5-hydroxymethylfurfural oxidase gene HMFO and codase thereof as well as application
A hydroxymethylfurfural oxidase and gene technology, applied in the direction of oxidoreductase, application, genetic engineering, etc., can solve the problem of not being able to complete the three-step oxidation of HMF at the same time, and achieve the effect of short catalytic reaction time and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Implementation example 1: 5-hydroxymethylfurfural oxidase gene sequence analysis
[0032] The sequencing results were analyzed by Basic Local Alignment Search Tool (BLAST) in the GenBank database, and Vector NTI Suite 8.0 software was used for multiple sequence alignment to analyze sequence information.
[0033] The obtained 5-hydroxymethyl furfural oxidase gene (named HMFO) has a coding region of 1632 bp in length, and its nucleotide sequence is shown in SEQ ID NO 1. HMFO encodes 543 amino acids and a stop codon. Its amino acid sequence is shown in SEQ ID NO 2. The theoretical molecular weight of the protein is 58686 Da, and the predicted isoelectric point is 5.96. Among the amino acids, 152 are charged, accounting for 34.45%. 65, accounting for 13.17%, 52 basic amino acids, accounting for 12.98%, polar amino acids, 102, accounting for 19.07%, and hydrophobic amino acids, 205, accounting for 36.44%. There are 58 in Ala, 56 in Gly, and 26 in Glu.
[0034] (1) SEQ ID No.1 inf...
Embodiment 2
[0056] Example 2 Cloning of the full-length gene of 5-hydroxymethylfurfural oxidase HMFO
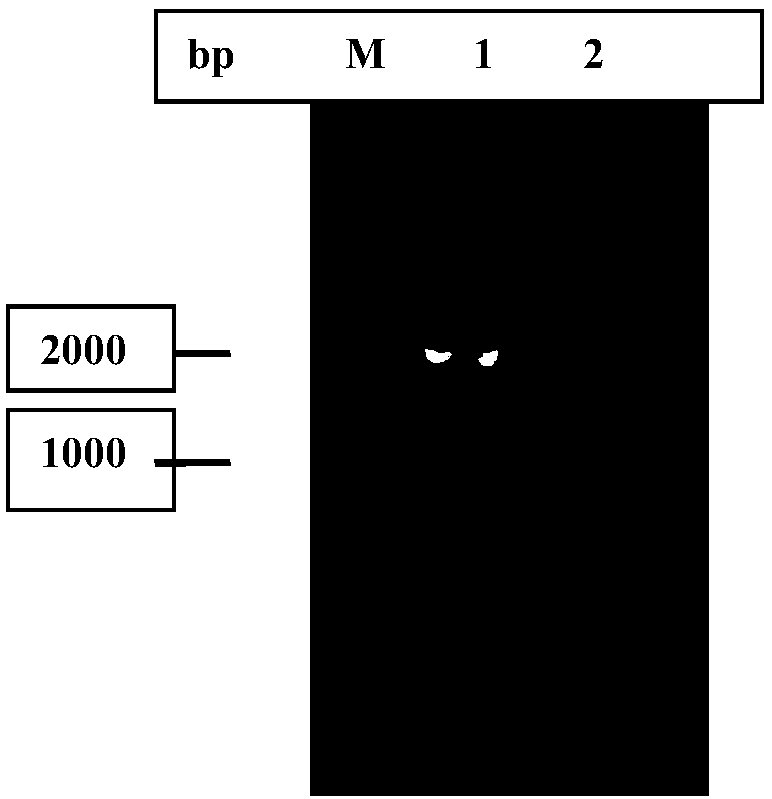
[0057] The genomic DNA of Pseudomonas nitroreduced bacteria was extracted according to the operation steps of the bacterial genomic DNA extraction kit (TaKaRa company). After performing multiple sequence alignment analysis on the 5-hydroxymethylfurfural oxidase gene sequence, primers F: 5'-AGTCCATATGACCACTCCTCGCTATGAC-3' and R: 5'-ATATAAGCTTGGCTCCGCCCAGAATG-3' were designed. The genomic DNA of Pseudomonas nitroreducing strain was used as a template for PCR amplification. The PCR reaction conditions were: 94°C for 4min, 1 cycle; 98°C for 10s, 68°C for 5s, each cycle reduced by 0.2°C, 72°C for 1min 45s, 35 cycles; 72°C for 10min, 1 cycle. The PCR product was detected by 1% agarose gel electrophoresis, and the product size was 1596 bp. Gel recovery kit (purchased from Axygen company) to purify PCR products.
Embodiment 3
[0058] Example 3 Construction of recombinant plasmid PET21a-HMFO
[0059] The purified PCR product and the empty vector pET21a were digested with restriction enzymes Nde Ⅰ and Hid Ⅲ, respectively. The digested product was purified by the gel recovery kit. Under the action of T4 DNA ligase, the purified digested product was subjected to After ligation, the ligation product is transformed into E. coli Top10 competent cells, and then coated on 100μg / ml ampicillin LB (yeast powder 5g / L, tryptone 10g / L, sodium chloride 10g / L, agar powder 15g / L) solid On the medium, incubate at 37°C for 12 hours. Take a single clone into a liquid LB medium containing 100 μg / ml ampicillin for culture, and extract the plasmid. The recombinant plasmid was digested with restriction enzymes Nde Ⅰ and Hid Ⅲ, and the digested products were detected by 1% agarose gel electrophoresis. The digested products with the correct band size were obtained. The constructed recombinant plasmid was initially proved to be ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com