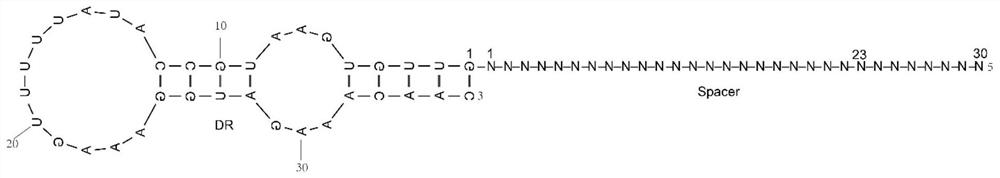VI-B type CRISPR/Cas13 gene editing system and application thereof
A VI-B, gene editing technology, applied in the field of gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] This embodiment analyzes and predicts the genes related to Lt1Cas13b described in the present invention.
[0039] (1) Materials: metagenomic sequencing data.
[0040] (2) Software: fastqc (v0.11.5), fastp (v0.19.8), SOAPnuke (v1.5.2), hista2 (v2.0.4), samtools (v1.9), iTools (v0.23), GeneMark (v3 .38), IDBA-UD(v1.1.3), Bowtie2(v2.3.5.1), CD-HIT(v4.6), hummer(3.1b2), BLAST(v2.3.0+),Ordination(v1.0 ), WilcoxonTest (v1.0), Environmental CorrAnalysis (v1.0)
[0041] (3) Detection method
[0042] The data source of this embodiment is metagenomic sequencing data, and all possible gene sequences in the metagenomic set are obtained by performing quality control, splicing and gene prediction on the metagenomic data.
[0043] For the raw data of the metagenomic group, the specific operation is as follows:
[0044] (a) First use fastqc to charge, check whether the data is original data or cleaned data, if it is original data, then use fastp to clean the original data and remov...
Embodiment 2
[0051] In this example, the obtained contig that may be related to the type 2 CRISPR-Cas system is analyzed, and the predicted protein and related elements related to the CRISPR-Cas system are obtained; that is, the contig obtained in Example 1 is related to CRISPR / Cas13 CRISPR sequence and Cas13 protein prediction.
[0052] (1) Materials: the contig sequence obtained in Example 1 that may be related to the CRISPR-Cas system.
[0053] (2) Software: CRISPR CasFinder (v4.2.19), python (v3.7.4), bedtools (v2.25.0).
[0054] (3) Detection method:
[0055] The specific operation is as follows:
[0056] (a) Use CRISPRCasFinder to analyze and predict the obtained contig that may be related to the type 2 CRISPRCasFinder system, and obtain Cas protein, Spacer, direct repeat and other CRISPR-Cas system related proteins and elements;
[0057] (b) Annotate the subclassification of candidate type VI protein families: collect the HMM files of Cas13a, Cas13b, Cas13c and Cas13d effector pr...
Embodiment 3
[0061] This embodiment is to predict the RNA secondary structure of the guide RNA molecule recognized by the Cas13b gene editing system of the present invention, and the obtained RNA secondary structure is as follows: figure 2 shown.
[0062] (1) Material: repeat sequence;
[0063] (2) Software: NUPACK (http: / / www.nupack.org / partition / new);
[0064] (3) Prediction method: By using the online application NUPACK to simulate the secondary structure of repeat RNA in vitro at 37°C, the figure 2 RNA secondary structures shown.
[0065] Depend on figure 2 It can be seen that the pre-crRNA forms a mature crRNA under the action of Lt1Cas13 nuclease, forming a guide RNA (guide RNA), and the guide RNA includes two stem-loop structures.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com