High-throughput quantitative detection kit for diarrhea pathogenic bacteria
A detection kit, quantitative detection technology, applied in the determination/inspection of microorganisms, methods based on microorganisms, resistance to vector-borne diseases, etc., to achieve the effect of strong reliability, strong specificity and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
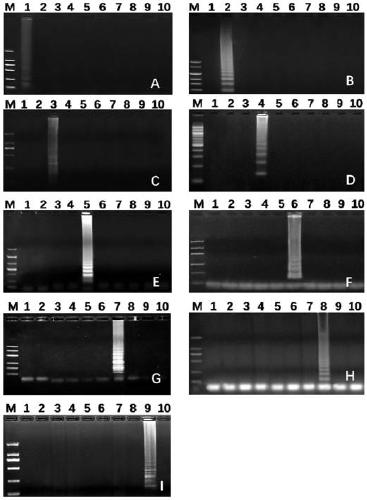
[0016] A high-throughput quantitative detection kit for diarrheal pathogens, including 9 kinds of LAMP detection primer sets for diarrheal pathogens, and its gene sequence is shown in Table 1 below:
[0017] Table 1
[0018] .
[0019] The above-mentioned gene target site design: select gene design primers that are specific between species and conserved within species because the external primer F3 also has the opportunity to bind and extend with F3c on the template to replace a complete FIP-linked complementary single strand. At this time, the F1c on the FIP can be complementary to the F1 on the single strand, forming a circular structure through self-base pairing. Similarly, the downstream primers BIP and B3 are similar to the synthesis of primers FIP and F3, which makes it possible to form a dumbbell-shaped single-stranded structure. At this time, under the action of DNA polymerase capable of strand displacement, the F1 segment at the 3'end will use itself as a template for DNA...
specific Embodiment 2
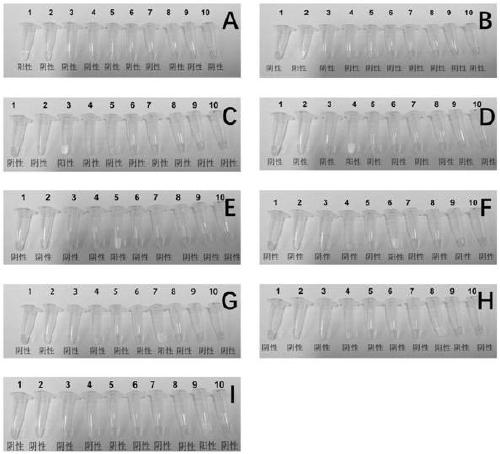
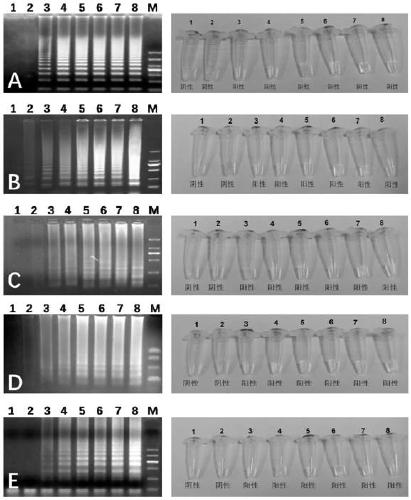
[0022] A high-throughput quantitative detection kit for diarrheal pathogens. The LAMP reaction system and the final concentration of each component are as follows: sample template DNA 1μL, 2.5μL 10× reaction buffer, 1.5μL 100mM MgSO 4 , 3.5μL of 10mM dNTP, 1μL of 8 U / μl Bst DNA polymerase, 1μL FIP / BIP primer set solution, 1μL F3 / B3 primer set solution and 4μL betaine, and then make up to 25μL with double distilled water; the FIP / BIP primer set solutions are listed in Table 1 respectively. FIP / BIP primer set solution of 9 bacteria, Vibrio cholerae, Staphylococcus aureus, Salmonella, Shigella, pathogenic Escherichia coli, Bacillus cereus, Proteus mirabilis and Vibrio parahaemolyticus, The final concentration of each primer is 40 μM, and the F3 / B3 primer set solutions are Listeria monocytogenes, Vibrio cholerae, Staphylococcus aureus, Salmonella, Shigella, and pathogenicity as shown in Table 1 above. F3 / B3 primer set solution of 9 bacteria of Escherichia coli, Bacillus cereus, Prot...
specific Embodiment 3
[0025] A method for high-throughput quantitative detection of diarrheal pathogens by using the detection kit of the second embodiment described above, including the following steps:
[0026] Take the sample to be tested, extract the sample template DNA according to the commercially available bacterial DNA extraction kit, take 1μL of the sample template DNA, add 2.5μL of 10× reaction buffer, 1.5μL of MgSO according to the composition of the LAMP reaction system 4 , 3.5μL dNTP, 1μL Bst DNA polymerase, 1μL FIP / BIP primer set solution, 1μL F3 / B3 primer set solution and 4μL betaine, then make up 25μL with double distilled water and mix well. Take 20μL of the mixed solution and add it to each loading hole of the sample detection chip (The primer set solutions are Listeria monocytogenes, Vibrio cholerae, Staphylococcus aureus, Salmonella, Shigella, pathogenic Escherichia coli, Bacillus cereus, Proteus mirabilis and Vibrio parahaemolyticus The primer set solution of 9 bacteria, the primer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com