Creatine amidinohydrolase, production thereof and use thereof
a technology of amidinohydrolase and creatine, which is applied in the field of creatine amidinohydrolase, can solve the problems of lower heat stability and greater km value of creatine, enzymes are not suitable for clinical testing, and are produced from various known cell lines. , to achieve the effect of small km value, superior reactivity to creatine, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
reference example 1
Isolation of chromosomal DNA
The chromosomal DNA of Alcaligenes.cndot.faecalis TE3581 was isolated by the following method.
The cells (FERM P-14237) were shake-cultured overnight at 30.degree. C. in a nutrient broth (150 ml) and the cells were collected by centrifugation (8000 rpm, 10 min). The cells were suspended in a solution (5 ml) containing 10% sucrose, 50 mM Tris-HCl (pH 8.0) and 50 mM EDTA, and a lysozyme solution (1 ml, 10 mg / ml) was added. The mixture was incubated at 37.degree. C. for 15 min. Then, 10% SDS solution (1 ml) was added. An equivalent amount (1 ml) of a choroform.cndot.phenol solution (1:1) was added to this mixture. The mixture was stirred and separated into an aqueous layer and a solvent layer by centrifugation at 10,000 rpm for 3 min. The aqueous layer was separated, and onto this aqueous layer was gently layered a 2-fold amount of ethanol. The content was slowly stirred with a glass rod to allow the DNA to wind around the rod.
This DNA was dissolved in 10 mM ...
reference example 2
Preparation of DNA fragment containing a gene encoding creatinine amidinohydrolase and recombinant vector containing said DNA fragment
The DNA (20 .mu.g) obtained in Reference Example 1 was partially cleaved with restriction enzymes Sau3AI (Toyo Boseki Kabushiki Kaisha) and 2-10 kbp fragments were recovered by sucrose density gradient centrifugation. Meanwhile, pBluescript KS(+) cleaved with restriction enzyme BamHI (Toyo Boseki Kabushiki Kaisha) was dephosphorylated with bacterial alkaline phosphatase (Toyo Boseki Kabushiki Kaisha). Then, the both DNAs were treated with T4DNA ligase (1 unit, Toyo Boseki Kabushiki Kaisha) at 16.degree. C. for 12 hr to ligate the DNA. Escherichia coli JM109 competent cell (Toyo Boseki Kabushiki Kaisha) was transformed with the ligated DNA and plated onto a creatine amidinohydrolase activity detection agar medium [0.5% yeast extract, 0.2% meat extract, 0.5% polypeptone, 0.1% NaCl, 0.1% KH.sub.2 PO.sub.4, 0.05% MgSO.sub.4 / 7H.sub.2 O, 1.15% creatine, 10...
example 1
Preparation of recombinant plasmid pCRH273 by mutating creatine amidinohydrolase gene
The region of from .beta.-galactosidase structural gene derived from the vector to the upstream region of the creatine amidinohydrolase structural gene of the insert DNA was deleted from the recombinant plasmid pCRH173 of Reference Example 2, using the synthetic DNA depicted in SEQ ID:No.3 and a commercially available mutation introduction kit (Transformer.TM.; Clonetech) to prepare recombinant plasmid pCRH173M. The detailed method for introducing the mutation was given in the protocol attached to the kit.
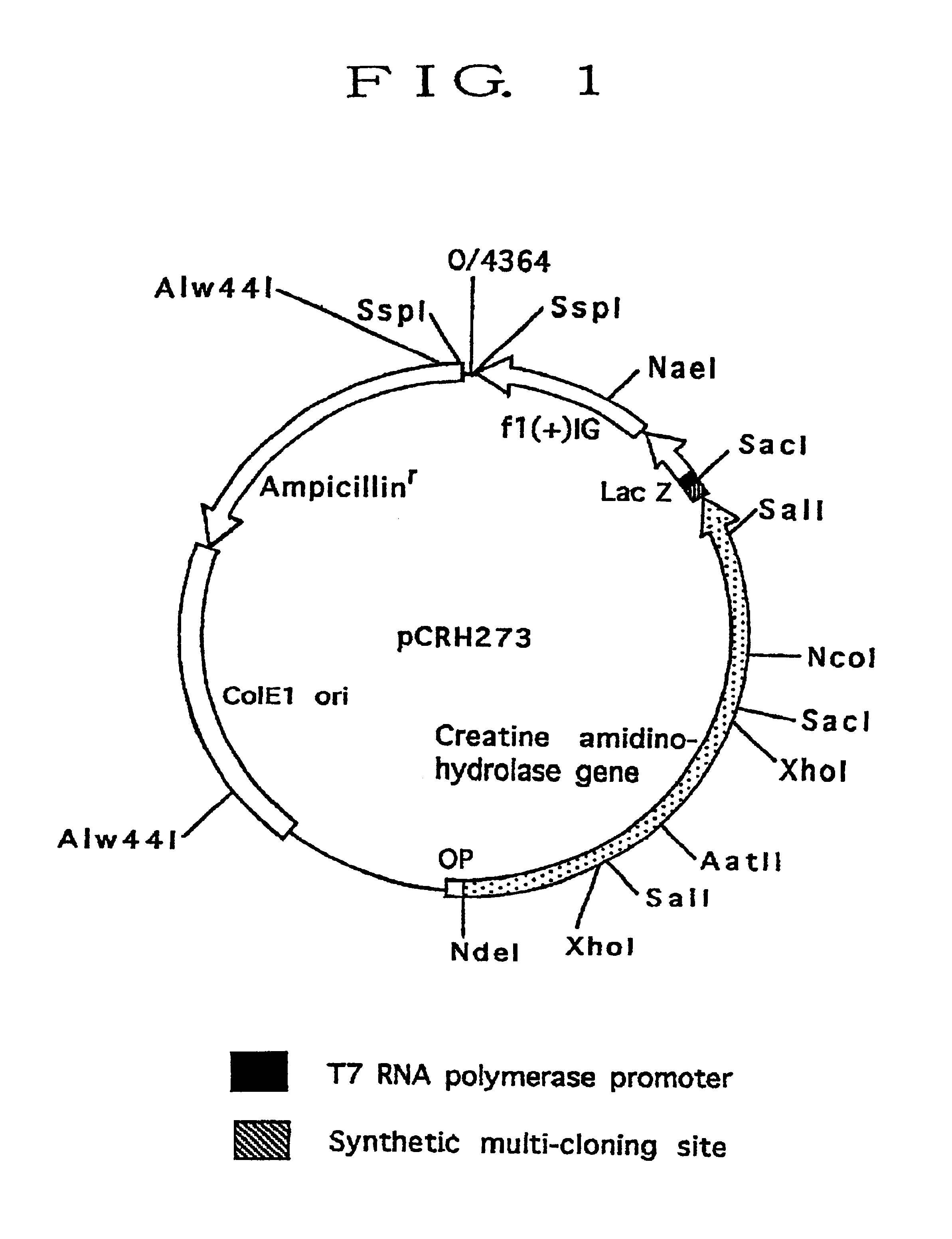
The pCRH173M was cleaved with restriction enzyme EcoRI (Toyo Boseki Kabushiki Katsha) and self-ligated to prepare pCRH273 (FIG. 1).
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com