Succinic acid genetic engineering bacterium and method for fermenting and producing succinic acid
A genetically engineered bacterium producing succinic acid technology, applied in the field of a succinic acid-producing genetically engineered bacterium and its fermentation to produce succinic acid, can solve the problems of wasting resources, polluting the environment, etc., and achieve improved production capacity and increased output And the effect of huge changes in production capacity and acid production characteristics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
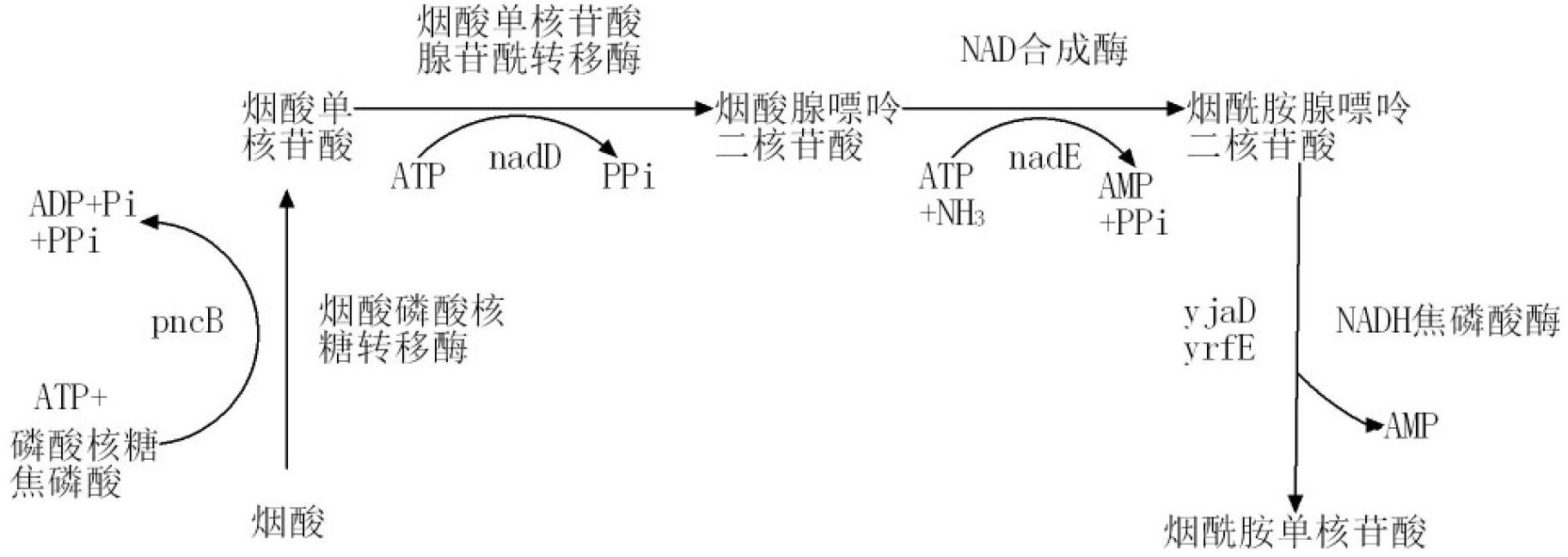
Image
Examples
Embodiment 1
[0050] This example illustrates the process of knocking out the phosphoenolpyruvate carboxylase ppc gene in the starting strain NZN111 by using homologous recombination technology to obtain a strain that eliminates apramycin resistance.
[0051] 1. Using LB medium, cultivate Escherichia coli NZN111 to OD at 37°C under aerobic conditions 600 =0.4~0.6, prepared to be electrotransfer competent.
[0052] 2. The plasmid pKD46 was electrotransformed into competent Escherichia coli NZN111. The electric shock conditions were: 200 Ω, 25 μF, electric shock voltage 2.3 kV, electric shock time 4-5 ms. Immediately after the electric shock, the cells were added to pre-cooled 1 mL SOC medium, cultured at 150 r / min, 30°C for 1 h, and then spread on the LB medium plate with ampicillin (amp) to screen out the positive transformant Escherichia coli NZN1 11 (pKD46).
[0053] 3. Add 10 mM L-arabinose to LB medium, induce plasmid pKD46 to express λ recombinase at 30°C, and make electroporation c...
Embodiment 2
[0067] This example illustrates the process of using the ppc gene-knockout strain obtained in Example 1 to knock out the ptsG gene in the PTS transport system by using homologous recombination technology again to obtain an apramycin-resistant strain.
[0068] 1. Using LB medium, cultivate the ppc gene-knockout strain obtained in Case 1 at 37°C under aerobic conditions to OD 600 =0.4~0.6, prepared to be electrotransfer competent.
[0069] 2. Electrotransfer the plasmid pKD46 into the competent cells. The electric shock conditions were: 200 Ω, 25 μF, electric shock voltage 2.3 kV, electric shock time 4-5 ms. Immediately after the electric shock, the cells were added to pre-cooled 1 mL SOC medium, cultured at 150 r / min, 30°C for 1 h, and then spread on the LB medium plate with ampicillin (amp) to screen out the positive transformant Escherichia coli NZN1 11 / Δppc(pKD46).
[0070] 3. Add 10 mM L-arabinose to LB medium, induce plasmid pKD46 to express λ recombinase at 30°C, and ...
Embodiment 3
[0084] This example illustrates the construction of an expression plasmid for overexpressing phosphoenolpyruvate carboxykinase.
[0085] 1. Construction of an expression plasmid for overexpressing phosphoenolpyruvate carboxykinase, the process comprising:
[0086] (1) Synthesize primers with SacI and XbaI restriction sites,
[0087] Upstream primer: 5'-CGAGCTCATGAACTCAGTTGATTTGACCG-3';
[0088] Downstream primer: 5'-GCTCTAGAGCATTCCGTCAATTAAAACAAG-3'.
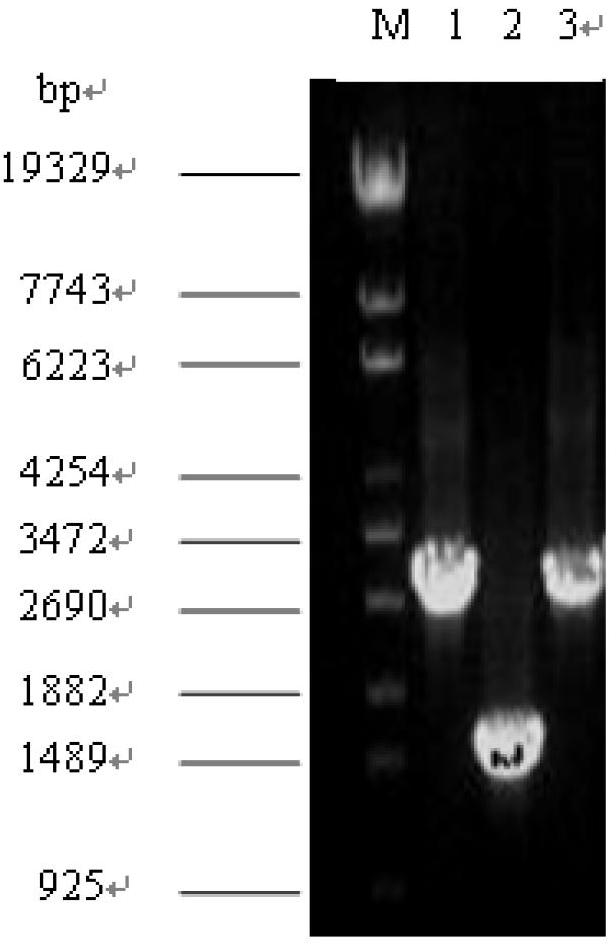
[0089] (2) Use the Bacillus subtilis genome as a template to amplify the target gene fragment by PCR. The reaction conditions are: 94°C, 5 min; (94°C for 45 s, 53°C for 45 s, 72°C for 100 s, 35 cycles); 72°C , 10 min. After purifying the amplified pck gene, the expression plasmid was digested with pTrc99a with SacI and XbaI respectively, and ligated to obtain the recombinant plasmid pTrc99a-pck. Double restriction electrophoresis identification of plasmid pTrc99a-pck as Image 6 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com