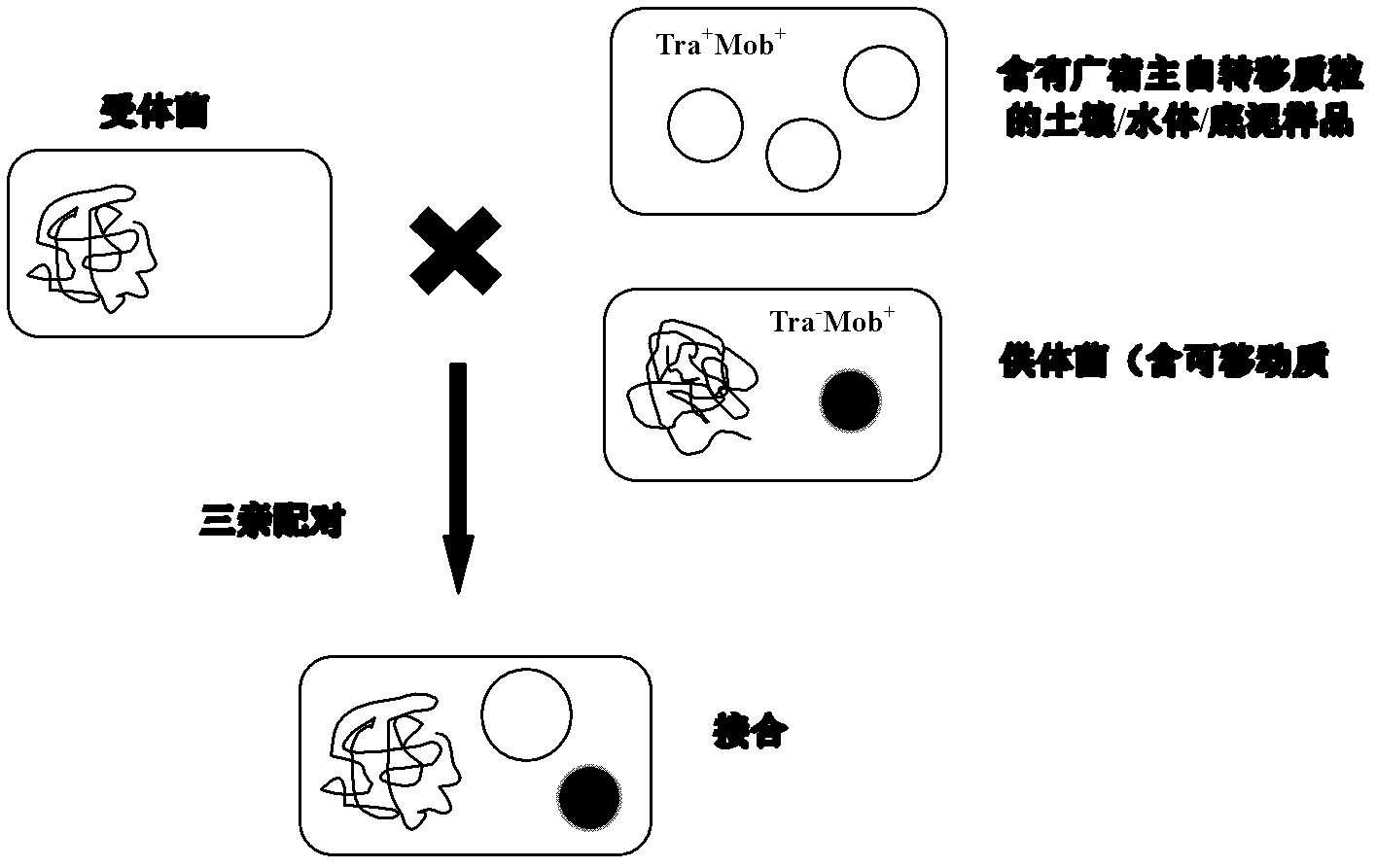
Method for screening self-transmissible broad host range plasmid carrying petroleum hydrocarbon degrading gene by utilizing triparental pairing conjugation and single carbon source of petroleum hydrocarbon
A technology of petroleum hydrocarbons and plasmids, applied in the field of biological technology and bioremediation of polluted soil
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] The surface layer (0-20cm) of paddy field soil (0-20cm) in the Shenfu irrigation area irrigated with petroleum sewage for more than 50 years was collected, passed through a 1mm sieve, and stored in a refrigerator at 4°C. Weigh 5g (dry weight) of the soil sample and add it to a 100ml Erlenmeyer flask filled with 25ml of normal saline (0.85% NaCl solution), shake it on a shaker for 1 hour; let the suspension stand for about 30min, and pour the supernatant into a 30ml sterile Centrifuge at 10,000rpm at 4°C for 10min in a centrifuge tube; resuspend the pellet in 3ml 1 / 10LB liquid medium, and vortex thoroughly.
[0056] Select a suitable combination of donor bacteria and recipient bacteria to ensure that the two have different genetic selection markers, and preferably come from different Proteobacteria subclasses. The donor bacteria and recipient strains frozen in glycerol tubes were taken out from the -70°C refrigerator, streaked on the LB solid plate containing the corresp...
Embodiment 2
[0061] Scrape the control group grown on the LB plate in Example 1 and the bacterial lawn of the matching group, resuspend the culture in 2ml sterile physiological saline, and dilute to 10% under sterile conditions with sterile physiological saline. -4 , take 0.1ml to dilute to 10 -3 -10 -4 The control group was coated with LB solid plates containing corresponding antibiotic selection markers, and 0.1ml was diluted to 10 0 -10 -1 The paired group was coated with LB solid plates containing corresponding double antibiotics (antibiotic resistance of recipient bacteria and antibiotic resistance of donor bacteria plasmid) markers, placed in a constant temperature incubator at 30°C, and cultured overnight.
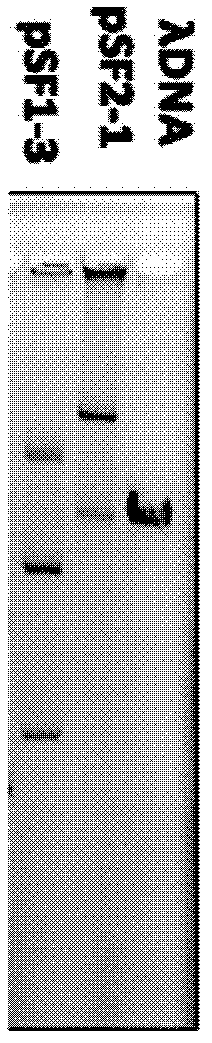
[0062] Pick a single colony of the zygote, insert it into 5ml of LB liquid medium containing the corresponding antibiotic, and cultivate overnight on a shaker at 30°C. SDS-alkaline hydrolysis was used to extract the plasmids of each zygote, and the 1% agarose gel electrophore...
Embodiment 3
[0064] Select zygote strains containing different plasmids in Example 2, inoculate in LB liquid medium, and cultivate to OD 600 =0.6-0.8, according to the mass percentage 10% inoculum size access to a certain petroleum hydrocarbon components (n-octane, benzene, toluene) and its main metabolic intermediates (salicylic acid, catechol) as the only substrate In the liquid inorganic salt medium of carbon source, place it on a 30° C. constant temperature shaker at 180 rpm for 5-10 days (cultivation time depends on different petroleum hydrocarbon components). Observe whether the strain containing the zygote can grow with these petroleum hydrocarbon components (measure the OD600 value of the cell density). Preparation method of liquid inorganic salt medium with petroleum hydrocarbon as the only carbon source: take 5g / L petroleum hydrocarbon component mother liquor (prepared with acetone or water), sterilize with 0.22μm filter membrane, measure 50-200μl mother liquor, add 50ml to steri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com