Beta-glucosidase mutant and application thereof
A glucosidase and amino acid technology, applied in the field of β-glucosidase mutants, can solve the problems of low activity and low β-glucosidase content, and achieve the effect of a wide range of applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Cloning of β-glucosidase gene and construction of its mutant
[0019] 1.1 Extraction of Aspergillus fumigatus total genomic DNA
[0020] Incubate Aspergillus fumigatus overnight, place an appropriate amount of bacteria in a centrifuge tube, centrifuge at 13000 rpm for 5 min, discard the supernatant; add 400 μl extraction buffer (100 mM Tris-HCl, 100 mM EDTA, 250 mM NaCl, 1% SDS) ; Then add 100mg quartz sand or glass beads, vigorously shake in the beading instrument for about 2min; after 20min at 65℃, add 200μl 10M NH4AC, ice bath for 10min; centrifuge at 13000rpm for 10min, take the supernatant; add 2 times the volume of absolute ethanol, Place at -20°C for 30 minutes; centrifuge at 13000 rpm for 10 minutes, discard the supernatant; wash twice with 70% ethanol; dry, add water to dissolve, and store at -20°C.
[0022] Using the total genomic DNA extracted in 1.1 as a template, PCR amplification was performed using primers BG-F and BG-R:
[002...
Embodiment 2
[0033] Example 2 Transformation and Screening
[0034] Pipette the spore suspension of Trichoderma reesei strain in the center of the PDA plate (9cm petri dish). When the colony grows over the entire petri dish, cut the medium of 1cm×1cm each, and place it in 120mL YEG+U liquid medium at 30℃ , 200rpm, culture for 14-16h.
[0035] Collect the mycelium with a sterile Miracloth filter cloth and wash it once with solution A. Transfer the washed mycelium to 40mL protoplastization solution under aseptic conditions, incubate at 30℃, 90rpm, for 1-2h, observe with a microscope Detect the progress of protoplast transformation.
[0036] Filter the above warm bath liquid with a sterile Miracloth filter cloth, and the obtained filtrate is the protoplast solution. Divide the protoplast solution into two 50 mL sterile disposable centrifuge tubes, and dilute the volume of each tube to 45 mL with solution B, centrifuge at 3000 rpm for 10 min, discard the supernatant to obtain a precipitate; use 5 m...
Embodiment 3
[0054] Example 3 Fermentation verification
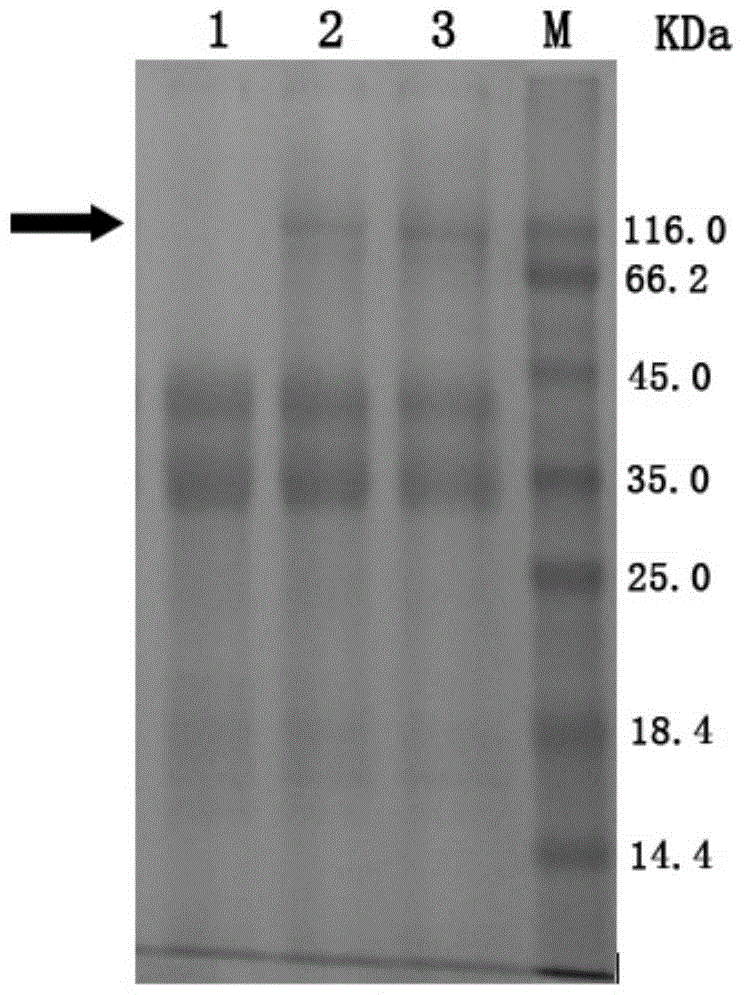
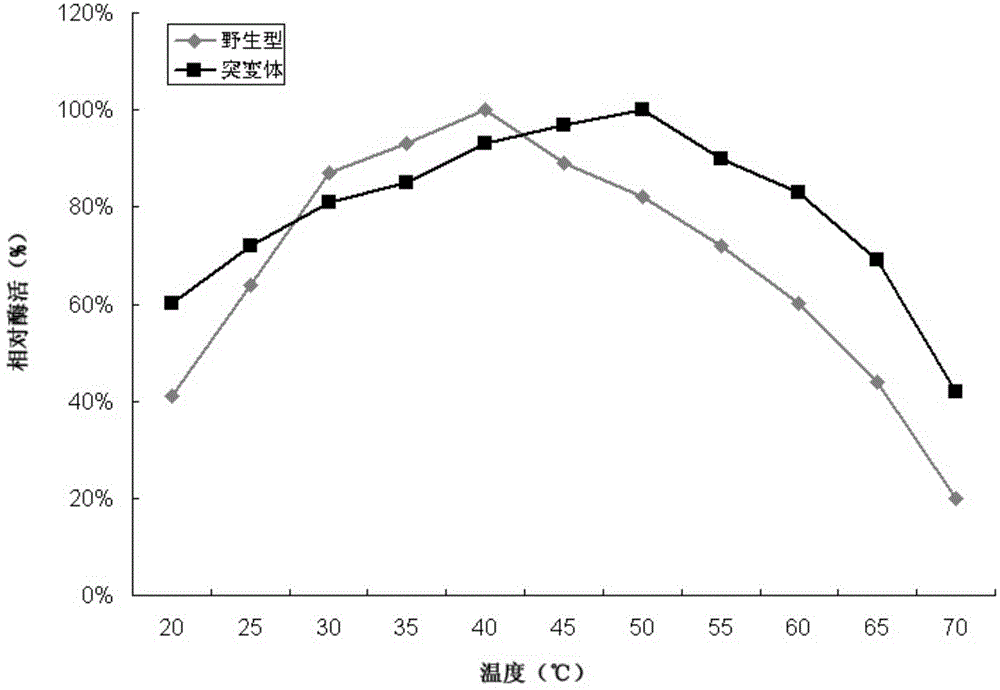
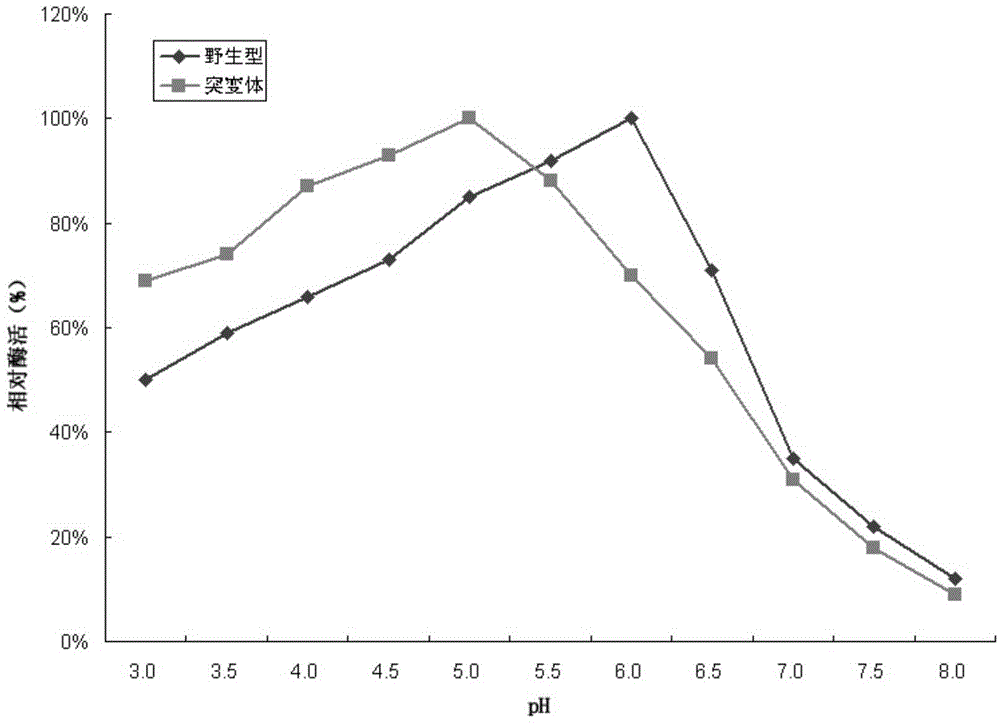
[0055] The host bacteria of Trichoderma reesei, Trichoderma reesei TR-BG5 and Trichoderma reesei TR-BG that have not been introduced into foreign genes were respectively inoculated into MM fermentation medium (1.5% glucose, 1.7% lactose, 2.5% corn steep liquor, 0.44%(NH 4 ) 2 SO 4 , 0.09% MgSO 4 , 2% KH 2 PO 4 , 0.04%CaCl 2 , 0.018% Tween-80, 0.018% trace elements) cultured at 30°C for 48 hours, and then at 25°C for 48 hours. The fermentation broth was centrifuged separately, and the supernatant was taken for SDS-PAGE electrophoresis detection. The result is figure 1 As shown, the theoretical molecular weight of the β-glucosidase derived from Aspergillus fumigatus in the present invention is about 95KDa, namely figure 1 The position indicated by the middle arrow, lane 1 is the host bacteria fermentation supernatant, which has almost no protein band at the position indicated by the arrow, while the T. reesei TR-BG in lane 2 and the T. r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com