Compositions and methods for detecting s-nitrosylation and s-sulfinylation
a technology of s-nitrosylation and s-sulfinylation, which is applied in the field of compositions and methods for detecting s-nitrosylation and s-sulfinylation, can solve the problems of unstable and transient modification, introduction of false positives, and insufficient investigation of the selectivity of organomercury enrichment, so as to reduce loading buffer, reduce sulfenic acid, and reduce the effect of sulfenic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
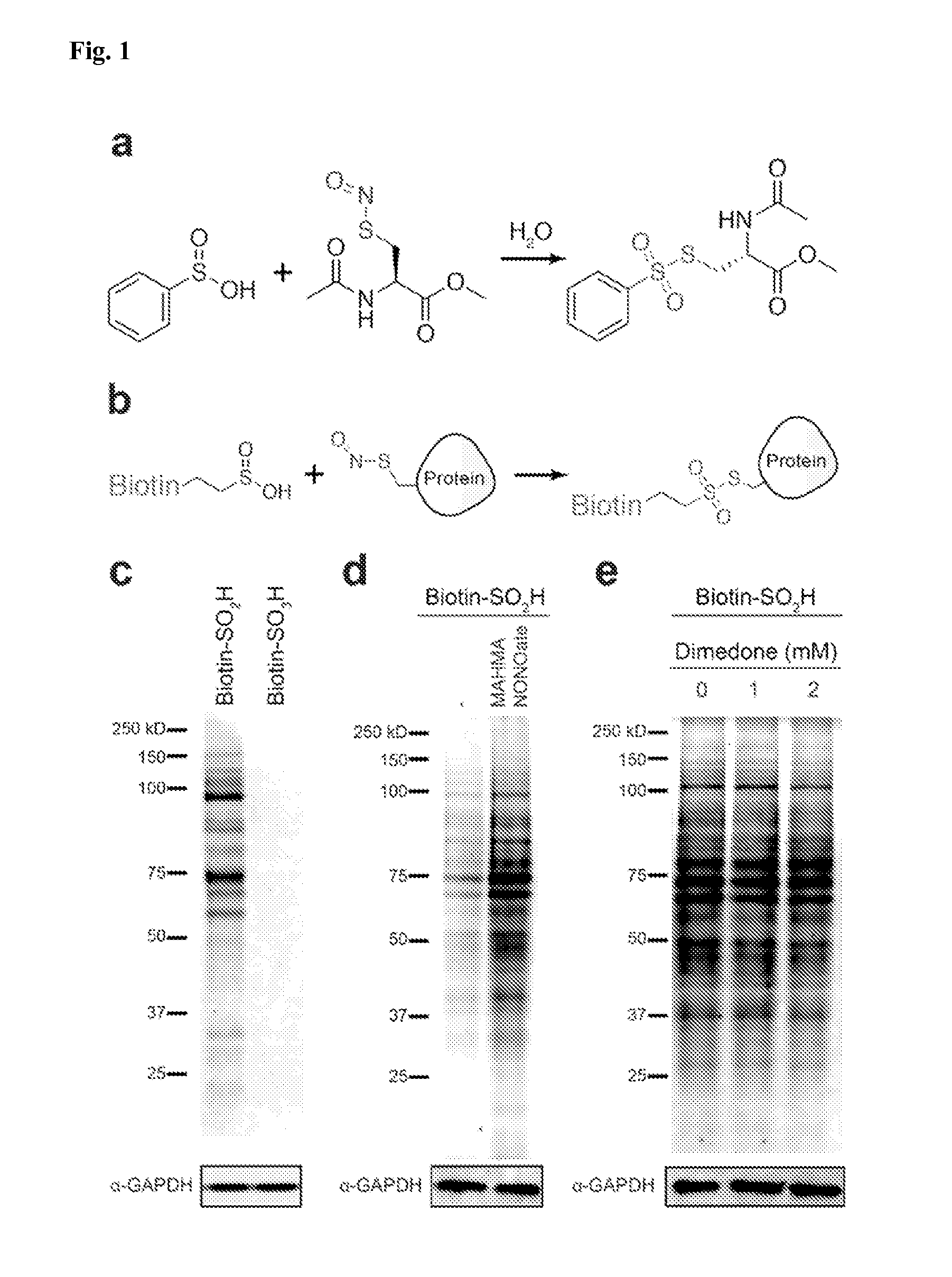
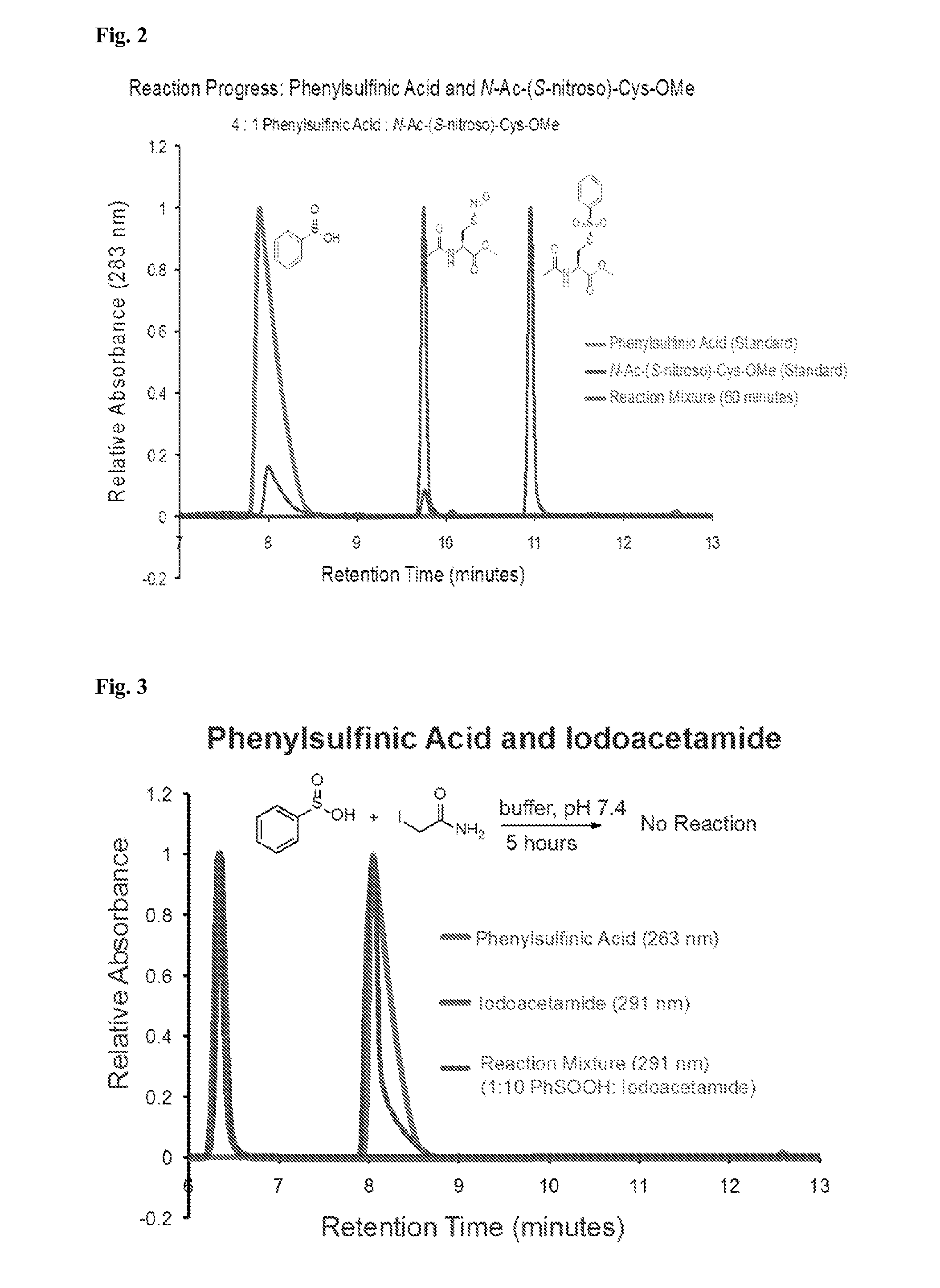
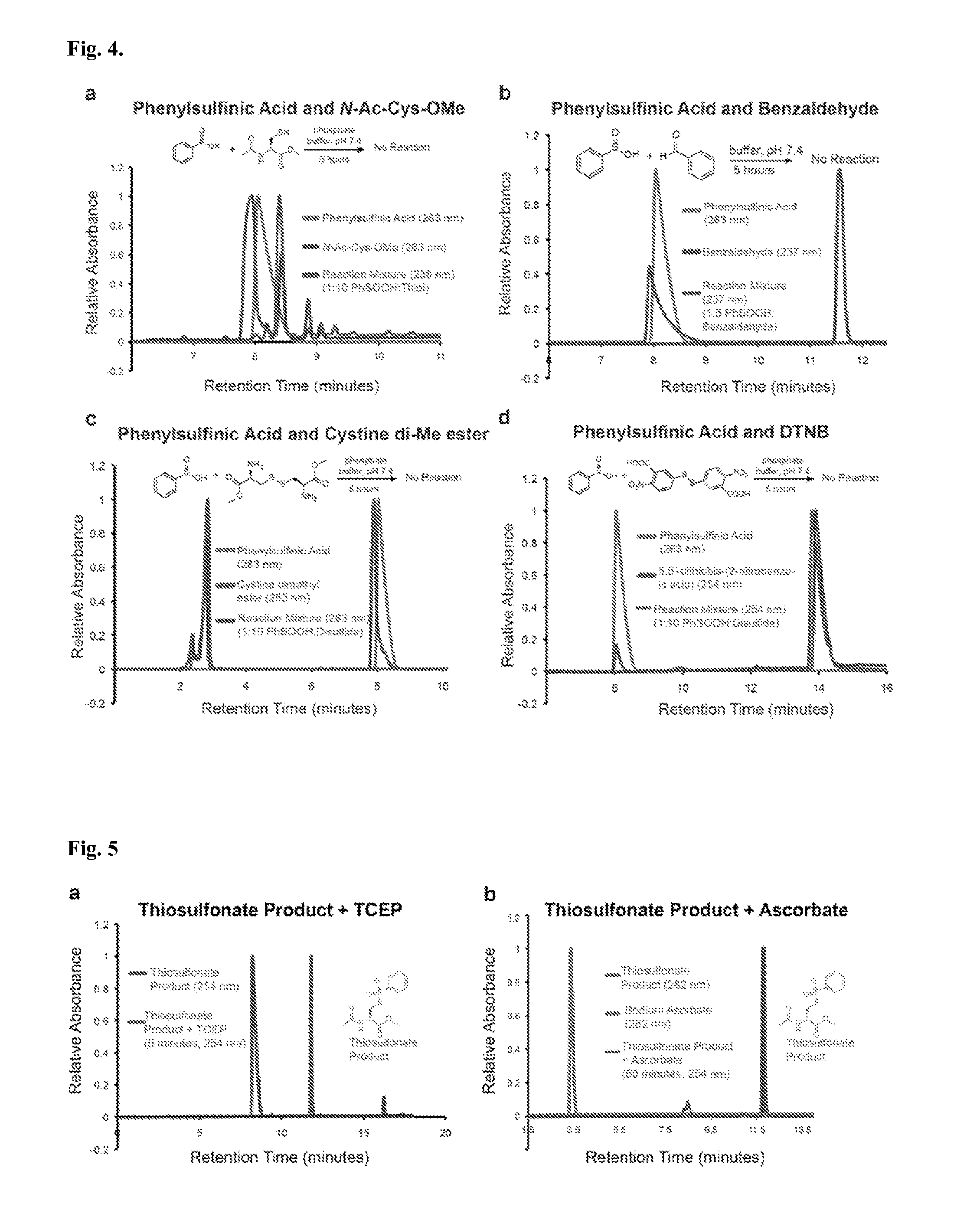
[0087]While exploring the interplay of cysteine post-translational modifications, a reported reaction between phenylsulfinic acid and S-nitrosocysteine was identified, leading to thiosulfonate formation in aqueous buffers at room temperature (see, e.g., Hart, T. W. Tetrahedron Letters 26, 2013-2016 (1985)) (FIG. 1a, FIG. 2). Thiosulfonates are readily exchangeable with thiols, serving as the basis for the cysteine capping agent methyl methanethiosulfonate (MMTS). To prevent such exchange, it was found that sulfinic acids do not react with iodoacetamide, enabling orthogonal alkylation of thiols without perturbing nitrosothiols or sulfinic acids (FIG. 3). Furthermore, it was found that sulfinic acids do not react with thiols (cysteine), disulfides (cystine or 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB)), or aldehydes (benzaldehyde) (FIG. 4). Additionally, the thiosulfonate product is unaffected by ascorbate, but reduced by tris(2-carboxyethyl)phosphine (TCEP) (FIG. 5). Most S-sulfinyl...
example ii
[0096]This example describes synthetic methods pertaining to Example I.
[0097]All compounds were purchased from Sigma-Aldrich, unless otherwise noted. NMR analysis was performed using a Varian 400 MHz NMR instrument. Small molecule high-resolution mass spectrometry was performed using an electrospray Agilent Q-TOF mass spectrometer (accuracy 1-5 ppm). Low-resolution mass spectrometry was performed using an electrospray Micromass LCT time-of-flight mass coupled to a HPLC pump with a rheodyne loop injector. Compounds were purified by normal phase silica column chromatography or by semi-prep High-Performance Column Chromatography (HPLC). HPLC purifications were performed using a Waters semi-preparative 1525 binary pump system coupled to a photodiode array detector, an autosampler, and an automatic fraction collector. Separations were carried out on an Atlantis prep T3 C18 column (10×250 mm), in 95 / 5 water / acetonitrile 0.1% formic acid for 2 minutes, followed by a 40 minute gradient incr...
example iii
[0098]This example describes materials and methods pertaining to Examples I and II.
Rate Constant Determination.
[0099]S-nitrosoglutathione (GSNO, Cayman) and sodium phenylsulfinate (Sigma-Aldrich) were used for rate-determination studies at four pH values: pH 1.0 (0.2 N HCl / KCl buffer), pH 4.0 (0.1 M Sodium acetate / Acetic Acid buffer), pH 7.0 (0.1 M potassium phosphate buffer) and pH 10.0 (0.1 M Sodium bicarbonate / Sodium hydroxide buffer). The purity of S-nitrosoglutathione was calculated as 94±1.3% by absorbance at 334 nm using the molar extinction coefficient of 900 M−1 cm−1. Assays were performed using a plate reader (Tecan Infinite F500) monitoring absorbance of 2 mM GSNO at 340 nm over a course of 90 minutes in the presence of varying concentrations of sodium phenylsulfinate. Additional experiments confirmed thiosulfonate stability for >5 hours at pH 1, 4, and 7, but hydrolysis at pH 10. GSNO was stable in 6 M urea / PBS for >1 hour. Absorbance data was imported into KaleidaGraph ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| retention time | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com