Method for remarkably improving fish genome editing efficiency
A genome editing and genome technology, applied in the biological field, can solve the problem of unequal distribution of injected nucleic acid and other problems, and achieve the effect of reducing invalid F1 generation individuals and reducing time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Directed knock-in of mloxP in the intron of zebrafish aldh1a2 gene
[0037] 1. Construction of a CRISPR / Cas9 system targeting the third and fourth introns of the zebrafish aldh1a2 gene
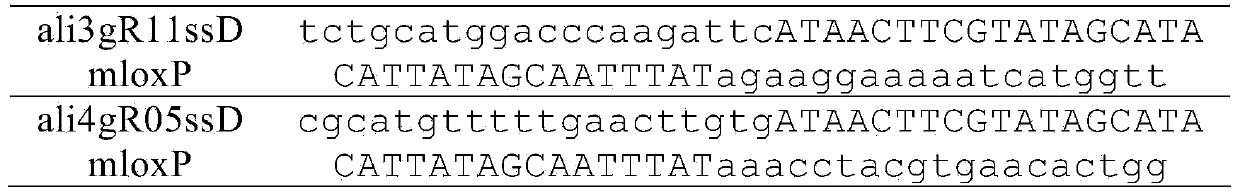
[0038] Table 1. Primer and template sequences used to construct sgRNA targeting aldh1a2
[0039]
[0040] Use primers ali3gRF11 (Seq ID No: 1) and sgRNAR (Seq ID No: 3) to amplify the plasmid template containing the sgRNA backbone, and the PCR conditions are: 95°C for 2 minutes, 35 cycles (94°C for 30 seconds, 56°C for 30 seconds , 72°C for 30 seconds), and a final 72°C extension for 5 minutes. The same method was used to amplify the plasmid template containing the sgRNA backbone (Seq ID No: 4) with primers ali4gRF5 (Seq ID No: 2) and sgRNAR (Seq ID No: 3). The above PCR products were transcribed into RNA with MEGAscript Kit (Ambion, USA), and the products were ali3gR11 and ali4gR5, respectively. The total RNA of zebrafish 24hpf embryos was extracted with TRIzol reagent,...
Embodiment 2
[0060] Example 2 Introducing an insertion-deletion mutation in the intron of the zebrafish aldh1a2 gene
[0061] 1. Construction of a CRISPR / Cas9 system targeting the third and fourth introns of the zebrafish aldh1a2 gene
[0062] Table 1. Primer and template sequences used to construct sgRNA targeting aldh1a2
[0063]
[0064] Use primers ali3gRF11 (Seq ID No: 1) and sgRNAR (Seq ID No: 3) to amplify the plasmid template containing the sgRNA backbone, and the PCR conditions are: 95°C for 2 minutes, 35 cycles (94°C for 30 seconds, 56°C for 30 seconds , 72°C for 30 seconds), and a final 72°C extension for 5 minutes. The same method was used to amplify the plasmid template containing the sgRNA backbone (Seq ID No: 4) with primers ali4gRF5 (Seq ID No: 2) and sgRNAR (Seq ID No: 3). The above PCR products were transcribed into RNA with MEGAscript Kit (Ambion, USA), and the products were ali3gR11 and ali4gR5, respectively. The total RNA of zebrafish 24hpf embryos was extracted ...
Embodiment 3
[0079] Embodiment 3 introduces insertion-deletion mutations in the yellow catfish mstna gene and mstnb gene
[0080]As two independent genome editing tools, zinc finger nuclease and transcription activator-like effector nuclease can respectively complete the recognition and cutting of specific sequences. In the experiment, you can choose to edit the genome separately or together according to the needs of the experiment. In order to make the present invention concise, in this embodiment, the zinc finger nuclease mRNA that specifically recognizes and cleaves the specified site sequence of the mstna gene, and the transcription activator-like effector nuclease mRNA that specifically recognizes and cleaves the specified site sequence of the mstnb gene Co-injection into yellow catfish embryos to carry out the present invention, but use the zinc finger nuclease mRNA that specifically recognizes and cuts the sequence of the designated site of the mstna gene or the transcription activa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com