A primer, probe and kit for specific detection of type 2 ungulate boca parvovirus
A parvovirus and ungulate technology, applied in the field of biological detection, can solve the problems of diagnosis, high mutation rate, gene insertion, etc., and achieve the effects of high accuracy, high sensitivity and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Design and synthesis of embodiment 1 primers and probes
[0038] Download all the complete gene sequences belonging to the type 2 ungulate boca parvovirus population from GenBank, combine the complete gene sequence (GeneBank No. KJ755666) obtained by our laboratory with DNAStar software for homology comparison, and use primerExpress3.0 software, Primers and TaqMan probes were designed, and the primers and probes were synthesized by Invitrogen. The length of the amplified target fragment is 80bp.
[0039] The nucleotide sequence of the upstream primer is shown as PboVF1;
[0040] The nucleotide sequence of the downstream primer is shown in PBoVR 1;
[0041] The nucleotide sequence of the probe used in conjunction with the above primers is shown as PboVP1. The 5' end of the probe is labeled with the reporter VIC fluorescent dye, and the other 3' end is labeled with the MGB quencher group.
Embodiment 2
[0042] The extraction steps of embodiment 2 total DNA are as follows:
[0043] Add 2-4ML of PBS solution or normal saline to the feces sample for homogenization, centrifuge at 12000r / min for 5min, and take the supernatant; add 2ML / g of PBS solution after the tissue sample is pulverized, homogenate, freeze-thaw 3-5 times, and centrifuge at 12000r / min After 5 minutes, the supernatant was taken for the following experiments. Extract viral DNA according to the instructions of Beijing Tiangen Biotechnology Company Viral Genomic DNA / RNA Extraction Kit (DP315) (the DNA extraction in this test is only based on the DP315 kit, and other commercial viral DNA extraction kits are applicable), directly use or Store in a -20°C refrigerator for later use, and store in a -70°C refrigerator for long-term storage.
Embodiment 3
[0044] Embodiment 3 Establishment of real-time fluorescent quantitative PCR amplification method
[0045] 1. Real-time fluorescence quantitative PCR reaction system
[0046] Use the total DNA as a template to carry out real-time fluorescent quantitative PCR reaction, that is, in a 20 μL reaction system containing: 10× fluorescent quantitative PCR buffer 2.0 μL, 2.0 μL dNTPs (2.5 mmol / L), 0.5 μL primer PboVF1 (10 μmol / L) , 0.5μL primer PBoVR1 (10umol / L), 1.0μL magnesium ion (50mmol / L), 0.5μL fluorescent probe (10umol / L), 1.0μL (10ng~50ng / μL) DNA template, 0.2μL Taq enzyme (5U ).
[0047] 2. Real-time fluorescent quantitative PCR reaction conditions
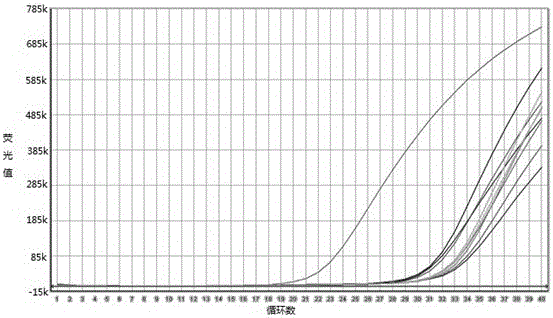
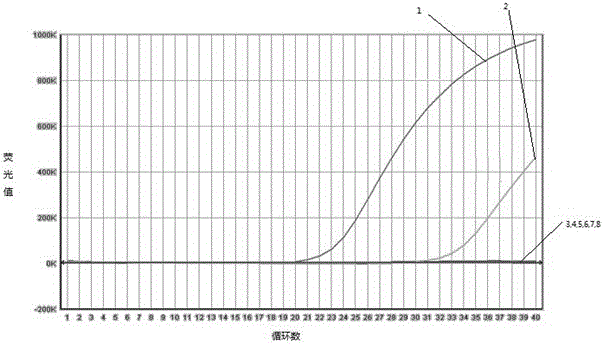
[0048] After putting the sample tube into the 7500 fluorescent PCR instrument of ABI company, set the following conditions to carry out: 95°C, 5min; 95°C, 10s; 60°C, 40s for a total of 40 cycles. Collect data after each cycle. After the reaction, judge the result according to the amplification curve and the standard curve.
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com