A method for improving 7-aca producing bacteria
A 7-ACA, bacteria-producing technology, applied in the biological field, can solve the problems of low toxicity, allergic reaction, high cost, and the yield has not reached industrialized production, and achieves the effects of convenient screening, increased yield, and large genetic differences.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
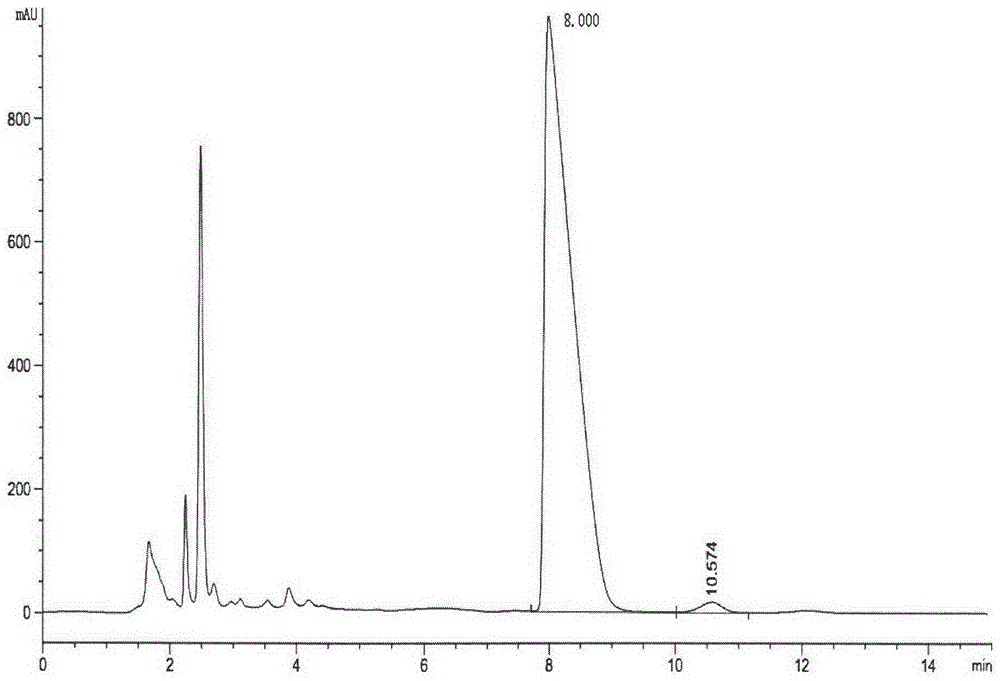
Examples
Embodiment 1
[0047] Embodiment 1ecs-50 gene and its function
[0048] 1. Error-prone PCR for the starting sequence and construction of product expression plasmid
[0049] 1.1 Perform error-prone PCR on the starting sequence ecs gene
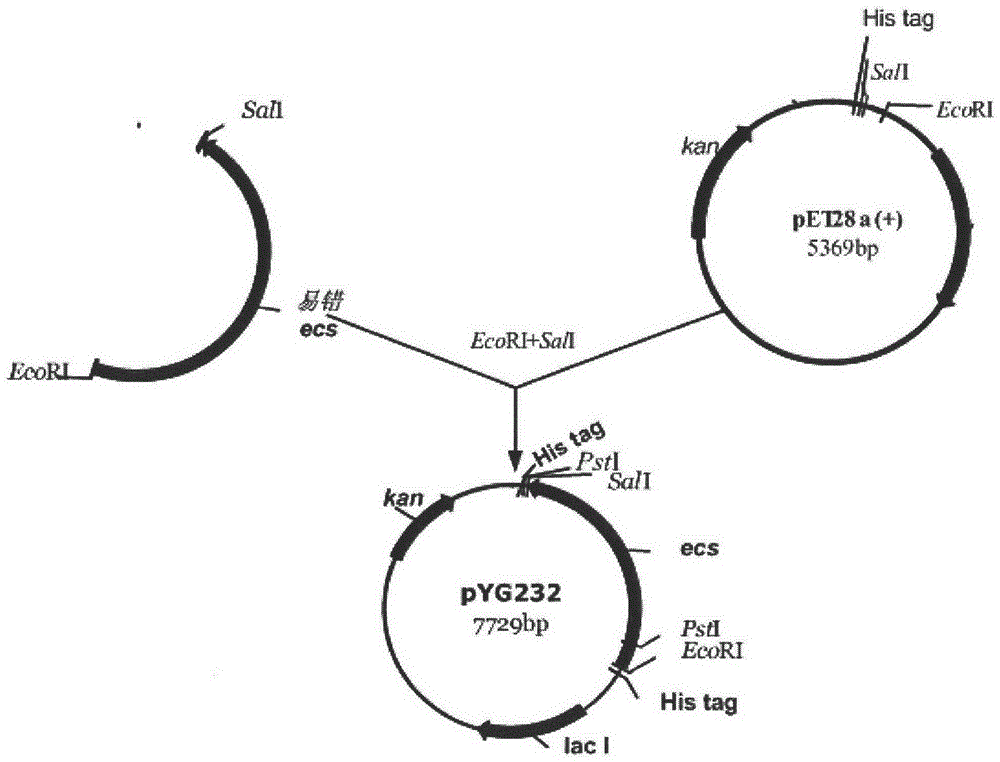
[0050] The starting sequence is obtained by ligation of segmented PCR products, and its gene sequence is shown in pyg232 in FIG. 8 . When using DNA polymerase to amplify the target gene, by adjusting the reaction conditions, increasing the concentration of magnesium ions or adding manganese ions, using low-fidelity DNA polymerase to change the mutation frequency during the amplification process, so as to increase the mutation frequency at a certain frequency. Randomly introduce mutations into the target gene to obtain random mutants of protein molecules. The PCR reaction system used is as follows:
[0051]
[0052] The sequences of primers P1 and P2 are:
[0053] P 1 : 5'-GCTT GTC GAC TCTAGATCAGTGGTG-3' (the underlined part is the SalI restriction s...
Embodiment 2
[0082] Example 2 Construction and Transformation of Expression Plasmid pYG236 Cephalosporium acremonium (Acremonium chrysogenum, ATCC11550)
[0083] 1. Construct the Cephalosporium acremonium expression plasmid pYG236 containing the resistance gene hygB and the exogenous gene ecs, use the Ptrp promoter to activate the ecs gene, and the pcpc promoter to activate the resistance gene hygB. The pYG236 plasmid construction process is as follows: use XbaI+EcoRI to double-digest the plasmid pYG232-50 to obtain the ecs-50 gene, from the pDH25 plasmid (Huang Dafang et al., Plasmid pDH25 carrying the hygromycin resistance gene to transform Botrytis cinerea research, China Bioengineering magazine 1990 05 period) with EcoRI+KpnI digestion to obtain the Ptrp promoter, the obtained ecs-50 gene and Ptrp promoter were connected with the pUC18 plasmid digested with KpnI+XbaI to obtain the plasmid pUC18+ecs-50+Ptrp plasmid. Digested with SalI+HindIII to obtain pcpc+hgh gene, connected with pUC1...
Embodiment 3
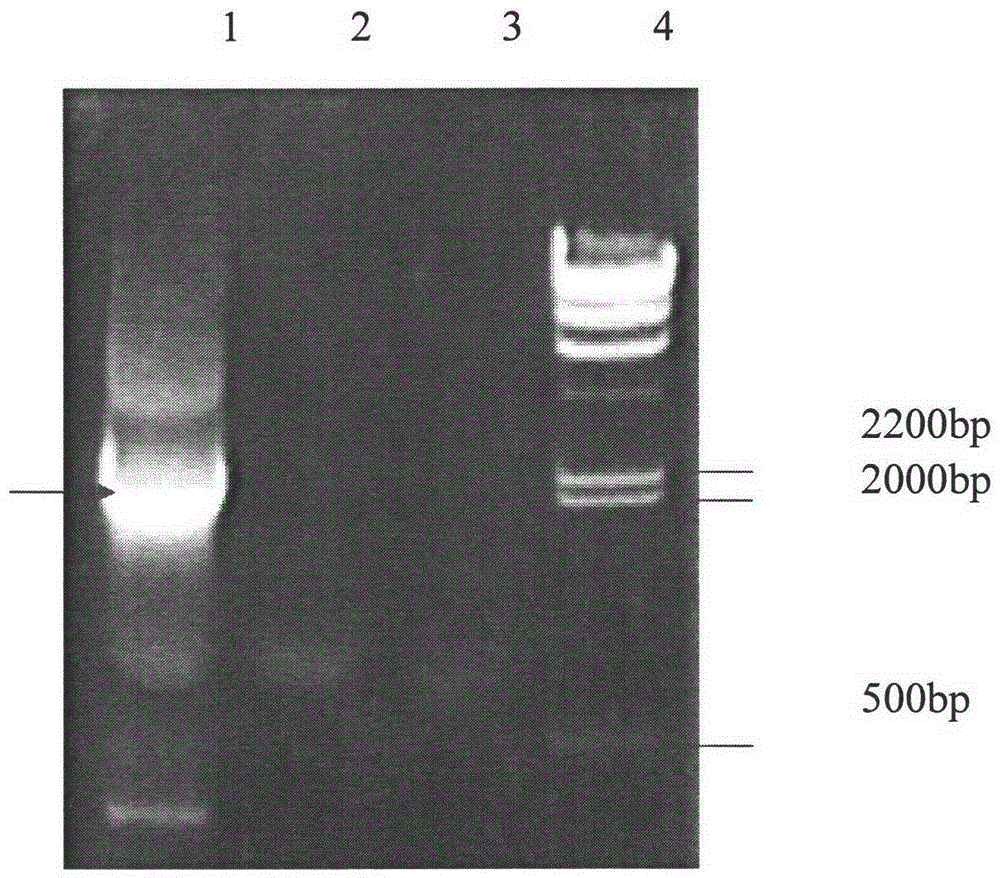
[0105] Example 3 PCR verification of transformant Acremoniumchrysogenum / pYG236
[0106] Pick Cephalosporium acremonium-resistant transformants from the regenerative transformation plate of Example 1 and inoculate into the basal medium containing 10 μg / ml hygromycin [wort juice 20% (w / v), maltose 4.0% (w / v) , polypeptone 1% (w / v), pH7.0, add purified agar powder 2% (w / v)] culture on the plate for 10-14 days (d), when the spores grow plump, spread it to Continue culturing on the slant of the basic medium for 10 to 14 days, then scrape off the spores from the slant, and extract genomic DNA by liquid nitrogen extraction for PCR verification.
[0107] 1. Liquid nitrogen extraction method to extract the genome of Cephalosporium acremonium:
[0108] (1) Scrape off a small amount of spores from the inclined surface and add an appropriate amount of liquid nitrogen to grind;
[0109] (2) Dissolve with 1ml extract after grinding, extract (Tris-HClpH7.50.2mol / ml, NaCl0.5mol / l, EDTA0.01m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com