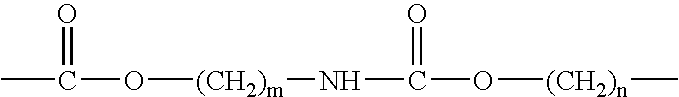
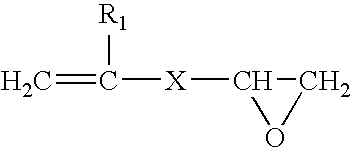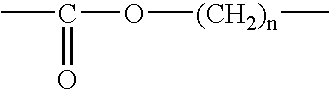Epoxide polymer surfaces
a polymer surface and epoxide technology, applied in the field of reagents and support surfaces, can solve the problems of uv crosslinking, affecting the stability of dna, and limiting the number of nucleic acids in the uv, so as to achieve stable hydrolysis, less undesirable side reactions, and convenient use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of 4-Benzoylbenzoyl Chloride (BBA-Cl) (Compound I)
[0081] 4-Benzoylbenzoic acid (BBA), 1.0 kg (4.42 moles), was added to a dry 5 liter Morton flask equipped with reflux condenser and overhead stirrer, followed by the addition of 645 ml (8.84 moles) of thionyl chloride and 725 ml of toluene. Dimethylformamide, 3.5 ml, was then added and the mixture was heated at reflux for 4 hours. After cooling, the solvents were removed under reduced pressure and the residual thionyl chloride was removed by three evaporations using 3×500 ml of toluene. The product was recrystallized from 1:4 toluene:hexane to give 988 g (91% yield) after drying in a vacuum oven. Product melting point was 92-94° C. Nuclear magnetic resonance (NMR) analysis at 80 MHz (1H NMR (CDCl3)) was consistent with the desired product: aromatic protons 7.20-8.25 (m, 9H). All chemical shift values are in ppm downfield from a tetramethylsilane internal standard. The final compound was stored for use in the preparation ...
example 2
Preparation of N-(3-Aminopropyl)methacrylamide Hydrochloride (APMA) (Compound II)
[0082] A solution of 1,3-diaminopropane, 1910 g (25.77 moles), in 1000 ml of CH2Cl2 was added to a 12 liter Morton flask and cooled on an ice bath. A solution of t-butyl phenyl carbonate, 1000 g (5.15 moles), in 250 ml of CH2Cl2 was then added dropwise at a rate which kept the reaction temperature below 15° C. Following the addition, the mixture was warmed to room temperature and stirred 2 hours. The reaction mixture was diluted with 900 ml of CH2Cl2 and 500 g of ice, followed by the slow addition of 2500 ml of 2.2 N NaOH. After testing to insure the solution was basic, the product was transferred to a separatory funnel and the organic layer was removed and set aside as extract #1. The aqueous layer was then extracted three times with 1250 ml of CH2Cl2, keeping each extraction as a separate fraction. The four organic extracts were then washed successively with a single 1250 ml portion of 0.6 N NaOH beg...
example 3
Preparation of N-[3-(4-Benzoylbenzamido)propyl]methacrylamide (BBA-APMA) (Compound III)
[0085] Compound II 120 g (0.672 moles), prepared according to the general method described in Example 2, was added to a dry 2 liter, three-neck round bottom flask equipped with an overhead stirrer. Phenothiazine, 23-25 mg, was added as an inhibitor, followed by 800 ml of chloroform. The suspension was cooled below 10° C. on an ice bath and 172.5 g (0.705 moles) of Compound I, prepared according to the general method described in Example 1, were added as a solid. Triethylamine, 207 ml (1.485 moles), in 50 ml of chloroform was then added dropwise over a 1-1.5 hour time period. The ice bath was removed and stirring at ambient temperature was continued for 2.5 hours. The product was then washed with 600 ml of 0.3 N HCl and 2×300 ml of 0.07 N HCl. After drying over sodium sulfate, the chloroform was removed under reduced pressure and the product was recrystallized twice from 4:1 toluene:chloroform usi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com