Saccharomyces cerevisiae gene engineering bacterium capable of utilizing xylose and glucose jointly as well as construction method and application of saccharomyces cerevisiae gene engineering bacterium
A technology of genetically engineered bacteria and Saccharomyces cerevisiae is applied in the field of using xylose saccharomyces cerevisiae strains and its construction, which can solve problems such as imbalance, carbon utilization of engineered bacteria and reduction of ethanol production, and achieve the effect of simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
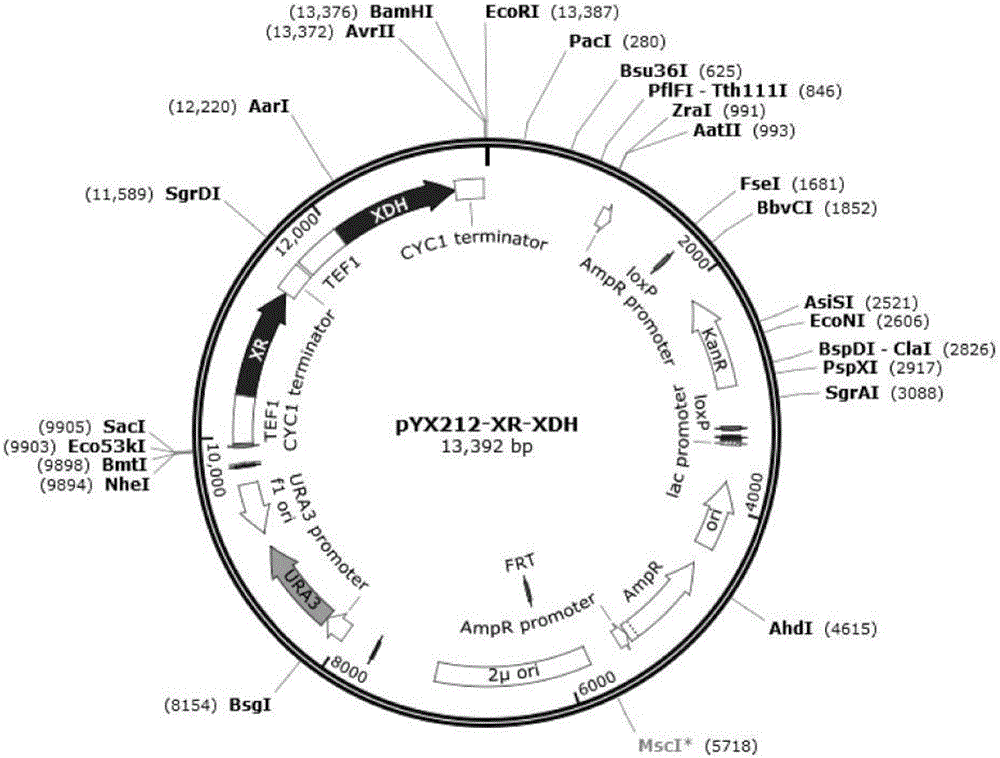
[0051] Using the xylose-utilizing Saccharomyces cerevisiae strain BY4741 (MATa; ura3; his3; leu2; met15) as the starting strain, the expression vector constitutive high-copy plasmid pYX212 was used to overexpress XR and XDH with TEF1p as the promoter to obtain the recombinant strain XR-XDH The Saccharomyces cerevisiae strain BY4741 (MATa; ura3; his3; leu2; met15) containing the empty plasmid is the control bacteria CON; the source strain Candida tropicalis 121 of XR and XDH is also used as the control bacteria (hereinafter abbreviated as Ctp strain).
[0052] Use the above three strains to do glucose aerobic fermentation experiments:
[0053] The seed medium is a synthetic medium YPD, and the fermentation medium is a YPD medium with an initial sugar concentration of 100 g / L. Pick an appropriate amount from a single colony in a 30% glycerol tube stored at -80°C with an inoculation loop or a pipette tip and inoculate it into 5ml of YPD liquid medium, culture at 30°C and 200rpm f...
Embodiment 2
[0060] Carry out glucose, xylose mixed sugar fermentation experiment to three bacterial strains among the embodiment 1. Seed culture method is the same as embodiment 1. The fermentation medium is 20g / L glucose, 40g / L glucose, 10g / L yeast powder, and 20g / L peptone. The detection method is the same as in Example 1.
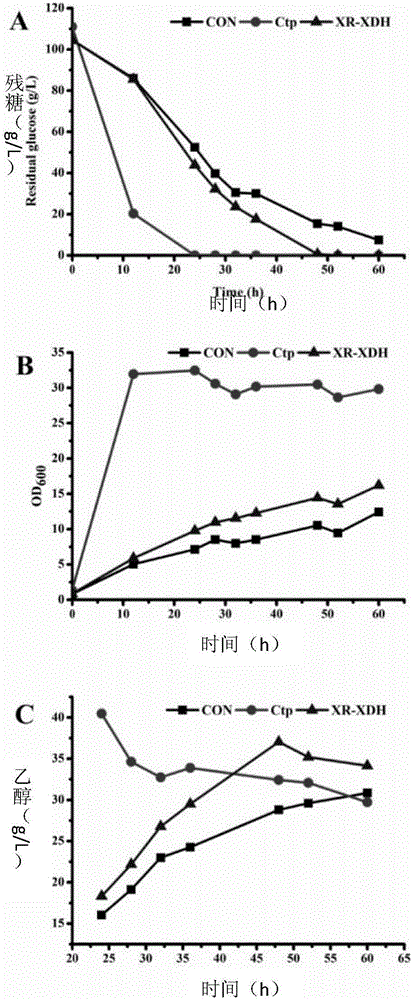
[0061] Experimental results such as Figure 4 as shown, Figure 4 Saccharomyces cerevisiae control strain CON, Candida tropicalis Ctp, Saccharomyces cerevisiae strain XR-XDH overexpressing XR and XDH with TEF1 as promoter, glucose and xylose mixed sugar fermentation experiments.
[0062] The results show that the Ctp strain can consume xylose in 3 days, but the CON and XR-XDH strains can hardly utilize xylose, and the bacterial growth and ethanol production are limited to the glucose utilization stage. After glucose is exhausted, they turn to use ethanol Used as a carbon source to sustain growth.
Embodiment 3
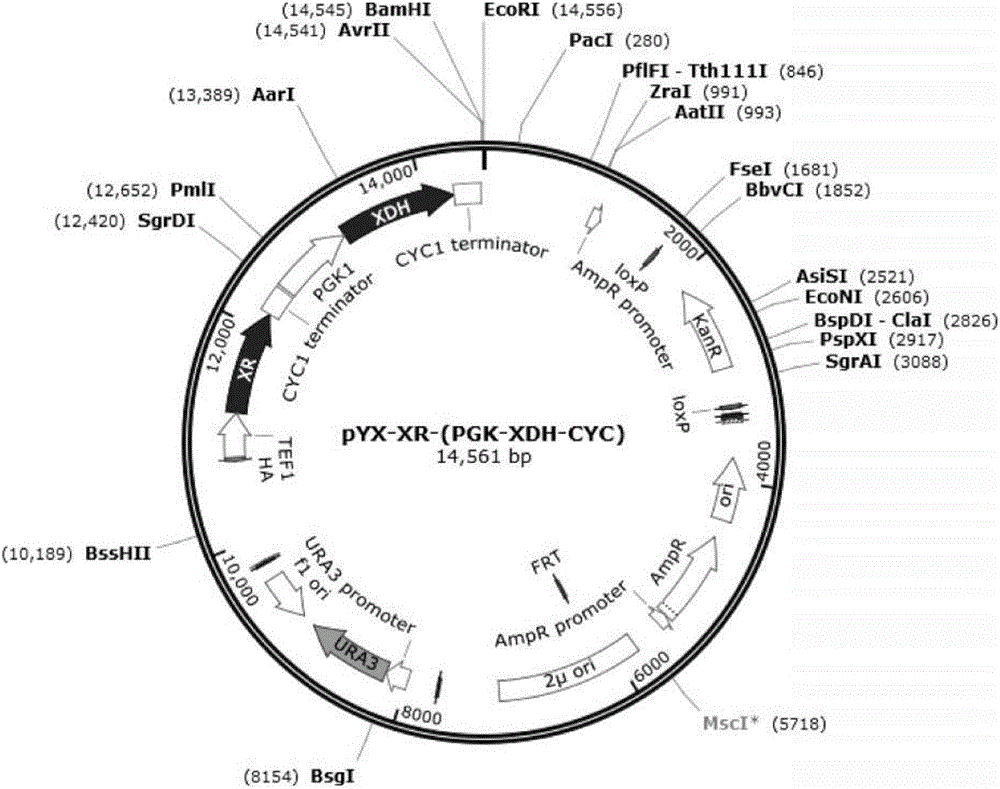
[0064] Xylulokinase XK and transaldolase TAL derived from Candida tropicalis were further overexpressed in the XR-XDH strain to obtain strains RH-TAL and RH-XK for glucose fermentation. The cultivation and detection methods are the same as in Example 1.
[0065] Experimental results such as Figure 5 as shown, Figure 5 The strain XR-XDH of XR and XDH with TEF1 as the promoter, the strain RH-TAL that further overexpresses the transaldolase derived from Candida tropicalis, and the strain XK that further overexpresses the xylulokinase derived from Candida tropicalis Glucose fermentation experiment of the strain RH-XK.
[0066] The results showed that overexpression of xylulokinase XK, a key enzyme in the non-oxidative part of the PPP pathway, could significantly increase the glucose consumption rate (RH-XK 28h vs. RH 48h), and the cell growth was significantly accelerated, but the ethanol production was reduced. Analysis The reason may be that overexpression of XK increases t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com