotide deamination activity, preferably an ADAR enzyme, to deaminate the target nucleotide in the target RNA molecule. The AONs carry at least one methylphosphonate-modified internucleosidic linkage on a position that would render the AON more stable in comparison to an AON not carrying that methylphosphonate modification at that position.
Owner:プロキューアールセラピューティクスツーベスローテンフェンノートシャップ
Adenine base editors with reduced off-target effects
PendingUS20220307003A1Expanded toolboxImprove efficiencyPolypeptide with localisation/targeting motifFusion with DNA-binding domainBiologyOrganic chemistry
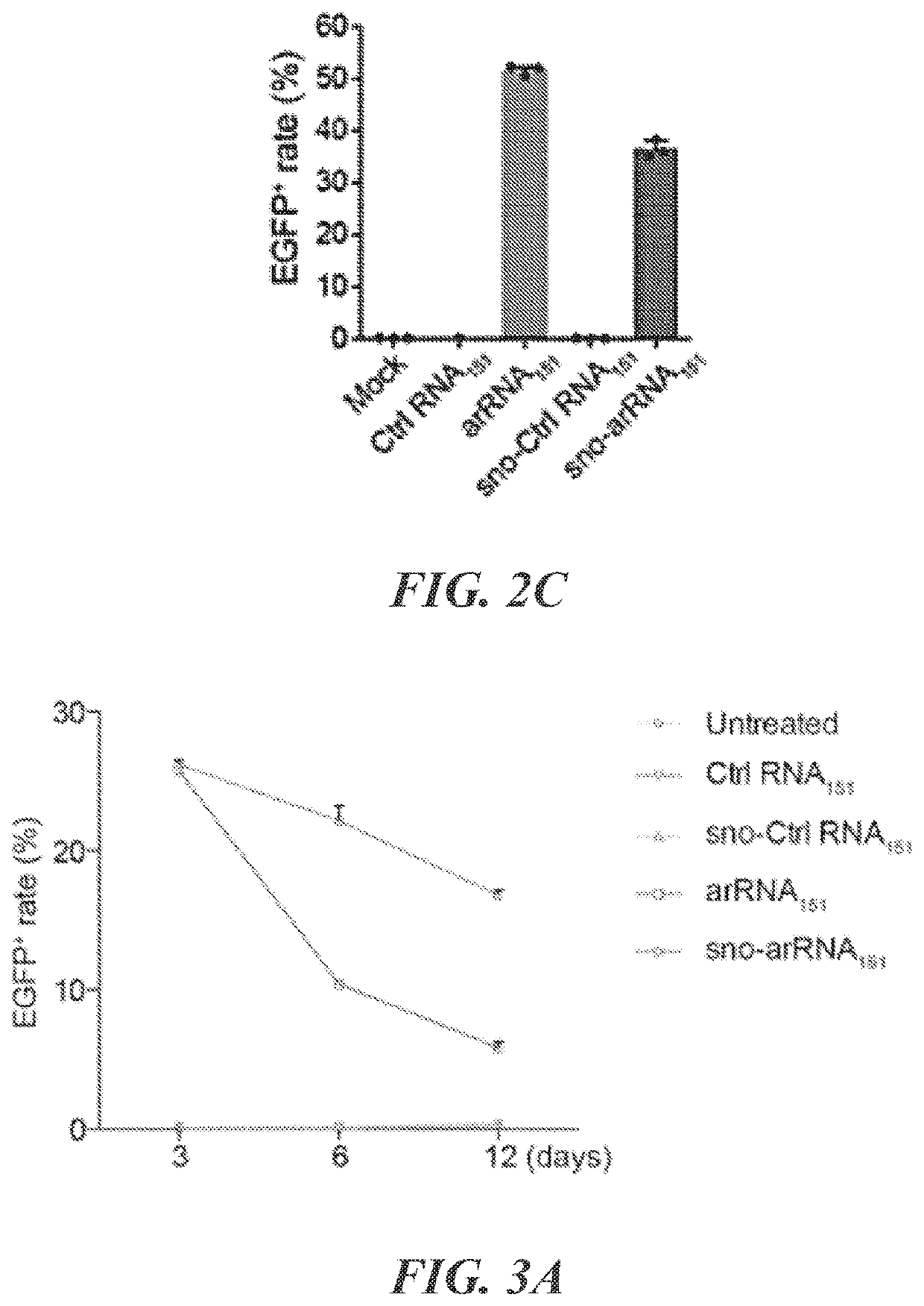
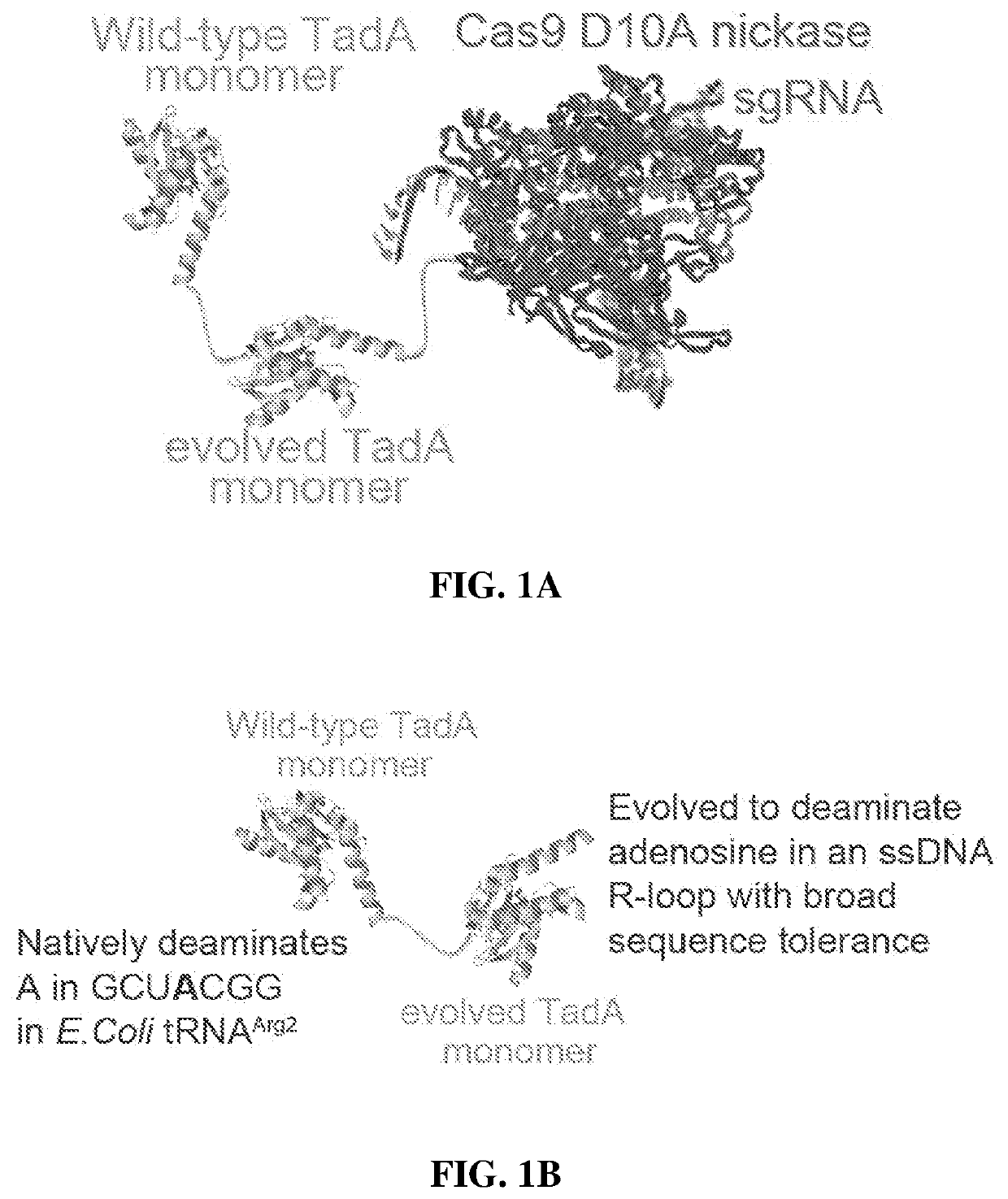
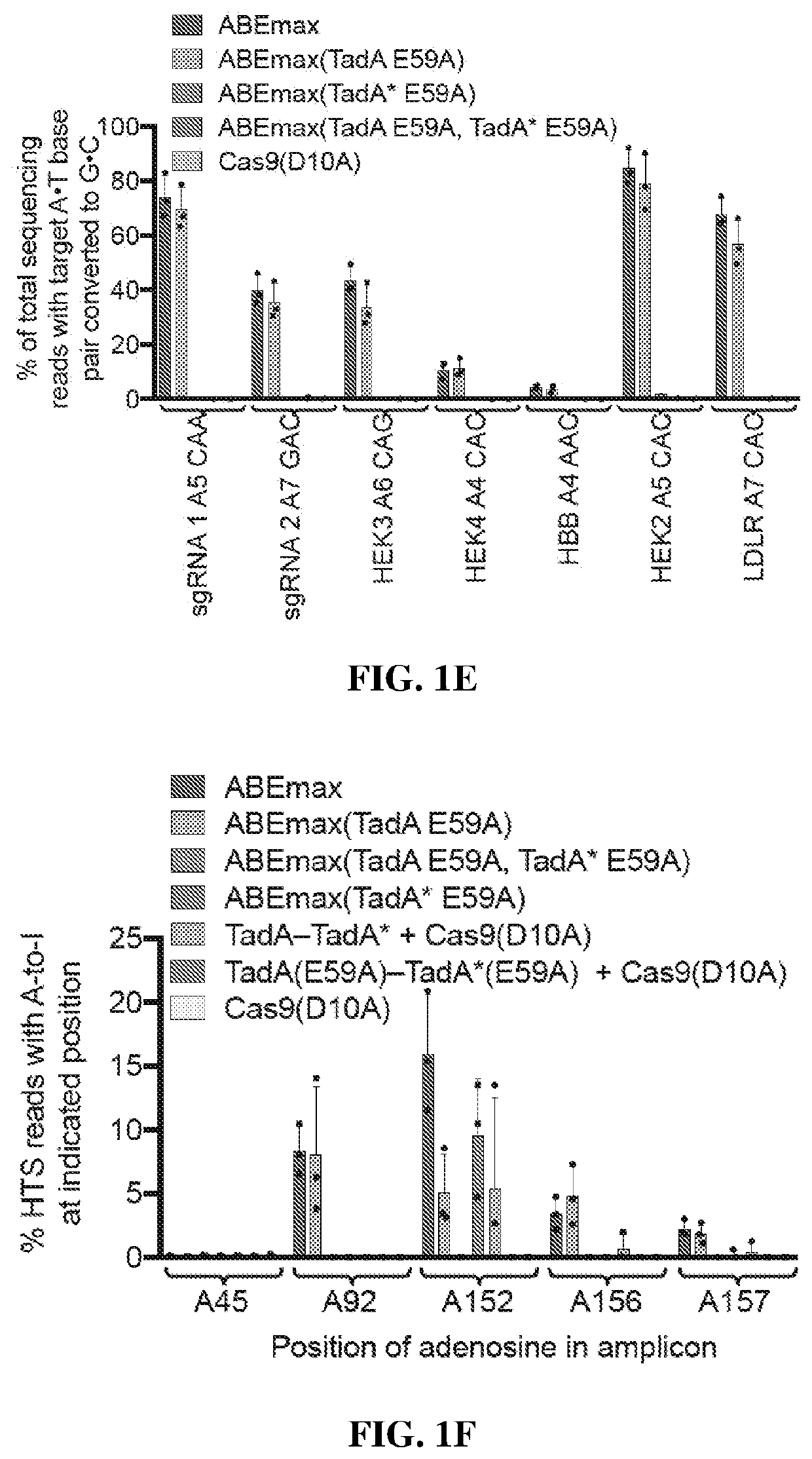
The present disclosure provides novel adenine base editors that retain ability to edit DNA efficiently but show greatly reduced off-target effects, such as reduced RNA editing activity, as well as lower off-target DNA editing activity and reduced indel by product formation. Also provided are base editing methods comprising contacting a nucleic acid molecule with an adenine base editor and a guide RNA that has complementarity to a target sequence. Further provided are complexes comprising a guide RNA bound to a base editor provided herein; and kits and pharmaceutical compositions for the administration of adenine base editor variants to a host cell.
Owner:THE BROAD INST INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com