Preparation and application of OCH1 genetic flaw type P. pastoris X-33 bacterial strain
A technology of defective and genetically engineered bacteria, applied in the field of construction and utilization of Pichia pastoris X-33 strain, can solve the problems of reduced activity, shortened half-life, enhanced immunogenicity of glycoproteins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
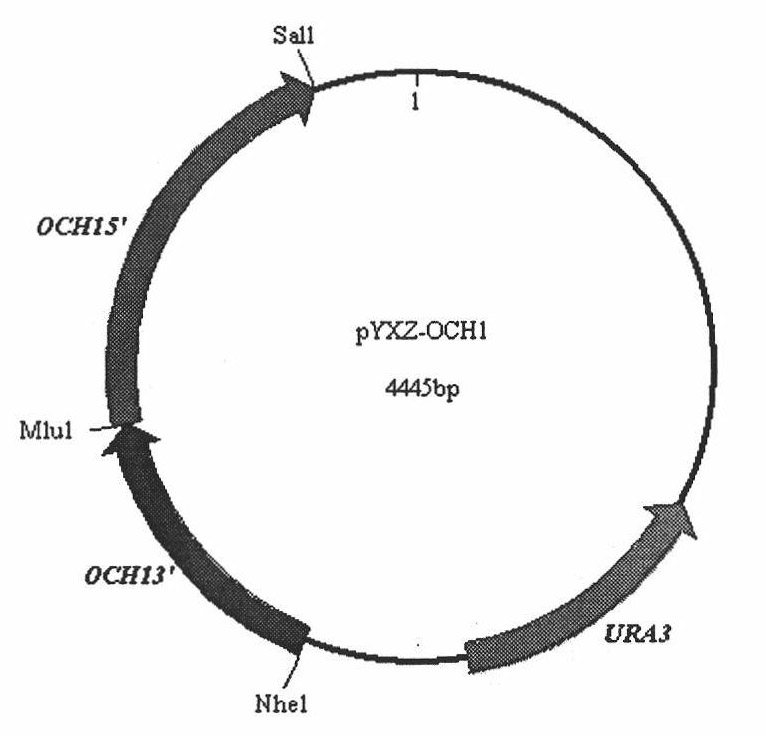
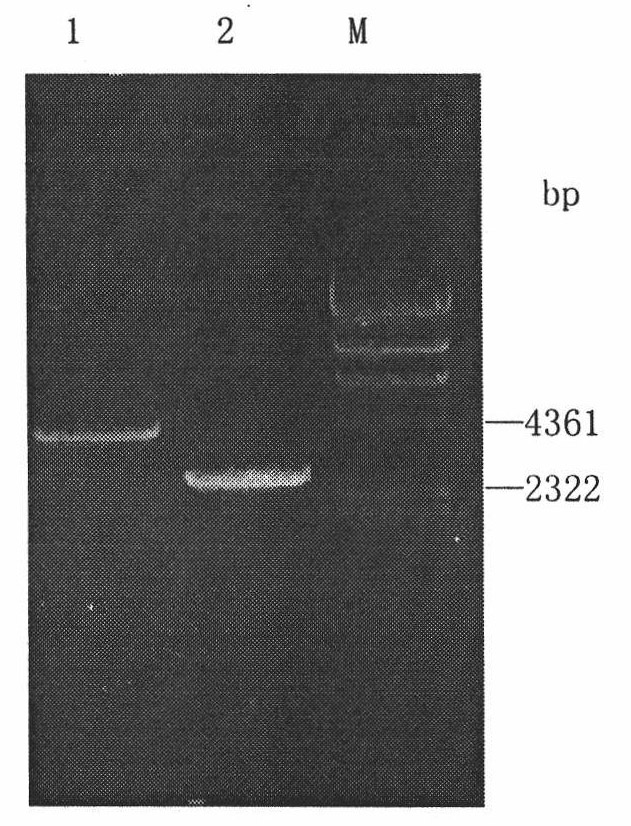
[0030] Knockout and identification of OCH1 gene: the knockout plasmid pYXZ-OCH1( figure 1 ) was sexualized with MluI and then electroporated into X-33 (Δura3) competent cells to undergo double exchange homologous recombination with the target. The knockout plasmid contains the URA3 marker gene, so the strains that can grow on MD medium are homologous recombination strains; at the same time, the och1 knockout bacteria grow well at 25°C but cannot grow at 37°C. Select the strains that meet the above two traits, Genomic PCR identification was performed. Since the sequence of about 1000bp in the reading frame of the OCH1 gene was replaced by a 2.8kb selection marker sequence, the external primers OCH5F and OCH3R were used to identify, and the PCR amplification product of X-33 bacteria was 2.8kb, and that of X-33 (Δoch1) bacteria was 4.4kb ( figure 2 ); (in)3R in the internal primers (in)5F and (in)3R is located in the replaced reading frame sequence, so the X-33 strain can amp...
Embodiment 2
[0032] Construction and expression of GM-CSF expression vector
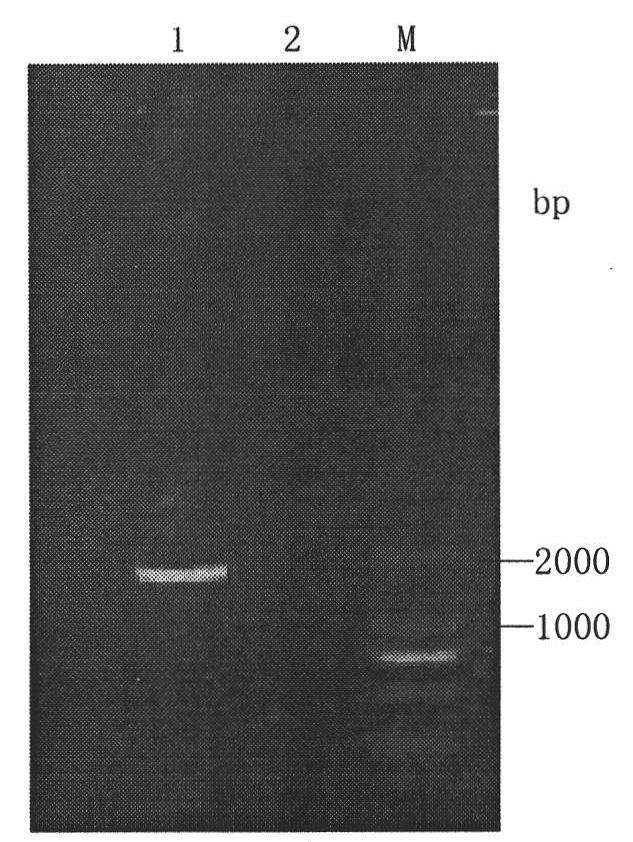
[0033] 1) Construction of the expression vector: the plasmid pUC19 / GM-CSF stored in our laboratory was digested with EcoRI and NotI, and the GM-CSF gene fragment was recovered (6xHis tag sequence was introduced at the end), and cloned into Pichia On the yeast expression vector pPICZaA, the expression vector pPICZaA / GM-CSF( Figure 4 ), positive clones were screened, identified by enzyme digestion and sequenced.
[0034] 2) Expression of GM-CSF in P. pastoris X-33 and X-33 (Δoch1): the expression vector pPICZaA / GM-CSF was linearized with the restriction endonuclease SacI and transferred into X-33 and X-33 respectively In -33 (Δoch1) competent cells, the bacterial solution after electric shock was applied to YPDZ (containing 100ug / mL, 300ug / mL, and 500ug / mL Zeocin respectively) medium, and X-33 transformants were cultured at 30°C for 3 -5d, X-33 (Δoch1) transformants were cultured at 25°C for about a week, and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com