Species-specific Quantitative PCR Primers and Its Design Method for Methylotrophic Bacillus
A methylotrophic and Bacillus technology, applied in the field of agricultural microorganisms, can solve problems such as information lag and genome data update, and achieve high accuracy and good quantitative specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1 is tested bacterial strain source
[0027] The methylotrophic bacillus NJAU-Z9 tested in the present invention (preserved in the General Microbiology Center of the China Microbiological Culture Collection Management Committee, the preservation date is April 11, 2016, and the preservation number is CGMCC NO.12342) and 6 strains Similar strains belonging to the genus Bacillus, all the bacteria are stored in the Jiangsu Key Laboratory of High-tech Research on Solid Organic Waste Recycling and Utilization, Nanjing Agricultural University (Table 1), and the whole genome sequence is uploaded to the NCBI Biotechnology Information Center with accession number NZ_CP022556.1 .
[0028] Table 1 The strains used for primer analysis and their corresponding GenBank accession numbers
[0029]
[0030]
Embodiment 2
[0031] Example 2 Primer sequence selection and cross-amplification verification test
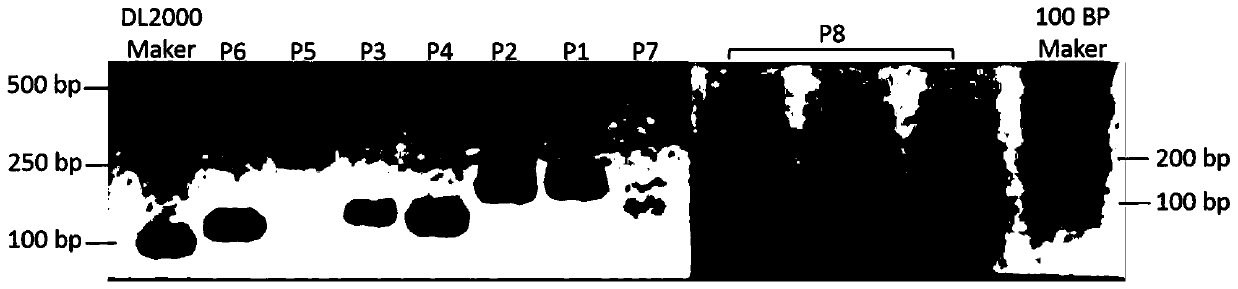
[0032] The invention compares the whole genome sequence in the database and searches for the specific sequence for NJAU-Z9. Step 1: Compare the whole genome sequence of NJAU-Z9 (NZ_CP022556.1.) with two authoritative databases, NCBI and EzTaxon, and select genome fragments larger than 500bp that do not match. The second step: Utilize Oligo 6 software to implement primer sequence design, set the product size to be 70-250bp, and the primer length is 22 ± 2bp, design 8 pairs of primers (Table 2) in total, and pass 8 pairs of primers to NJAU- Z9 genomic DNA was amplified sequentially, and a preliminary judgment was made on the primers according to the quality of the bands ( figure 1 ). In addition, the genomic DNA of 6 strains of other similar species and the DNA of the target strain were used as primers P1-P8 to cross-amplify the templates sequentially, and a pair of reported Bacillus primers...
Embodiment 3
[0038] Construction and quantitative PCR of embodiment 3 plasmid
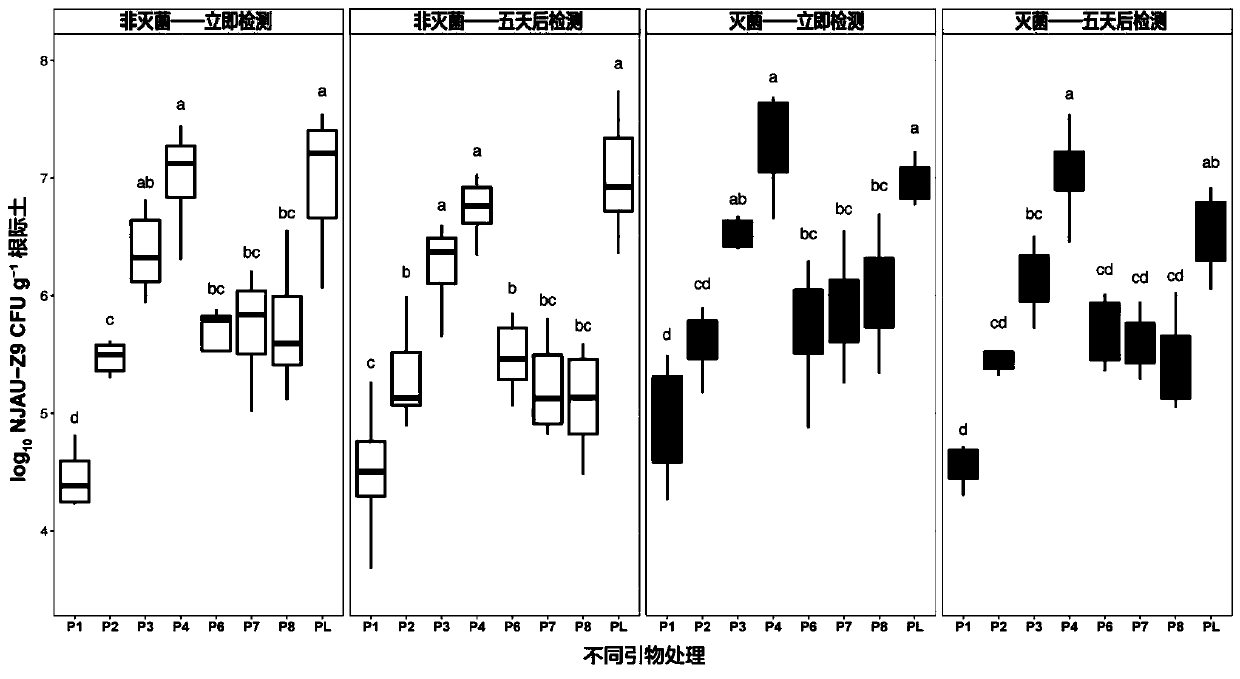
[0039] The fragments of 8 pairs of primer amplification products designed from the selected target gene region in the methylotrophic bacillus NJAU-Z9 genome were cloned into the pMD19-T vector (TaKaRa), and the plasmid was transformed into Escherichia coli Top10 cells, Common PCR amplification was performed to verify the amplified fragment, and sequenced by Nanjing GenScript Biotechnology Co., Ltd. The DNA concentration of the plasmid was measured using a spectrophotometer (NanoDrop 2000, Thermo Scientific Inc., USA). Use on 7500 real-time PCR system (Applied Biosystems, USA) Premix Ex TaqTM (TaKaRa) was used for gradient qPCR amplification in a 20 μl reaction system. Plasmids with different inserts were used to prepare a 10-fold dilution series (triplicate), using sterile water as a negative control. After determining the cycle threshold (CT) value for each sample, a standard curve was generated by plottin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com