A kind of complete set of siRNA for suppressing clusterin gene expression and application thereof
A suite of, gene-targeting techniques applied in the field of molecular biology to address issues such as increased response
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
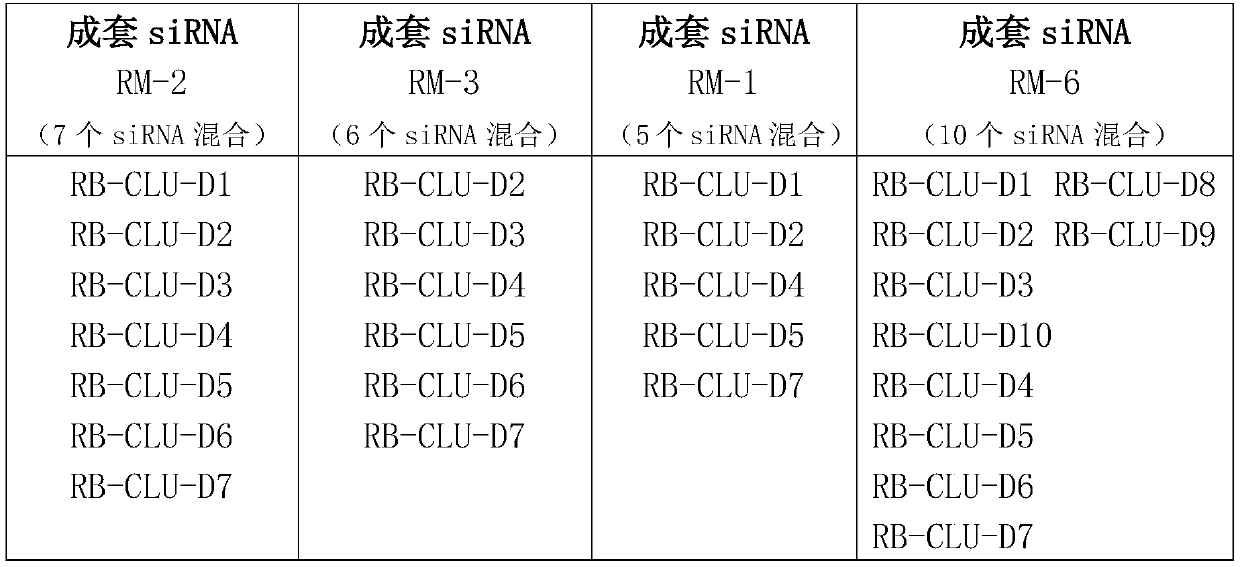
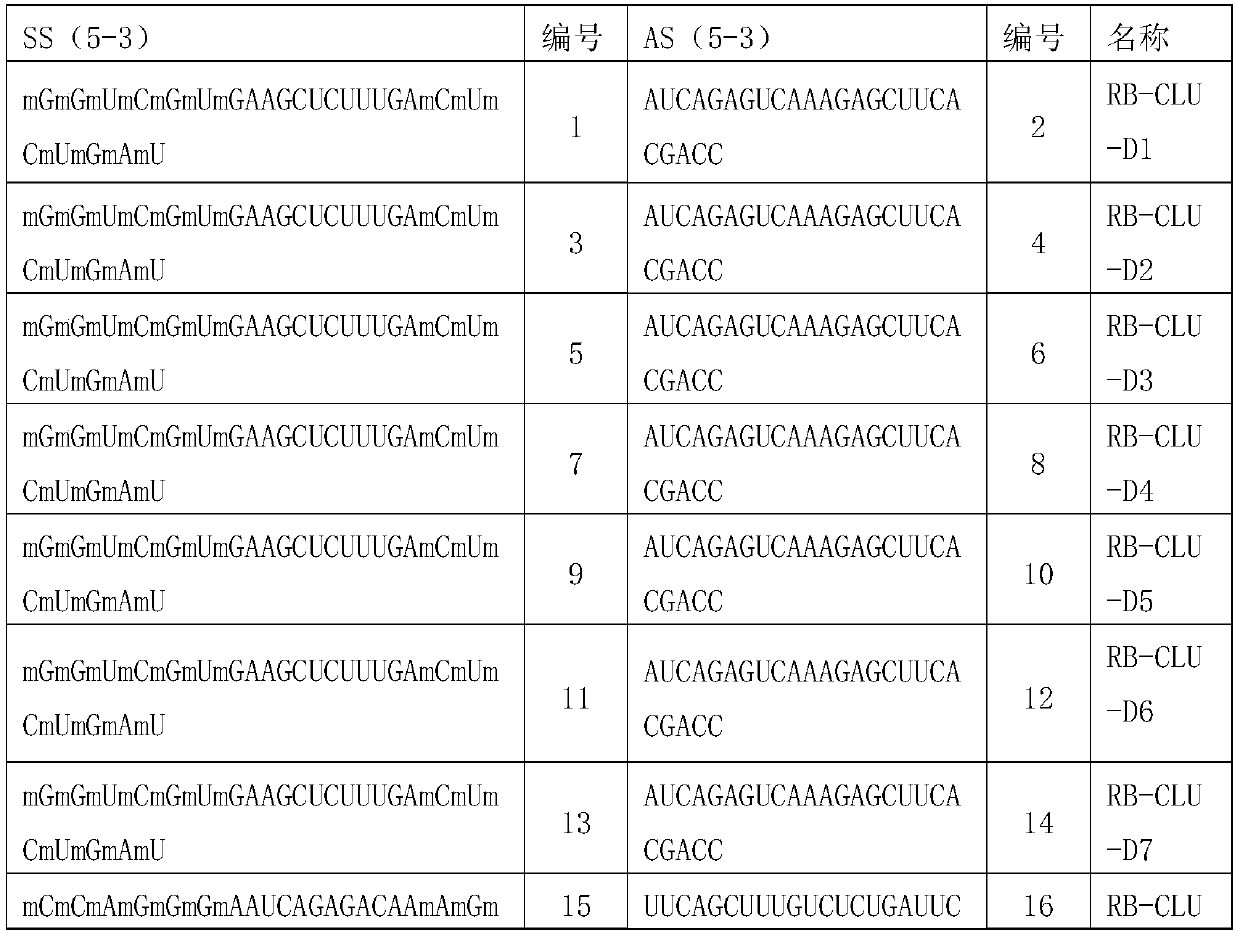
[0077] Embodiment 1, the preparation of complete set of siRNA
[0078] 1. Design principles
[0079] All single siRNAs designed target the target gene CLU (as shown in Table 1), and the design method refers to Elbashir et al.2002; Paddison et al.2002; Reynolds et al.2004; Ui-Tei et al.2004 et al. S.M., Harborth, J., Weber, K., and Tuschl, T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199–213; Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing inmammalian cells. Genes & Dev. 16:948–958; Reynolds, A., Leake, D., Boese, Q., Scaringe, S ., Marshall, W.S., and Khvorova, A. 2004. Rational siRNA design for RNAinterference. Nat. Biotechnol. 22:326–330; Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T. , Ohki-Hamazaki, H., Juni, A., Ueda, R., and Saigo, K. 2004. Guidelines for the selection of highly effective siRNA sequences for mam...
Embodiment 2
[0095] Embodiment 2, the study of complete set of siRNA inhibiting target gene expression
[0096] 1. Comparison of the inhibitory effects of a complete set of siRNA RM-2 in different cell lines
[0097] Four different cell lines (293T, HeLa, A549 and HUVEC (ATCC) were inoculated in cell culture plates and cultured for 24 hours to observe the cells, and the cells were in good condition to start transfection.
[0098] Table 4 Source of Cell Lines
[0099] cell line name source 293T human embryonic kidney cells ATCC HeLa cervical cancer cells ATCC A549 Non-small cell lung cancer cells ATCC HUVECs human umbilical vein endothelial cells ATCC
[0100] 50μL riboFECT transfection system: 5μL riboFECT TM CP Reagent (Guangzhou Ruibo Biotechnology Co., Ltd., C10511-05), 5 μL of the complete siRNA group RM-2 prepared in Example 1 (the total concentration of all siRNAs is 100 nM) and 40 μL riboFECT TM CP Buffer (Guangzhou Ruibo Biotec...
Embodiment 3
[0131] Embodiment 3, in vitro stability determination
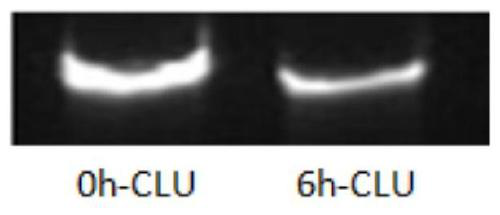
[0132] Dilute siRNA RB-CLU-D6 to 5 μM with RNase-free water, add an equal volume of fresh rat serum (product of Shanghai Yuanmu Biotechnology Co., Ltd.), and incubate at 37°C for 6 hours, take samples for electrophoresis to observe the siRNA integrity.
[0133] The result is as figure 1 As shown, the siRNAs of the present invention are stable in serum and are expected to have better potency in vivo.
[0134] For other RB-CLU-Dx (x is D1-D10), the experimental results are the same, and the specific figures are omitted.
[0135]
[0136]
[0137]
[0138]
[0139]
[0140]
PUM
| Property | Measurement | Unit |
|---|---|---|
| control rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com