Detection method suitable for molecular identification and authentic right identification of crop variety
A technology of identity identification and detection method, which is applied in the field of detection of molecular identity identification and identification of crop varieties, can solve the problems of lack of a comprehensive and systematic detection method, achieve high variety discrimination ability, highlight detection efficiency, and operability strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
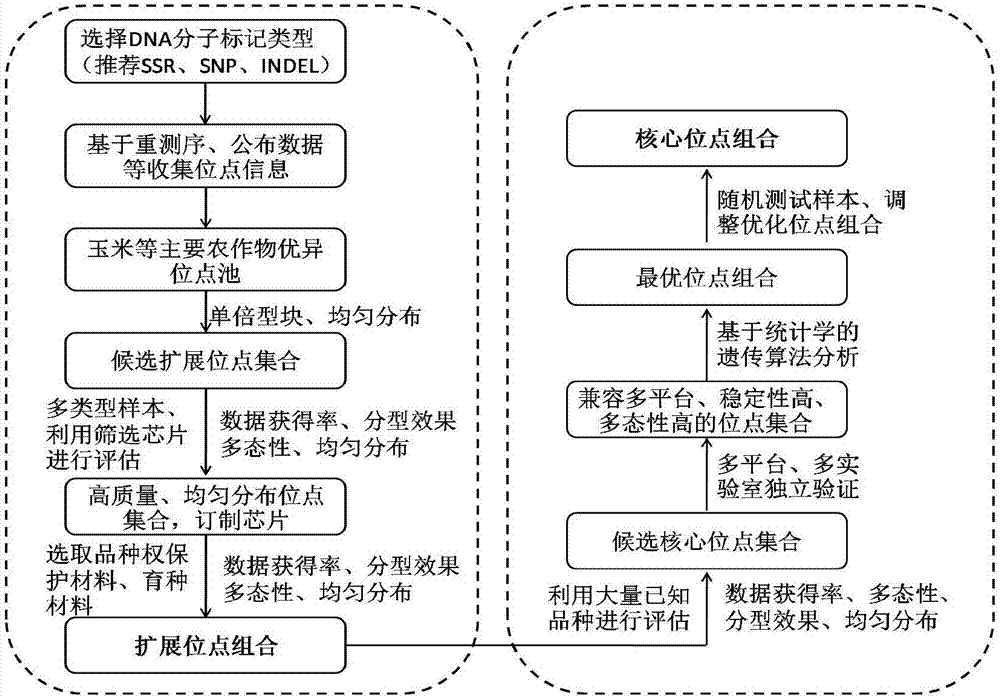
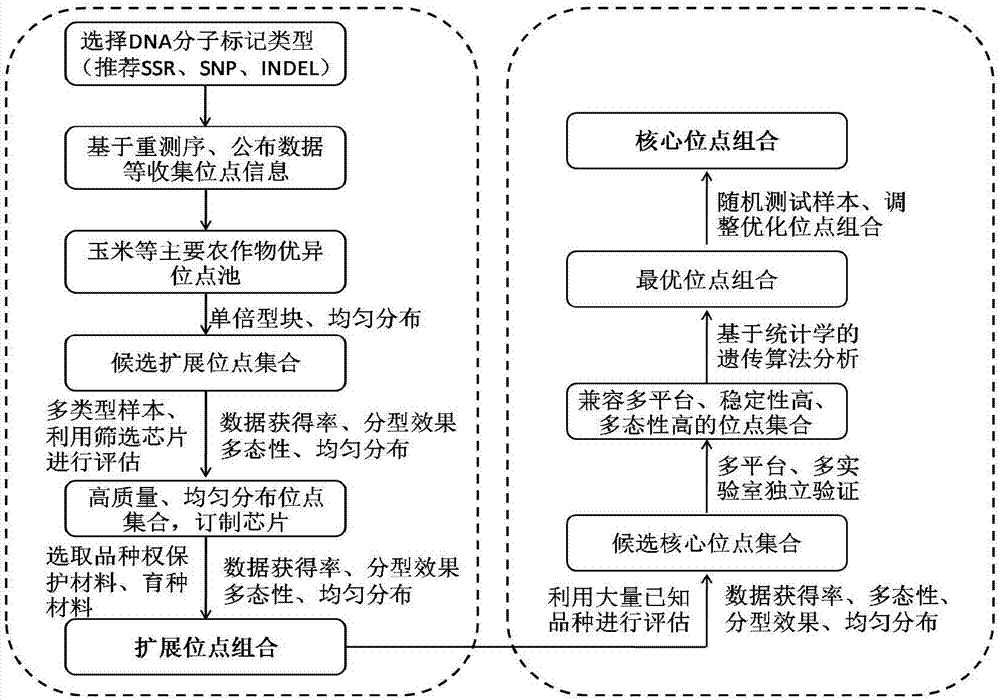
[0035] Example 1 The core of maize (diploid plant) variety identification and the acquisition of extended SNP marker site combinations
[0036] 1. Determination of the expansion site combination:
[0037] (1) Maize excellent SNP locus pool: select representative maize inbred line samples at home and abroad (covering 9 types of germplasm groups, as well as tropical, sweet waxy, local varieties, etc.) for deep genome-wide sequencing (10×), from Mining and locating SNP sites in high-quality resequencing data, and determining single-copy and dimorphic SNP sites based on bioinformatics methods; combining SNP sites in published chips or SNP sites in haplotype maps to form relatively A comprehensive maize excellent SNP site pool (the number of sites is greater than 20 million).
[0038] (2) A set of candidate loci suitable for maize variety identification: the above-mentioned excellent SNP locus information was submitted to two American chip production companies, Affymetrix and Illu...
Embodiment 2
[0046] Example 2 The core of wheat (hexaploid) variety identification and the acquisition of extended SNP site combinations
[0047] 1. Determination of the expansion site combination:
[0048] (1) Wheat excellent SNP locus pool: Based on the physical map information of genetic variation loci in the whole wheat genome, comprehensively analyze the distribution, copy number, homology and polymorphism level of SNP loci within and between subgenomes , to screen specific markers that have the ability to identify homologous chromosomes and are highly conserved within cultivars, and establish a pool of excellent marker sites for chromosome-specific SNPs in subgroup A, subgroup B, and subgroup D of hexaploid wheat.
[0049] (2) A set of candidate loci suitable for identification of wheat varieties: the information of the above-mentioned excellent SNP loci was submitted to two American chip production companies, Affymetrix and Illumina, for evaluation. Chip design evaluation and biolo...
Embodiment 3
[0056] The construction of embodiment 3 corn DNA fingerprint database
[0057] (1) Reference varieties: Jing 724 and Jing 92, the two parents of corn hybrid Jingke 968, were selected as reference varieties. The homozygosity of the samples was detected by the combination of extended loci, and the homozygosity reached more than 99.9%.
[0058](2) DNA preparation of maize varieties: each sample was mixed with 30 green leaves of individual plants, ground with liquid nitrogen, and the specific steps of DNA extraction were carried out in accordance with the standards for molecular identification of maize DNA (Wang Fengge et al., 2014). Use ultraviolet spectrophotometer and agarose electrophoresis to detect the quality of the extracted DNA respectively. Agarose electrophoresis shows that the DNA band is single and there is no degradation; the ultraviolet spectrophotometer detects that A260 / 280 is between 1.8-2.0; A260 / 230 is greater than 1.8 . Dilute the DNA to form a working soluti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com