Method and detection kit for detecting helicobacter pylori drug resistance gene
A technology of Helicobacter pylori and drug resistance gene, applied in the field of molecular biology detection of Helicobacter pylori in digestive tract diseases, can solve problems such as failure of eradication treatment, achieve high specificity, high sensitivity, and reduce the effect of antibiotic abuse
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] 1. Collection of saliva
[0040] After obtaining the informed consent of the patients, the oral saliva of the patients was collected before gastroscopy in the morning before upper gastrointestinal symptoms were prepared.
[0041] 2. Genomic DNA extraction from saliva
[0042] Genomic DNA was extracted from saliva using a rapid genomic DNA preparation kit (Shanghai Bocai Biotechnology Co., Ltd.) and used as a template for the following polymerase chain reaction (PCR) amplification. The specific experimental operation process is as follows:
[0043] (1) Processing of specimens
[0044] 1) 25-100 mg tissue samples were cut into small pieces with a scalpel, then crushed with liquid nitrogen, or homogenized with a tissue homogenizer. Collect the powder into a 1.5mL centrifuge tube and suspend with 400μL Disgestion Buffer.
[0045] 2) Take 1 mL of fresh saliva, add 2 mL of PBS (or saline), and soak at room temperature for 2-5 hours.
[0046] (2) Add 3 μL of Proteinase K ...
Embodiment 2
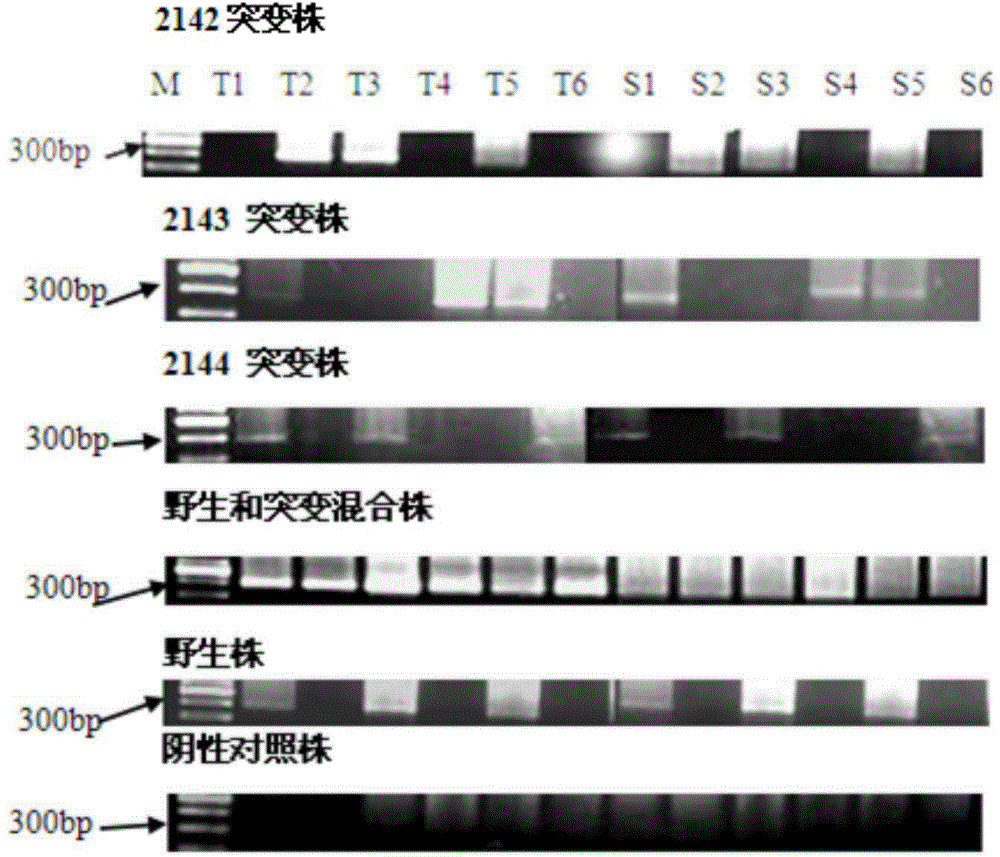
[0092] Example 2 The saliva and gastric mucosal tissues of 60 patients with upper gastrointestinal symptoms and positive rapid urease test were collected. Genomic DNA was extracted from saliva and gastric mucosal tissue using a rapid genomic DNA preparation kit (Shanghai Bocai Biotechnology Co., Ltd.), and detected by two PCR methods. The first is the common PCR method, using genomic DNA in saliva (or gastric mucosal tissue) as a template, and using allele-specific primers for direct PCR amplification. The second method is the nested PCR method, that is, the first round of PCR uses genomic DNA in saliva (or gastric mucosal tissue) as a template, and the outer primers of nested PCR are used for amplification; the second round of PCR uses the first round of PCR products as templates , amplified using allele-specific primers. After the PCR, the common PCR products and nested PCR products were electrophoresed in 1.5% agarose gel, and the presence of specific amplification bands w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com