A kind of penicillin v acylase mutant, coding sequence, recombinant expression vector, genetic engineering bacteria and application
A technology of acylase mutants and penicillin V, applied in the direction of genetic engineering, application, plant gene improvement, etc., can solve the problems of less batches, long reaction time, restrictions on popularization and application, etc., and achieve enhanced stability, catalytic Vitality-enhancing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
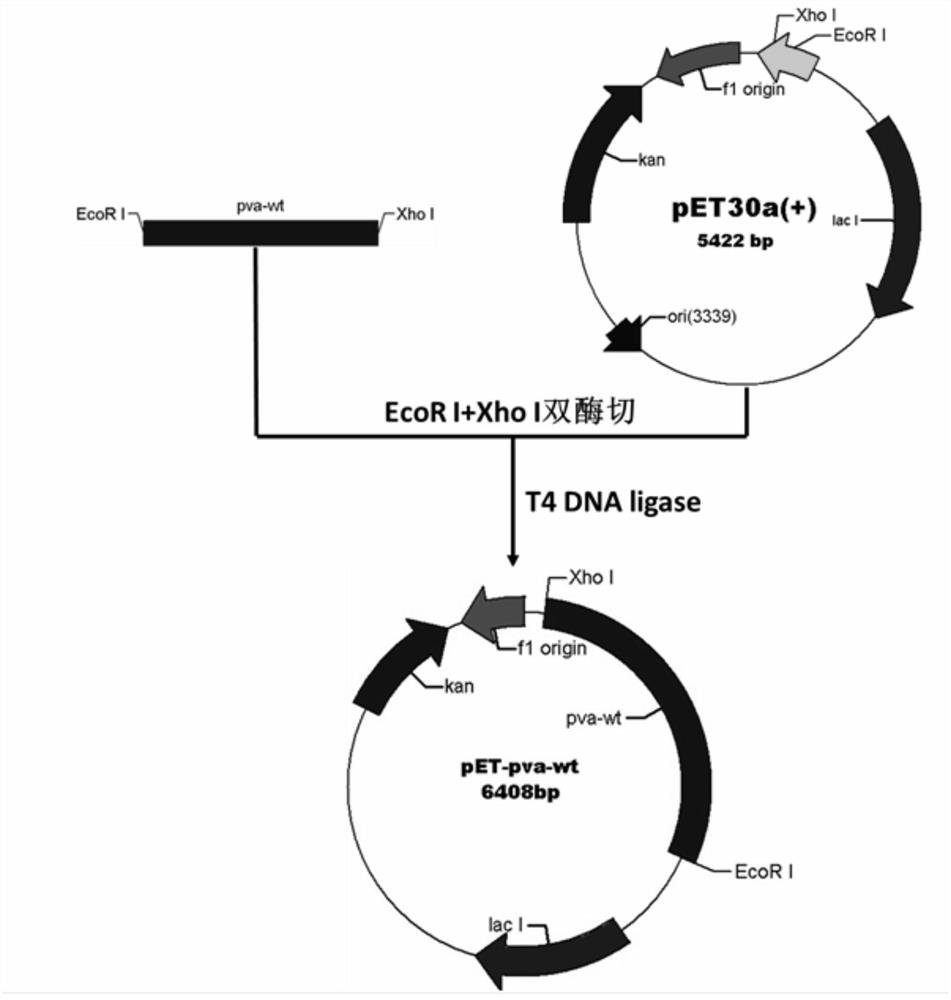
[0068] Example 1: Construction, expression and purification and immobilization of recombinant protein of wild-type PVA recombinant genetically engineered bacteria
[0069] 1-1 Construction of wild-type PVA prokaryotic expression vector and construction of recombinant genetically engineered bacteria
[0070] The wild-type PVA gene and amino acid sequence used in the present invention are derived from Bacillus sphaericus (GeneBank accession number: P12256.1). The coding gene is optimized through the codon bias of Escherichia coli and the whole gene synthesis is carried out, and the wild-type The penicillin V acylase is named PVA-WT, and its coding gene is named pva-wt. See SEQ ID NO: 1 and SEQ ID NO: 2 for the nucleotide sequence and amino acid sequence.
[0071] refer to figure 1 , the wild-type PVA coding gene pva-wt synthesized by the whole gene and the prokaryotic expression vector pET30a(+) were subjected to EcoR I and Xho I double enzyme digestion respectively, and after ...
Embodiment 2
[0084] Embodiment 2: the preparation of high activity PVA mutant
[0085] 2-1 Construction of PVA mutant library
[0086] In order to improve the activity of wild-type PVA, the inventors used the recombinant expression vector pET-pva-wt as a DNA template, wherein the primers were T7 universal primers (SEQ ID NO: 9 and 10), and constructed a random mutation by error-prone PCR body library, and by adjusting the error-prone PCR reaction system Mg 2+ and Mn 2+ Concentration and concentration of dCTP and dTTP oligonucleotides, so that the base mismatch rate of the mutant library is 5 / 1000, that is, to ensure that a mutant has 1 to 3 amino acid mutations, the specific process of constructing the mutant library is as follows .
[0087] Error-prone PCR reaction system:
[0088]
[0089]
[0090] The error-prone PCR product obtained above was subjected to electrophoresis and gel-cut recovery and purification, and the purified product and the prokaryotic expression vector pET3...
Embodiment 3
[0098] Example 3: Preparation of high substrate concentration tolerance PVA mutants
[0099] Since the activity of wild-type PVA is inhibited by the concentration of penicillin V, in order to obtain a mutant resistant to high concentration of penicillin V and improve its catalytic efficiency, the inventors performed directed evolution mutation again on the basis of the above-mentioned mutant PVA-1, according to the above-mentioned The method in the examples was used to construct a mutant library and perform screening (increasing the penicillin V potassium salt concentration (W / V) from 10% to 20%). About 20,000 clones were screened from the above-mentioned mutant library, and 5 mutants with obvious color changes and higher values than PVA-1 were obtained. After re-screening by shaking flasks, a clone with 2 times higher activity than PVA-1 was obtained, named PVA-2, and the sequencing results showed that PVA-2 had an amino acid at the 91st position relative to wild-type penic...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com